Abstract
Infectious bursal disease virus (IBDV), a double-stranded RNA (dsRNA) virus belonging to the Birnaviridae family, is an economically important avian pathogen. The IBDV capsid is based on a single-shelled T=13 lattice, and the only structural subunits are VP2 trimers. During capsid assembly, VP2 is synthesized as a protein precursor, called pVP2, whose 71-residue C-terminal end is proteolytically processed. The conformational flexibility of pVP2 is due to an amphipathic α-helix located at its C-terminal end. VP3, the other IBDV major structural protein that accomplishes numerous roles during the viral cycle, acts as a scaffolding protein required for assembly control. Here we address the molecular mechanism that defines the multimeric state of the capsid protein as hexamers or pentamers. We used a combination of three-dimensional cryo-electron microscopy maps at or close to subnanometer resolution with atomic models. Our studies suggest that the key polypeptide element, the C-terminal amphipathic α-helix, which acts as a transient conformational switch, is bound to the flexible VP2 C-terminal end. In addition, capsid protein oligomerization is also controlled by the progressive trimming of its C-terminal domain. The coordination of these molecular events correlates viral capsid assembly with different conformations of the amphipathic α-helix in the precursor capsid, as a five-α-helix bundle at the pentamers or an open star-like conformation at the hexamers. These results, reminiscent of the assembly pathway of positive single-stranded RNA viruses, such as nodavirus and tetravirus, add new insights into the evolutionary relationships of dsRNA viruses.
Infectious bursal disease virus (IBDV) infects young chickens, causing a highly contagious disease that targets the precursors of antibody-producing B cells (4). This avian disease is economically relevant to the poultry industry worldwide (45). IBDV belongs to the genus Avibirnavirus in the family Birnaviridae. The IBDV virion is spherical, with a diameter of ∼650 to 700 Å, and contains two double-stranded RNA (dsRNA) segments, of 3.2 and 2.8 kbp (segments A and B, respectively), and five encoded mature proteins (VP1 to VP5) (15). Most of these proteins are encoded by segment A, which has two partially overlapping open reading frames; the first one encodes a dispensable nonstructural protein, VP5 (17 kDa), and the second encodes three proteins synthesized as a polyprotein (110 kDa). Segment B has a single open reading frame encoding VP1 (98 kDa), the RNA-dependent RNA polymerase (47). The viral polyprotein is cotranslationally processed by the viral protease VP4 (5, 17), rendering the VP2 precursor, termed pVP2 (512 residues; 54.4 kDa), VP3 (29 kDa), and VP4 (27 kDa). Most of the pVP2 C-terminal region is further processed at three Ala-Ala bonds, which are secondary VP4 targets (positions 487, 494, and 501) (38). The resulting intermediate pVP2 is again cleaved, by an unknown mechanism, between residues 441 and 442, giving the mature VP2 polypeptide (47 kDa) (Fig. 1). The released C-terminal peptides remain inside the capsid (14). VP2 (and a residual amount of pVP2), representing ∼50% of the total protein, is the structural component in the capsid, and VP3 (∼40%) is a multifunctional protein that interacts with pVP2, dsRNA, VP1, and itself (28, 29).
FIG. 1.
Schematic topology of VP2. Domains P, S, and B are colored red, blue, and green, respectively. Major secondary structural elements are shown (with α-helices as cylinders and β-strands as arrows; 310 helices were omitted). Initial and final positions of the α-helices of domain B are labeled. The αNter helix (containing residues 1 to 10) is ordered only in a VP2 subunit of the asymmetric unit in the virion capsid (12). The 71-residue C-terminal sequence (yellow) of the precursor pVP2 (512 residues), which is absent in mature VP2 (441 residues), contains the amphipathic α-helix (the 443-452 sequence) acting as a molecular switch (39). The viral VP4 protease targets are indicated by small arrows; cleavage between residues 441 and 442 is performed by an unknown mechanism (large arrow).
IBDV has a relatively complex icosahedral capsid (protein mass, ∼37 MDa) based on a single T=13 levo lattice (6, 12, 39) and formed by 260 VP2 trimers making 12 pentamers and 120 hexamers. From strict geometric considerations, VP2 trimers adopt five distinct conformations (a to e); however, contacts between VP2 trimers are all quasi-equivalent. Differences in subunit interactions and conformations are variable in all icosahedral capsids analyzed to date. The distinct conformational states may be controlled by alternatives between order and disorder of flexible regions in the protein (loops and N and C termini), ds- and ssRNA, metal ions, pHs, or combinations of all these (23). These factors, referred to as molecular switches, may be insufficient, especially when large and/or complex viruses are considered. Complex capsids require one or more auxiliary proteins (scaffold, accessory, and proteolytic proteins) acting as morphogenic factors to trigger structural changes (16, 32).
Recent studies of the IBDV capsid have resulted in progress towards understanding the molecular factors involved in assembly. Expression of mature VP2 alone results in the spontaneous assembly of icosahedral T=1 subviral particles (SVP) with an all-pentamer capsid, whereas pVP2 or intermediate pVP2 variant expression leads to the assembly of tubular structures with a hexagonal lattice (9, 39). The pVP2 C-terminal domain, a 71-residue sequence (Fig. 1), is responsible for allowing the formation of multiple VP2 conformations; specifically, this ability resides within the amphipathic α-helix at residues 443 to 452 (39). VP3, the other major structural protein, also participates in the inherent polymorphism of pVP2 through interaction with the pVP2 C-terminal end (36), working as a canonical scaffolding protein. In the absence of VP3, efficient T=13 capsid-like assembly can be achieved but requires a His tag bound covalently at the VP2 N terminus (chimeric protein His-VP2-466; 466 residues), which probably mimics VP3 function during virion assembly (39). Comparison of VP3 and His tag sequences revealed some degree of similarity between the VP3 C-terminal residues (DEDLE) and the acidic region of the His tag (DYDIPTTE).
