Abstract
The limited efficiency of in vivo gene transfer by replication-deficient retroviral vectors remains an obstacle to achieving effective gene therapy for solid tumors. One approach to circumvent this problem is the use of replication-competent retroviral vectors. However, the application of such vectors is at a comparatively early stage and the effects which virus strain, transgene cassette position, and target cell can exert on vector spread kinetics, genomic stability, and transgene expression levels remain to be fully elucidated. Thus, in this study a panel of vectors allowing the investigation of different design features on an otherwise genetically identical background were analyzed with respect to these readout parameters in cultures of both murine and human cells and in preformed tumors in nude mice. The obtained data revealed that (i) Moloney murine leukemia virus (Mo-MLV)-based vectors spread with faster kinetics, drive higher levels of transgene expression, and are more stable than equivalent Akv-MLV-based vectors; (ii) vectors containing the transgene cassette directly downstream of the envelope gene are genomically more stable than those containing it within the 3′-long terminal repeat U3 region; and (iii) the genomic stability of both strains seems to be cell line dependent.
Simple retroviruses are seen as ideal tools for introducing therapeutic genes into cancer cells in vivo, since they can transduce only actively dividing cells (21). During more than a decade of clinical studies, however, it has become apparent that replication-defective retroviral (RDR) vectors can infect only a fraction of cells comprising a solid tumor when applied using currently available delivery techniques (25). Attention, therefore, has begun to turn to the development of replication-competent retroviral (RCR) vectors which spread from cell to cell and deliver the therapeutic gene throughout the entire solid tumor mass. For such vectors to be efficacious, of course, it is imperative that the therapeutic gene is stably maintained within the retrovirus genome for at least as many replication cycles as are required for transduction of almost the complete tumor.
RCR vectors based upon murine leukemia virus (MLV) (1, 2, 4-7, 9, 10, 13, 16-20, 26, 29-36) represent the most promising candidates for human gene therapy, not least due to the fact that several hundred clinical trials over the last decade have provided detailed information regarding the safety profile of MLV-based vectors, to an extent which is not available for vectors based on other retroviruses (8).
RCR vectors have been generated containing transgenes placed at several different locations in the MLV genome, of which the most common and widely applicable designs have involved insertion of a transgene cassette into various positions of the U3 region of the MLV long terminal repeat (LTR) (1, 2, 4, 6, 7, 13, 26, 31) or immediately downstream of the envelope (env) gene in the 3′ untranslated region of the MLV genome (5, 7, 16-20, 30, 33-36). Alternative designs have included insertion of the enhanced green fluorescence protein (eGFP) gene into the proline-rich region of the env gene (9, 29) and insertion of a gene encoding a secreted protein after the signal peptide of the env gene (10, 32); however, use of these vectors is restricted to specific therapeutic applications requiring secreted gene products or gene products functional in the context of a fusion protein.
Of all the designs published to date, the two which are seemingly the most promising and widely applicable for cancer gene therapy both employ an internal ribosomal entry site (IRES) to direct expression of the transgene in infected cells (13, 19), a strategy which should allow stable propagation of any therapeutic gene of suitable size over several replication cycles. One variation of this strategy uses the encephalomyocarditis virus (EMCV) IRES placed 21 bp downstream of the 5′ end of the U3 region in an Akv-MLV vector to mediate transgenic mRNA translation (13); the other employs the EMCV IRES-transgene cassette inserted immediately downstream of the 3′ end of the 4070A amphotropic env gene in a Moloney MLV (Mo-MLV)-based RCR vector (19). Such vectors, for example, have been shown, by delivering and expressing the cytosine deaminase (CD) gene in experimental gliomas in vivo, to provide a considerable survival benefit to the treated animals (34-36).
The use of MLV-based RCR vectors for cancer gene therapy is therefore highly promising; however, the vector development process is still at a comparatively early stage. It is hence of great importance, at this juncture, to gain more knowledge regarding the effects of basic design features such as virus strain and transgene cassette position upon vector performance parameters such as replication kinetics, genomic stability, and transgene expression levels. In order for the effects of changes in individual design features to be evaluated and delineated from one another, it is essential that they are investigated on otherwise identical backgrounds and that the same assay system is applied in each case.
To this end, we have directly compared Mo-MLV-based and Akv-MLV-based vectors in which an identical IRES-eGFP transgene cassette was positioned either directly after the 4070A env gene or 30 bp downstream of the 5′ end of the 3′-LTR U3 region by carrying out serial infection cycles in both murine (NIH 3T3) and human (HEK293 and U87-MG) cell lines. Moreover, by using these vectors to infect preformed human tumors in nude mice, we were able to show that differences in the basic design of vectors also exert major effects on their spread kinetics and genomic stability in an in vivo model with direct relevance to cancer gene therapy.