The atomic structure of the T=13 virion was recently solved by X-ray crystallography to 7 Å (12), and that of the T=1 SVP capsid was resolved to 3 (12) and 2.6 Å (21, 27). The VP2 subunit is folded into three domains, termed the projection (P), shell (S), and base (B) domains (Fig. 1). Domains S and P are β-barrels, oriented such that the β-strands are tangential and radial to the particle surface, respectively. The B domain consists of N- and C-terminal α-helices that line the inner capsid surface. Domains S and B together are unexpectedly similar to the capsid proteins (CPs) of positive-sense ssRNA viruses, such as noda- and tetraviruses (12). In this study, we investigated different IBDV-related structures at or close to subnanometer resolution, combined with VP2 atomic models. The availability of three-dimensional models close to the native state allows us to define major conformational changes of specific secondary structural elements of VP2 that regulate the formation of pentamers and hexamers during the assembly process.
MATERIALS AND METHODS
Purification of virions and pVP2 deletion mutant protein-derived capsids.
IBDV strain Soroa was purified from QM7 quail muscle cells (39) and stored in PES buffer [25 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.2, 150 mM NaCl, and 20 mM CaCl2]. H5 insect cells were infected with the appropriate recombinant baculoviruses (39), and T=1 SVP and His-VP2-466 capsids were purified as described previously (9).
Cryo-electron microscopy (cryo-EM).
Capsid samples (5 μl) were applied to one side of a holey carbon film, washed twice with water, blotted, and plunged into liquid ethane following standard procedures (9, 39). Micrographs were recorded under low-dose conditions (∼10 e−/Å2) in a Tecnai G2 electron microscope operating at 200 kV and equipped with a field emission gun, using a nominal magnification of ×50,000 (calibrated using the 40.5-Å axial spacing of the phage T4 tail sheath).
Image analysis.
General image processing operations were performed using Bsoft (http://www.niams.nih.gov/rcn/labbranch/lsbr/software/bsoft/), Xmipp (http://xmipp.cnb.csic.es/), and Spider (http://www.wadsworth.org/spider_doc/spider/docs/). For selected micrographs analyzed (129 for IBDV particles, 82 for His-VP2- 466 capsids, and 23 for T=1 SVP), defocus values determined with Bshow (http://www.niams.nih.gov/rcn/labbranch/lsbr/software/bsoft/bshow/bshow.html) ranged from 0.6 to 3.0 μm (first zeros of the contrast transfer function [ctf] at spacings of 11 to 28 Å). A Zeiss PhotoScan TD scanner was used to digitize suitable micrographs at 7 μm/pixel (1.4 Å at the specimen), and micrographs were binned to give 14 or 21 μm/pixel during initial refinement steps. X3d (11) was used to manually extract particle images, including 15,793 images for IBDV particles, 1,977 images for His-VP2-466 capsids, and 26,386 images for T=1 SVP. Initial estimates of the origin and orientation angles were determined for a base set of particles, using polar Fourier transform procedures (2), taking previous maps, appropriately scaled at a 20-Å resolution, as a starting model (39). A new density map was calculated and used for all subsequent orientation and origin refinements. Phases were ctf corrected by flipping them in the required lobes of the ctf with the Bsoft bctf routine. Enhancement of the high-resolution Fourier amplitudes was based on X-ray data from SVP (21) and IBDV (12) capsids and was carried out with Spider. Amplitude decay was calculated using the spatial frequency components from the cryo-EM maps (with an effective resolution determined by Fourier shell correlation [FSC]) and corresponding X-ray maps. The decay profile of the cryo-EM maps was then adjusted to match the profile of the X-ray maps, and the fitted function was applied to the cryo-EM maps in the frequency range from 245 Å to the maximum resolution achieved. A soft low-pass filter was then applied.
Three-dimensional density maps were calculated using Fourier-Bessel methods (13). The final reconstructions combined 9,483, 988, and 23,754 images for IBDV, His-VP2-466, and SVP capsids, respectively. Resolution was assessed by FSC calculated between independent half-data-set maps, applying a correlation limit of 0.5 (or 0.3). The resolutions calculated for IBDV, His-VP2-466, and T=1 SVP capsids were 11.6 Å (10.4 Å), 15.4 Å (14.0 Å), and 7.8 Å (7.2 Å), respectively (Fig. 2).
FIG. 2.
Assessment of the resolutions of the T=1 SVP, virion capsid, and His-VP2-466 capsid reconstructions. FSC resolution curves were calculated for the T=1 SVP capsid (blue line), the T=13 virion capsid (red line), and the T=13 His-VP2-466 capsid (green line). Each set of particle images was subdivided randomly into two subsets, and independent reconstructions were computed from these data. The resolutions where the correlations drop below 0.5 and 0.3 are indicated. For the 0.3 threshold, the value for IBDV was 10.4 Å, that for the His-VP2-466 capsid was 14.0 Å, and that for T=1 SVP was 7.2 Å (for the 0.5 threshold, the values were 11.6, 15.4, and 7.8 Å, respectively).
Density maps and atomic models were visualized with UCSF Chimera software (http://www.cgl.ucsf.edu/chimera/), except for Fig. 8A and B, which were made with PyMol (http://pymol.sourceforge.net).
FIG. 8.
Structural conformations of pVP2 amphipathic α-helix. (A and B) Inner surfaces of a pentamer (A) and a hexamer (B). Surfaces are represented with electrostatic potentials calculated with the GRASP program, showing the distributions of negative (red) and positive (blue) charges. Note the narrow channel (diameter, ∼20 Å) formed by five α3 helices around the fivefold axis, in contrast with the broader channel at the quasi-sixfold axis (diameter, ∼35 Å). The insets indicate the two classes of contacts between adjacent VP2 trimers, namely, bent at the fivefold axis (two pentameric trimers in red) or flat at the local sixfold axis (a pentameric red trimer and a hexameric green trimer). (C and D) Side views of ribbon diagrams of a VP2 pentamer (C) and a hexamer (D). For simplicity, the hexamer front half has been removed. The amphipathic α-helices are shown in blue. VP2 chains are shown in magenta, except for the last visible C-terminal amino acid, which is represented as an orange sphere. At the pentamer (C), αNter helices (yellow) are shown fitted as described in the legend to Fig. 6G to I. At the hexamer (D), αNter helices (green) are unchanged with respect to the original X-ray model.