MATERIALS AND METHODS
Recombinant viral constructs.
To generate plasmid pACEU3-GFP, the IRES-eGFP cassette was excised from plasmid pACE-GFP (17) by using BsiWI and NotI and the vector backbone was blunted using T4 DNA polymerase and religated. The excised IRES-eGFP cassette was also blunted and subsequently inserted into the religated vector, which was linearized with NheI and blunted. To construct plasmid pAkv4070A-eGFP, in which the ecotropic env gene is replaced by the amphotropic env gene, primers akvenpolext (5′-GAGAGCGTTGAACGCGCCATGATGGTTTTATCGGGGGCGT-3′) and akvenvforward (5′-AGCAAGAGTCGTGTCCAAGA-3′) were used to amplify a 681-bp sequence of plasmid pAkvEn-GFP (7) containing a unique SspI restriction site and primers pace4070ext (5′-ACGCCCCCGATAAAACCATCATGGCGCGTTCAACGCTCTC-3′) and pace7000rev (5′-TGTTCCTGTCCGGTACAGTC-3′) were employed to amplify a 666-bp sequence of plasmid pACE-GFP containing a unique BsrGI site by using PCR techniques. For fusion of the two fragments an overlap extension PCR was employed (12), resulting in a 1,347-bp product, which was then digested with SspI and BsrGI restriction enzymes and cloned into the large SspI/BsrGI fragment of plasmid pAkvEn-GFP, resulting in plasmid pAkv4070sub. Then, the 3,050-bp BsrGI fragment of plasmid pACE-GFP containing the amphotropic MLV 4070A env gene, the EMCV IRES, and the eGFP gene was inserted into plasmid pAkv4070sub linearized with BsrGI, resulting in plasmid pAkv4070A-eGFP. The integrity of plasmid pAkv4070A-eGFP was verified by sequencing.
In order to circumvent TRIM5α restriction factor-mediated resistance of human cell lines to Akv-based vectors, the arginine coding sequence AGA at codon position 110 of the capsid gene was changed to GAA, coding for a glutamine residue, by overlap extension PCR mutagenesis (12). To do so, primers mutagoutside (5′-CGATAATGGCGGACCTCTCA-3′) and mutbwd (5′-CTAGGTGGTTCCTACCTTCTTGGGTGGTGTA-3′), as well as primers mutagoutsideC (5′-AAGCCGACCTGTCACTTAGC-3′) and mutationfwd (5′-TACACCACCCAAGAAGGTAGGAACCACCTAG-3′), were used to PCR amplify upstream and downstream regions flanking the target sequence. Primer sequences that are underlined indicate the Arg-to-Glu codon exchange. The resulting DNA fragments were subsequently fused by overlap extension PCR using primers mutagoutside and mutagoutsideC, resulting in a 1,347-bp-long product, which was then digested with SacI and DraIII. Plasmid pAkv4070A-eGFP was digested with SacI, resulting in two fragments of 6,910 bp and 6,447 bp in length. The 6,910-bp fragment was further digested with SacII and DraIII, and the resulting 5,811-bp fragment was ligated with the 1,099-bp fusion fragment to obtain subclone pS1. Then, the SacI-linearized vector pS1 was ligated to the 6,447-bp fragment of pAkv4070A-eGFP to obtain plasmid pAkvB4070A-eGFP.
Cell culture, virus production, and titer measurement.
Human HEK293 cells (ATCC CRL-1573), mouse NIH 3T3 cells (ATCC CRL-1658), and the human glioma cell line U87-MG (ATCC HTB-14) were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Virus vector stocks were produced by transfection of HEK293 cells using the calcium phosphate coprecipitation method (11). Briefly, 2.5 × 106 cells were seeded in 10-cm dishes and transfected 24 h later with 8 μg of plasmids pACE-GFP, pACEU3-GFP, pAkv4070A-eGFP, and pAkvB4070A-eGFP, respectively. Virus-containing supernatant was collected 48 h following transfection and filtered through a 0.45-μm filter before aliquoting and storage at −20°C. Vector stocks were titrated by serial dilution of vector-containing supernatant and subsequent infections of NIH 3T3, HEK293, and U87-MG cells. Twenty-four hours postinfection zidovudine was added to a final concentration of 50 μM to prevent virus replication and spread, and 24 h later infected cells were analyzed by fluorescence-activated cell sorting (FACS) as described previously (20).
For the generation of high-titer vector stocks via ultracentrifugation viral vectors were pelleted at 50,000 × g for 1.5 h at 4°C using an SW40 swinging bucket rotor (Beckman) as previously described for the concentration of lentiviral vectors (22), and then resuspended in serum-free DMEM to 100th of the initial volume, aliquoted, and stored at −80°C.
Serial infection cycles with replicating vectors.