Atomic model manipulation and fitting into cryo-EM maps.
The crystal structures of the T=13 virion capsid and the T=1 SVP containing the first 441 VP2 residues have been determined to 7-Å and 3-Å resolutions, respectively, by Coulibaly et al. (12) (PDB entries 1WCE and 1WCD). Recently, we determined the crystal structure of the closely related T=1 SVP containing the first 452 amino acids of pVP2 at a 2.6-Å resolution (21) (PDB entry 2GSY). Situs (48; http://situs.biomachina.org/) and Uro (34) were used to fit the atomic structures of VP2 derived from SVP (PDB entries 2GSY and 1WCD) and the IBDV T=13 asymmetric unit (PDB entry 1WCE) to cryo-EM maps. For the T=1 SVP, both programs yielded essentially the same fits. Uro fitting was carried out on the entire cryo-EM map; each VP2 chain (PDB entry 1WCD) in the complete X-ray structure was treated independently, and icosahedral symmetry was applied. The excellent matching of the original and fitted X-ray models demonstrated that the shell and protruding spikes are stable structures. Situs fitting was carried out on local volumes encompassing the target density (segmentation of the volume corresponding to a trimer or a pentamer was done with Spider), and each X-ray model (comprising three or five VP2 chains) was treated as a single rigid body. For T=13 capsids, a similar approach was performed. Uro yielded essentially identical results with the three X-ray models used for the T=13 cryo-EM maps, with 13 independent VP2 monomers, 4 independent trimers (a to d) and a monomer (e), and the entire asymmetric unit, with a root mean square deviation between C-α positions of 1.8 Å. Situs fitting was carried out on densities comprising the asymmetric unit, a pentamer, and the two classes of hexamers for IBDV and His-VP2-466 capsids.
The T=1 SVP cryo-EM density encompassing helix α4 at the C-terminal end of VP2 (residues 434 to 439) was revealed in a difference map. Since helix α4 in the X-ray model did not superimpose with its corresponding rod-like density in the cryo-EM map, the following two equivalent X-ray models were used: SVP made by VP2-441 (12) (PDB entry 1WCD), in which helix α4 is not visible, and SVP made by VP2-452 (21) (PDB entry 2GSY) after deleting the position information for the α4 helix. Identical difference maps were obtained by subtracting either X-ray model from the cryo-EM reconstruction. Next, a region in the difference volume encompassing a VP2 trimer was used to fit the α4 helix by the colores routine of Situs. Situs generated the same set of three nearly identical, symmetry-related results. Correlation coefficients between atomic models and cryo-EM density maps were calculated with Uro and Spider as described previously (44) and were as follows: for T=1 SVP, 89%; for the T=13 virion capsid, 85%; and for the T=13 His-VP2-466 capsid, 80%.
The helix αNter (residues 1 to 10) in pentamers of the T=13 capsids did not superimpose with their corresponding rod-like densities in the cryo-EM map. T=13 X-ray and cryo-EM maps were less comparable in the resolution range analyzed, and numerous differences existed between them (see Results). For those reasons, the helix αNter was manually fitted as a rigid body to the nearest empty density.
Connecting loops between helices α4 and α3 at the C terminus (for T=1 SVP) and α1 and αNter at the N terminus (for T=13 capsids) were modeled using implementation of the Cyclic Coordinate Descent algorithm (8) included in S-flexfit (http://www.biocomp.cnb.uam.es/biocomp/public/Software/S_flexfit_web/), and geometry idealization was performed with the REFMAC5 program (33).
Electrostatic surface potentials for the VP2 pentamer and hexamer (lacking domain P) were calculated with Grasp (35). For this purpose, the 2.6-Å atomic coordinates of VP2 from a T=1 SVP were superimposed on the C-α carbon skeletons of the VP2 subunits conforming the pentamers and hexamers, determined at 7 Å. The side chain coordinates of the nonmatching residues were generated with the server MaxSprout (http://www.ebi.ac.uk/maxsprout/index.html).
The pVP2 amphipathic α helix (443-GFKDIIRAIR-452) was generated by using residues 241-EFRDIIDATR-250 of the Leishmania mexicana triose phosphate isomerase (PDB entry 1AMK), which have a nearly identical amphipathic character, as a template. The correct VP2 sequence was then replaced using the graphic program O (25).
Protein structure accession numbers.
Density maps have been deposited in the Macromolecular Structure Database at the European Bioinformatics Institute, with accession numbers EMD-1237 for T=1 SVP, EMD-1238 for IBDV, and EMD-1239 for His-VP2-466.
RESULTS
Structure of T=1 SVP.
A cryo-micrograph of T=1 SVP assembled from a 456-amino-acid pVP2 variant is shown in Fig. 3A. The final density map of the T=1 capsid was calculated to a 7.2-Å resolution (Fig. 3B and C). The molecular architecture of the dodecahedral capsid is essentially as previously described (6, 9, 12); the most prominent feature is the presence of 20 trimeric protrusions arranged equivalently (Fig. 3C). At this resolution, numerous rod-like and, less clearly, sheetlike densities are apparent (Fig. 3B and C). Secondary structural elements were identified by docking the closely related 2.6-Å VP2 X-ray map (containing the first 452 residues; PDB entry 2GSY) (21) into the cryo-EM map (Fig. 3D to F). Using two quantitative model-fitting programs, Situs (48) and Uro (34), the results were almost identical. Independently, secondary structural elements were partially identified with Helixhunter (22) as well as with Sheetminer and Sheettracer (26; not shown). The concordance of the two maps is clear in the matching of P and S domain β-barrels in the corresponding regions of the cryo-EM map, which shows β-sheets of both domains (Fig. 3E and F).
FIG. 3.
T=1 SVP cryo-EM reconstruction. (A) Representative cryo-electron micrograph of T=1 SVP capsids. Bar = 200 Å. (B) Central section of the SVP map. Red arrows point to the densities (red circles) where the α4 helix was fitted. Bar = 25 Å. (C) Shaded surface representation of the complete SVP, at 7.2-Å resolution, with a highlighted VP2 trimer (white). Bar = 25 Å. (D) SVP 50-Å-thick slab. VP2 (PDB entry 2GSY) secondary structure elements are color coded by domain (red, P; blue, S; and green, B). (E) VP2 trimer X-ray model (PDB entry 2GSY) fitted into the corresponding density in the 7.2-Å cryo-EM map. The VP2 chain (452 residues) lacks 7 N-terminal and 11 C-terminal residues. The C-terminal α4 helices from adjacent VP2 chains that project towards the base of the threefold axis are not shown. Arrows indicate the cut directions to show P and S domain fits in panel F. (F) Transversal sections through P and S domains to visualize the fit of the two β-sheets. The map is contoured at 2σ above the mean density (i.e., less density is visible), showing the physical separation of the two β-sheets and a fictitious cavity (the hydrophobic core) between them.
Conformational flexibility of the VP2 C-terminal α-helix.
VP2 domain B (PDB entry 2GSY) is formed mainly by α-helices from N- and C-terminal extensions facing the T=1 SVP inner surface. Whereas the N-terminal α1 and C-terminal α2 and α3 helices fitted remarkably well in the cryo-EM structure, the C-terminal α-helix α4 did not have a corresponding density in the cryo-EM map (Fig. 4A; also see Fig. 1). At the achieved resolution in our cryo-EM map, disordered VP2 segments remain invisible, as in the equivalent X-ray map. In the 2.6-Å-resolution map, helix α4 of each VP2 molecule is projected towards the threefold axis of a neighboring VP2 trimer, mediating domain swapping (21) (Fig. 4A, inset). The cryo-EM reconstructed volume shows a discernible vacant rod-shaped density located at the base of each VP2 molecule, adjacent to helix α4 of a neighboring VP2 molecule (Fig. 3B, red arrows, and 4A, black arrows). This rod-shaped density was the strongest feature in a difference map calculated between our cryo-EM map and a density map for a truncated version of the SVP X-ray structure in which helix α4 was manually deleted (data not shown). In addition, we considered that the VP2 structure in the vitrified T=1 SVP was analyzed under conditions closer to the native state than the crystallized one (e.g., in the presence of 17% polyethylene glycol and 3% isopropanol) and assumed that the α4 folding was maintained in both maps.
FIG. 4.
Conformational flexibility of the VP2 C-terminal α-helix. (A) Bottom view (facing the inner surface of the particle) of the X-ray model of the VP2 trimer fitted into the T=1 SVP cryo-EM map. α-Helices (1 to 4) of domain B (green) are indicated for a VP2 chain. α3 and α4 helices of three neighboring VP2 chains (magenta) that project towards the threefold axis of the trimer are also shown (labeled for a neighboring VP2 chain). Black arrows point to the three cryo-EM densities that remained empty after fitting of the original VP2 X-ray model (PDB entry 2GSY). The schematic diagram in the inset indicates bottom and side views of four VP2 trimers after filtration to 15 Å. Note the α-helix swapping (bottom view) and bent contacts (side view) between T=1 SVP VP2 trimers. (B) Same as panel A, but the C-terminal α4 helices of neighboring VP2 chains (shown in yellow and contoured in magenta) were remodeled to completely fill the cryo-EM density. Curved black arrows indicate that the conformational flexibility essentially involves bending the loop preceding the α4 helix. The C-terminal α4 helices of VP2 chains at the threefold axis are shown unaltered (green).
The proximity between helix α4 of a neighboring VP2 molecule and the rod-like density suggests the plausible possibility of fitting helix α4 into this rod, thus maintaining the domain swapping detected in the X-ray map (Fig. 4B) (see Materials and Methods). The proposed movement involves allowed rigid-body rotations of the C-α angles in the loop preceding helix α4. We cannot rule out the possibility that this movement is undergone by helix α4 of the same chain (rather than by that from the neighboring chain), although in this case the movement required for the α-helix is much greater than in the former possibility. In any case, both alternatives imply that the C-terminal α4 helix is flexible. This conformational flexibility would be critical when temporally bound peptides are attached at the end of the α4 helix, as in the precursor pVP2.
Structure of the T=13 virion capsid.
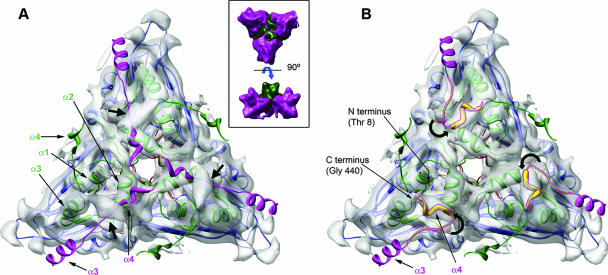
Although the IBDV capsid X-ray structure has been described at 7-Å resolution, detailed analysis of our cryo-EM data set allowed the observation of important differences, especially in flexible regions. The T=13 capsid density map was calculated to a resolution of ∼10 Å (Fig. 5A). The IBDV capsid surface consists of 260 protruding VP2 trimers, arranged in five distinct conformations (a to e), on the outer surface. The asymmetric unit is formed by four trimers (a to d), termed the G4 triangle (Fig. 5A, white triangle), and one trimer e subunit. In the T=13 lattice, trimer e bridges three G4 triangles to make the 20 flat faces of the icosahedral capsid. We used Uro to position the VP2 X-ray map from the 7-Å crystal structure of the IBDV capsid (12) within the virion cryo-EM map (Fig. 5A) and Situs to analyze local volumes. Besides the excellent matching of P and S domains, the inner surface of the cryo-EM virion map showed densities beneath each B domain with no corresponding masses in the VP2 atomic models (Fig. 5B, shown in yellow). Since all VP2 N-terminal segments (comprising about 10 to 12 residues) were invisible in the X-ray structure (Fig. 5B, inset), except those of the pentameric VP2 chains (Fig. 5B, inset, F chain), we interpreted these densities as disordered VP2 N-terminal segments. Comparison of full IBDV and empty His-VP2-466 capsids supported our interpretation (see below). In addition, fitting five VP2 chains into a segmented virion pentamer showed that the N-terminal α-helix (containing residues 1 to 10) (Fig. 1), which is disordered in SVP VP2, was the only secondary structural element that was outside the cryo-EM density map (Fig. 6A, cyan areas). This N-terminal α-helix, denoted αNter, fitted correctly in the nearest empty cryo-EM density by a rotation in the downstream loop (segments 11 to 13) and was positioned parallel to the inner surface of the pentamer (Fig. 6B and C, orange helices). The N-terminal αNter helix in the T=13 capsid therefore exhibits conformational flexibility, like the C-terminal α4 helix in the T=1 SVP capsid (Fig. 6D to F).
FIG. 5.
Fit of the IBDV X-ray model into the virion cryo-EM map. (A) Complete capsid virion (yellow), rendered at ∼10 Å, with the superimposed X-ray IBDV structure (magenta). The five VP2 trimer types are indicated by letters a to e; the white triangle indicates the limits of a G4 triangle. The locations of fivefold, threefold, and twofold axes are also indicated. Bar = 100 Å. (B) View from inside the capsid of the asymmetric unit X-ray model fitted into the corresponding region of the cryo-EM map. White surfaces show densities according to the X-ray model, displayed at a contour slightly lower than that of the cryo-EM map. Even at this threshold for the filtered atomic model, with P and S domains being well matched, the solid yellow densities correspond to empty cryo-EM virion densities; these densities probably arise from disordered VP2 N-terminal segments. A scheme of the asymmetric unit, seen from the inner surface, is shown in the inset. The first and last residues visible in the X-ray model are indicated for each of the 13 VP2 chains (A to M).
FIG. 6.
VP2 pentamer X-ray model fitted into virion, SVP, and His-VP2-466 capsids. (A) A VP2 pentamer (PDB entry 1WCE; VP2 chain F lacks five C-terminal residues from position 441) was fitted into the corresponding T=13 virion pentameric cryo-EM density. The αNter helix is shown in cyan; after applying the rotation movement in the downstream loop, the αNter helix is shown in orange, as fitted to the closest empty cryo-EM density. Black arrows indicate directions of movement. (B and C) Detailed bottom (B) and side (C) views of the T=13 virion shell innermost region at the fivefold axis. αNter helix positions are color coded as in panel A. In these views, only the C-terminal α3 helices are shown (magenta). (D) A VP2 pentamer (PDB entry 2GSY) was fitted into the corresponding T=1 pentameric cryo-EM density. The C-terminal α4 helix is shown in magenta; after applying appropriate rotation movement in the preceding loop, the α4 helix is shown in yellow and fitted to the closest rod-shaped cryo-EM density. Black arrows show directions of movement. (E and F) Detailed bottom (E) and side (F) views of the T=1 shell innermost region at the fivefold axis. Helix α4 positions are color coded as in panel D. In these views, only the N-terminal α1 helices are shown (green). (G) A VP2 pentamer (as in panel A) was fitted in the corresponding T=13 His-VP2-466 pentameric cryo-EM density. For clarity, the helical bundle density (see text) has been removed computationally. The αNter helix is shown in cyan; after applying rotation movement to the downstream loop, the αNter helix is shown in orange and fitted into the closest empty cryo-EM density. Black arrows indicate directions of movement. (H and I) Detailed bottom (H) and side (I) views of the T=13 His-VP2-466 shell innermost region at the fivefold axis. αNter helix positions are color coded as in panels A and G. Only C-terminal α3 helices are shown (magenta).
The molecular swapping mediated by helix α4 in the T=1 SVP is not present in T=13 particles, as shown both in the 7-Å X-ray structure of intact virions and in our 10-Å cryo-EM map. This feature can be explained by the coplanarity of neighboring VP2 trimers in the T=13 shell asymmetric unit (Fig. 5B), whereas VP2 trimers in the T=1 SVP form a dihedral angle of ∼144°, facilitating α4 helix swapping (Fig. 4A, inset).
Structure of the His-VP2-466 capsid.
The chimeric protein His-VP2-466, containing an N-terminal His tag and the nonprocessed amphipathic α-helix (see scheme in Fig. 1), assembles into empty T=13, T=7, and T=1-like particles (39). We fitted the IBDV capsid X-ray model into the T=13 cryo-EM map determined at a 14-Å resolution (Fig. 7A and B). The outer surfaces of the IBDV and His-VP2-466 capsids were almost superimposable, whereas the inner surfaces showed clear differences (39). As inferred above for virion capsids, we also detected similar extra densities beneath each VP2 domain B. These extra densities should therefore correspond to the common segments between CPs that form both T=13 capsids, i.e., the disordered N-terminal segment of VP2 chains in the asymmetric unit and, to a lesser extent, some C-terminal residues (Fig. 5B, inset).
FIG. 7.
Fit of the virion asymmetric unit into the chimeric His-VP2-466 cryo-EM map. (A) Inner surface view of the X-ray model of the asymmetric unit fitted into the corresponding region of the T=13 His-VP2-466 cryo-EM map. Cyan surfaces indicate 4 of the 132 (12 pentameric and 120 hexameric densities) difference densities calculated by subtracting IBDV from the His-VP2-466 capsid. Cyan arrows indicate the difference densities used to model and fit five and six amphipathic α-helices (segment 443-452 of pVP2 [blue]) at a fivefold axis (bottom left) and a quasi-sixfold axis (top right), respectively. (B) Same as panel A, but showing a side view section of the shell. Note that the S and P domains match with their corresponding densities. The model used for the amphipathic α-helix is shown (inset).
The main structural difference is related to the densities distributed at the local sixfold (120) and fivefold (12) axes in the His-VP2-466 capsid, which are absent in the IBDV capsid (Fig. 7A and B, cyan helices). These densities were mostly attributable to the C-terminal amphipathic α-helix (443-GFKDIIRAIR-452) (Fig. 7B, inset), since His tags tend to form a long loop or random coiled regions (18, 30). We consequently modeled and manually fitted the amphipathic α-helices in these extra densities following two different arrangements, namely, a compact bundle of five α-helices at the fivefold axis and an open star-like ring of six α-helices at the local sixfold axis (Fig. 7A and B, shown in blue). A bundle of five amphipathic α-helices not only accounts for the egg-like densities at the fivefold axes but is also stabilized by interactions among the hydrophobic surfaces of the helices, which would therefore be protected from the local hydrophilic environment (Fig. 8A). Following a similar approach to that used for the SVP and IBDV pentamers, we found that the VP2-466 chain helix αNter also displays conformational flexibility; αNter helices were reoriented, making an ∼45° angle with the capsid inner surface (Fig. 6G to I). This change is probably necessary to accommodate the densities corresponding to the helical bundle and the His tag.
The following two major reasons support the modeling of the amphipathic α-helices at the local sixfold axis with an “open” conformation: (i) connectivity arms with appropriate dimensions to fit an α-helix were visualized directly at the quasi-sixfold axis in the cryo-EM map (Fig. 7A and B), and (ii) inspection of the electrostatic potential at the inner surface shows the hydrophobic local environment that would favor this conformation by interacting with the hydrophobic side of the amphipathic helix (Fig. 8B). In this pseudo-atomic model of the His-VP2-466 capsid, the pentameric helical bundle protrudes inwards, since α3 helices of the VP2 domain B form a pentameric channel with a small, ∼20-Å diameter (Fig. 8A and C), whereas amphipathic α-helices are partially included in a broader central region on the inner hexamer surface (Fig. 8B and D). These distinct spatial arrangements establish different relationships between adjacent VP2 trimers; VP2 trimers building the same pentamer are tilted (as in T=1 particles and native T=13 virions), making bent contacts between them (dihedral angle, ∼144°), whereas hexameric VP2 trimers are oriented with domain P almost orthogonal to the outer capsid surface, making flat contacts (dihedral angle, ∼180°) (Fig. 8A and B, insets).
DISCUSSION
Structural polymorphism and transient conformations are common in multiprotein assemblies, such as those described during cyclic reactions of chaperones (37), dynamic biomachines such as ribosomes (31) and spliceosomes (41), and virus capsid assembly and maturation (42). The IBDV assembly pathway is based on a transient molecular switch responsible for the inherent structural polymorphism of pVP2; for this reason, no vestige remains in the mature capsid indicating how this event occurred. The results indicate that the spatial conformation of the (p)VP2 C-terminal region defines whether a pentamer or a hexamer is formed. Once the C-terminal region has performed this function, it is removed, probably as a safety mechanism, to render the conformational state irreversible.
Proteolytic processing of the capsid protein precursor.
The IBDV polyprotein is a fusion of precursor pVP2 with the viral protease (VP4) and the scaffolding protein (VP3). pVP2 undergoes sequential C-terminal processing events mediated by VP4. The resulting pVP2 intermediate is further cleaved by an unknown mechanism to render the mature VP2. The progressive processing of pVP2 thus seems to avoid the natural tendency of mature VP2 to assemble into all-pentameric T=1 SVP. An effective assembly pathway that avoids the dominance of aberrant assemblies would be provided by a postassembly maturation step in which virions are initially assembled as a procapsid that matures through structural transitions.
Biochemical studies of infectious pancreatic necrosis virus, a closely related birnavirus, showed the existence of a provirion (46) that matures by pVP2 processing. Postassembly maturation has been described for noda- and tetraviruses, which assemble as provirions with T=3 (40) and T=4 (7) lattices, respectively, composed of a single α-protein. Provirion maturation is relatively slow and takes place through autocatalytic cleavage of the α-protein (49) mediated by a single aspartic acid residue. This yields the mature CP β and the γ-peptide and leads to rearrangement of the capsid subunits. Considering the functional links (1) and the unexpectedly high structural similarity between CPs of IBDV and noda- and tetraviruses (12), as well as their similarities in the assembly pathway, we postulate that VP2 autoproteolysis is the final processing event that renders the mature VP2 protein. Furthermore, VP2 has two aspartic acid residues (Asp391 and -431), either of which could act as a catalytic residue, as in the CPs of nodavirus provirions. These residues are in close proximity to VP2 Ala441, located on the interior of the protein shell and inaccessible to cellular enzymes.
Pentamer assembly.
Although SVP pentamer assembly is independent of any other viral component, the corresponding virion and His-VP2-466 capsid pentamers are built in a more physiological context, and additional factors must be considered. Comparison of almost identical pentamers from the various analyzed structures suggests that the relatively large pVP2 C-terminal domain would interfere with the fivefold contacts. This interpretation is supported by the analysis of T=1-like capsids made by His-VP2-466, whose behavior was refractory when we attempted to identify their orientations (lack of icosahedral symmetry). The structure of the T=13 His-VP2-466 capsid indicated that the VP2 C-terminal domain is processed sufficiently to allow contacts for those VP2 chains at the fivefold axis. In virion assembly, this rapid processing would preclude the interaction with VP3, resulting in α-helical bundle formation. As a result, VP2 molecules associate as a pentamer in which contacts between neighbor trimers are bent. Finally, amphipathic α-helices would be removed. This is reminiscent of basic concepts from the well-established nodavirus assembly system (40). The different lattice geometry (T=13 versus T=3), together with increased complexity, indicates a more sophisticated assembly pathway for IBDV. The nodavirus capsid is formed by 60 trimers, and the CP forms two different contacts, as follows: (i) those related by icosahedral twofold symmetry axes form 60 flat contacts with a 180° dihedral angle, and (ii) those related by quasi-twofold symmetry axes form 120 bent contacts with a 144° dihedral angle. Flat contacts are due primarily to insertion of an ordered RNA duplex and, to a lesser extent, to the presence of an ordered N-terminal peptide arm in the groove between subunits (19). Kinetic studies suggested that 120 bent subunits are cleaved at a higher rate than the remaining 60 flat subunits (20). In the T=13 lattice, the contacts between VP2 trimers at the fivefold axis are similarly bent, with a dihedral angle of ∼140°. Rapid processing would lead to helical bundle formation that, once again, is similar to the pentameric helical bundle made by the γ-peptide in the nodavirus capsid (10). In this model, VP2 pentamers could act as nucleating centers for capsid assembly after joining additional VP2 trimers with flat contacts (see below).
The conformational flexibility of the α4 helix in SVP VP2 probably has a key role, when VP2 is still assembled as a pVP2 intermediate, for helical bundle formation and/or removal at the final assembly step. In the virion, most VP2 C-terminal ends remain disordered, suggesting a relaxed conformational state once the last proteolytic event has occurred.
Hexamer assembly.
Coplanar contacts are formed by VP2 trimers within a G4 triangle and by the e trimer. In our model, insertion of the VP2 amphipathic α-helix into the highly hydrophobic grooves that form the broad cavity at the quasi-sixfold axes would preclude the bending of these contacts. The amphipathic α-helix has an Asp residue (Asp446) between its hydrophobic and enriched basic residue sides, which could further stabilize the dominant hydrophobic interactions (Fig. 7B, inset, and 8B). Amphipathic α-helix behavior would resemble that of the ordered N-terminal peptide arm of the nodavirus capsid. Previous analysis of pVP2 polypeptides with variable-length C-terminal extensions showed that the longer the C-terminal domain, the more probable is hexamer formation, which would produce primarily tubular structures (39). Accordingly, the C-terminal domain cleavage would be slower and thus stabilized by its interaction with VP3.
Our analysis was restricted to His-VP2-466 capsid particles similar in size to IBDV virions from a sample containing heterogeneous assemblies in equilibrium. These particles probably represent an arrested morphogenetic intermediate and may constitute a set of conformationally metastable particles. This interpretation is reflected in the density arms around the quasi-sixfold axis, which are not identical, and one of which is almost completely absent (Fig. 7A).
Conclusions.
We show a scheme that summarizes our model for the IBDV assembly pathway in Fig. 9. Structural analysis of CP topology, virion architecture, and assembly between evolutionarily distant viruses revealed unexpected similarities (3). IBDV capsid assembly is controlled by scaffolding (VP3) and proteolytic (VP4 and possibly VP2) proteins that degrade short CP-specific sequences in consecutive events. This, together with the VP2 C-terminal flexibility, triggers interactions of the amphipathic α-helix for VP2 assembly into hexamers or pentamers. Interactions via amphipathic α-helices may be a common feature of macromolecular complex activation (24) and, as found for HIV assembly (43), may serve as an attractive target for future antiviral strategies. Our data for the IBDV assembly pathway reinforce the recently suggested evolutionary relationships between birnavirus and positive-strand ssRNA viruses.
FIG. 9.
IBDV assembly model. Mature VP2 is originally synthesized as a precursor, pVP2. The removable C-terminal extension (71 residues) contains the molecular switch, an amphipathic α-helix (segment 443-452), and is represented as a cylinder followed by a random coil. (A) When pVP2 (or shorter pVP2 variants) is expressed in a recombinant baculovirus system in the absence of other viral components, irregular helical tubes (all-hexamer structures) are formed (left); nonetheless, if the initial product is mature VP2, T=1 all-pentamer capsids form spontaneously (right). (B) The processing rate of the pVP2 C-terminal extension determines whether the C-terminal region (shown as a loop) of VP3 (green) can interact with pVP2. If processing is rapid, amphipathic α-helices associate to form a bundle, and a pentamer is formed with bent contacts between VP2 trimers (for simplicity, only a monomer is shown). In contrast, slow processing allows a longer, stable interaction between VP2 and VP3 C termini. Wavy lines indicate the dynamism of the VP3 interaction with VP2; six VP3 chains temporally stabilize hexamer formation between VP2 trimers with flat contacts, but the interaction ratio between pVP2 intermediates and VP3 might be higher.
Acknowledgments
We thank T. Baker, J. Conway, J. B. Heymann, and A. C. Steven for sharing reconstruction software and programming assistance, M. Valle for critically reading the manuscript, and C. Mark for editorial help.
D.L.B. was supported by an FPI fellowship from the Spanish Ministry of Education (MEC). J.R.C. was the holder of a contract from the Ramón y Cajal Programme (MEC). This work was supported by grants BFU2005-06487 and BIO2002-00517 from the Spanish Dirección General de Investigación (MEC) and by the Intramural Research Program of the NIH, Center for Information Technology. Support from EU contract LSHG-CT-2004-502828 is acknowledged.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Ahlquist, P. 2005. Virus evolution: fitting lifestyles to a T. Curr. Biol. 15:R465-R467. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 4.Becht, H. 1980. Infectious bursal disease virus. Curr. Top. Microbiol. Immunol. 90:107-121. [DOI] [PubMed] [Google Scholar]
- 5.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis ω virus (NωV). J. Mol. Biol. 299:573-584. [DOI] [PubMed] [Google Scholar]
- 8.Canutescu, A. A., and R. L. Dunbrack, Jr. 2003. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 12:963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castón, J. R., J. L. Martínez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodríguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, R. H., V. S. Reddy, N. H. Olson, A. J. Fisher, T. S. Baker, and J. E. Johnson. 1994. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway, J. F., B. L. Trus, F. P. Booy, W. W. Newcomb, J. C. Brown, and A. C. Steven. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 111:222-233. [DOI] [PubMed] [Google Scholar]
- 12.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 13.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos. Trans. R. Soc. Lond. B 261:221-230. [DOI] [PubMed] [Google Scholar]
- 14.Da Costa, B., C. Chevalier, C. Henry, J. C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobos, P., B. J. Hill, R. Hallett, D. T. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dokland, T. 2000. Freedom and restraint: themes in virus capsid assembly. Struct. Fold Des. 8:R157-R162. [DOI] [PubMed] [Google Scholar]
- 17.Feldman, A. R., J. Lee, B. Delmas, and M. Paetzel. 2006. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J. Mol. Biol. 358:1378-1389. [DOI] [PubMed] [Google Scholar]
- 18.Ferrer-Orta, C., A. Arias, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212-47221. [DOI] [PubMed] [Google Scholar]
- 19.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garriga, D., J. Querol-Audí, F. Abaitua, I. Saugar, J. Pous, N. Verdaguer, J. R. Castón, and J. F. Rodríguez. 2006. The 2.6-angstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J. Virol. 80:6895-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, W., M. L. Baker, S. J. Ludtke, and W. Chiu. 2001. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308:1033-1044. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. E. 1996. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. USA 93:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, C. A., F. S. Willard, M. R. Jezyk, Z. Fredericks, E. T. Bodor, M. B. Jones, R. Blaesius, V. J. Watts, T. K. Harden, J. Sondek, J. K. Ramer, and D. P. Siderovski. 2005. Structure of Gα(i1) bound to a GDP-selective peptide provides insight into guanine nucleotide exchange. Structure 13:1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, T., and M. Kjeldgaard. 1997. Electron density map interpretation, p. 173-208. In C. Carter and R. Sweet (ed.), Macromolecular crystallography, vol. B. Academic Press, London, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 26.Kong, Y., X. Zhang, T. S. Baker, and J. Ma. 2004. A structural-informatics approach for tracing β-sheets: building pseudo-C(α) traces for β-strands in intermediate-resolution density maps. J. Mol. Biol. 339:117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. C., T. P. Ko, C. C. Chou, M. Yoshimura, S. R. Doong, M. Y. Wang, and A. H. Wang. 2006. Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution: implications in virion assembly and immunogenicity. J. Struct. Biol. 155:74-86. [DOI] [PubMed] [Google Scholar]
- 28.Maraver, A., R. Clemente, J. F. Rodríguez, and E. Lombardo. 2003. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J. Virol. 77:2459-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maraver, A., A. Oña, F. Abaitua, D. Gonzalez, R. Clemente, J. A. Ruiz-Díaz, J. R. Castón, F. Pazos, and J. F. Rodríguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy, A. A., N. A. Peterson, R. Knijff, and E. N. Baker. 2004. Crystal structure of MshB from Mycobacterium tuberculosis, a deacetylase involved in mycothiol biosynthesis. J. Mol. Biol. 335:1131-1141. [DOI] [PubMed] [Google Scholar]
- 31.Mitra, K., and J. Frank. 2006. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu. Rev. Biophys. Biomol. Struct. 35:299-317. [DOI] [PubMed] [Google Scholar]
- 32.Morais, M. C., M. Fisher, S. Kanamaru, L. Przybyla, J. Burgner, B. A. Fane, and M. G. Rossmann. 2004. Conformational switching by the scaffolding protein D directs the assembly of bacteriophage φX174. Mol. Cell 15:991-997. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov, G. N., A. A. Vagin, A. Lebedev, K. S. Wilson, and E. J. Dodson. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55:247-255. [DOI] [PubMed] [Google Scholar]
- 34.Navaza, J., J. Lepault, F. A. Rey, C. Alvarez-Rua, and J. Borge. 2002. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D 58:1820-1825. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 36.Oña, A., D. Luque, F. Abaitua, A. Maraver, J. R. Castón, and J. F. Rodríguez. 2004. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322:135-142. [DOI] [PubMed] [Google Scholar]
- 37.Ranson, N. A., D. K. Clare, G. W. Farr, D. Houldershaw, A. L. Horwich, and H. R. Saibil. 2006. Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nat. Struct. Mol. Biol. 13:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez, A. B., and J. F. Rodríguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 39.Saugar, I., D. Luque, A. Ona, J. F. Rodriguez, J. L. Carrascosa, B. L. Trus, and J. R. Caston. 2005. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic alpha helix as a molecular switch. Structure 13:1007-1017. [DOI] [PubMed] [Google Scholar]
- 40.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50:381-446. [DOI] [PubMed] [Google Scholar]
- 41.Stark, H., and R. Luhrmann. 2006. Cryo-electron microscopy of spliceosomal components. Annu. Rev. Biophys. Biomol. Struct. 35:435-457. [DOI] [PubMed] [Google Scholar]
- 42.Steven, A. C., J. B. Heymann, N. Cheng, B. L. Trus, and J. F. Conway. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ternois, F., J. Sticht, S. Duquerroy, H. G. Krausslich, and F. A. Rey. 2005. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat. Struct. Mol. Biol. 12:678-682. [DOI] [PubMed] [Google Scholar]
- 44.Valle, M., A. Zavialov, J. Sengupta, U. Rawat, M. Ehrenberg, and J. Frank. 2003. Locking and unlocking of ribosomal motions. Cell 114:123-134. [DOI] [PubMed] [Google Scholar]
- 45.van den Berg, T. P., N. Eterradossi, D. Toquin, and G. Meulemans. 2000. Infectious bursal disease (Gumboro disease). Rev. Sci. Tech. 19:509-543. [PubMed] [Google Scholar]
- 46.Villanueva, R. A., J. L. Galaz, J. A. Valdes, M. M. Jashes, and A. M. Sandino. 2004. Genome assembly and particle maturation of the birnavirus infectious pancreatic necrosis virus. J. Virol. 78:13829-13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Einem, U. I., A. E. Gorbalenya, H. Schirrmeier, S. E. Behrens, T. Letzel, and E. Mundt. 2004. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J. Gen. Virol. 85:2221-2229. [DOI] [PubMed] [Google Scholar]
- 48.Wriggers, W., and S. Birmanns. 2001. Using Situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133:193-202. [DOI] [PubMed] [Google Scholar]
- 49.Zlotnick, A., V. S. Reddy, R. Dasgupta, A. Schneemann, W. J. Ray, Jr., R. R. Rueckert, and J. E. Johnson. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 269:13680-13684. [PubMed] [Google Scholar]