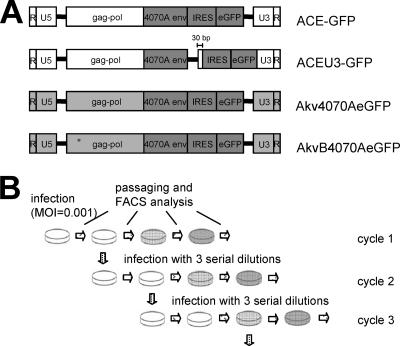
Cells (2 × 105 NIH 3T3, 1 × 106 HEK293, and 3 × 105 U87-MG cells) were seeded per well of a six-well plate on the day prior to infection. To initiate infection cycle 1, cells were transduced at a multiplicity of infection (MOI) of 0.001, as previously determined by individually titrating the respective vectors on each cell line. Two days later the cells were split 1:3 and one-third were analyzed for eGFP expression by FACS, one-third were subjected to DNA analysis, and the remaining cells were replated. Four days postinfection, supernatant from infected cells was serially diluted up to 1:100,000, depending on the replication kinetics of the respective vector, calculated from the previous infection cycle, to initiate infection cycle 2 (Fig. 1B). Cells infected with an MOI allowing exponential vector spread up to 4 days postinfection were used for virus harvest for the next cycle, cycle 3. Multiple serial infection cycles were continued in this way until no more than 2% of infected cells following serial passaging produced the eGFP protein.
FIG. 1.
MLV-based RCR vector constructs and experimental setup. (A) Mo-MLV-based (white vector backbone) and Akv-MLV-based (light gray vector backbone) vectors contain an IRES-eGFP cassette inserted either immediately adjacent to the 3′ end of the env gene or in the 3′-LTR U3 region. A glutamine-to-arginine transition at codon position 110 of the gag gene of vector AkvB4070A-eGFP is denoted by an asterisk. (B) Experimental setup for testing the genomic stability of vectors. Infection cycle 1 was initiated by inoculating cells with an MOI of 0.001, passaging cells 2 days later, and subjecting them to FACS analysis, followed by passaging and FACS analysis every 2 or 3 days until the percentage of eGFP-expressing cells no longer increased from one passage to the next. Four days postinfection, fresh cells were inoculated with three different dilutions of cell-free virus-containing supernatant to initiate infection cycle 2. Cells infected with a dilution factor resulting in infection levels of between 1% and 10% in FACS analyses 2 days postinfection were chosen for further analysis. This process was repeated for multiple serial infection cycles.
PCR analysis.
Genomic DNA was extracted from infected cells using the DNeasy tissue kit (QIAGEN), according to the manufacturer's recommendations. PCR amplification of DNA from infected cells was performed using primers hybridizing to the amphotropic env gene and in the 3′ LTR (5′-GGCCAAGGATGGTTCGAAGG-3′ and 5′-GCGCGAACAGAAGCGAGAAG-3′ in the case of vectors ACE-GFP and ACEU3-GFP and 5′-GAGACAGGCCAAGGATGGTT-3′ and 5′-CCTAGTGCTTGACCACAGAT-3′ in the case of vectors Akv4070A-eGFP and AkvB4070A-eGFP, respectively). PCR was performed for 30 cycles, and PCR products were analyzed by agarose gel electrophoresis. The quality of the extracted DNA was verified by performing a glyceraldehyde-3-phosphate dehydrogenase-specific PCR (25 cycles) using primers 5′-ATTTGCAGTGGCAAAGTGGAGATT-3′ and 5′-TATTTCTCGTGGTTCACACCCATC-3′.
Quantification of viral RNA, proviral DNA, and cellular DNA.
Viral RNA from supernatant of infected cells and genomic DNA from infected cells were extracted at infection cycle 1 using the viral RNA kit and the DNeasy kit (QIAGEN), respectively, according to the manufacturer's instructions. Extracted viral RNA was treated with Turbo DNase (Ambion) according to the manufacturer's instructions. Real-time PCR and real-time reverse transcription-PCR (RT-PCR) were performed using the eGFP-specific primers 5′-GCAGTGCTTCAGCCGCTAC-3′ and 5′-AAGAAGATGGTGCGCTCCTG-3′ and probe 6-carboxyfluorescein-5′-ACCACATGAAGCAGCACGACTT-3′-6-carboxytetramethylrhodamine, the 4070Aenv-specific primers 5′-GTAGCGGTCGTGGGCACTTATA-3′ and 5′-CTTATGTTGGGAAGTGGCCGTA-3′ and probe 6-carboxyfluorescein-5′-CATTCCACCGCTCCGGCCAACT-3′-6-carboxytetramethylrhodamine, and primers and probes specific for cellular rRNA coding sequences as previously described (14), respectively. Statistical analyses were performed using the one-sided Student t test for independent samples.
In vivo infection, quantification of vector spread, and histological analysis.
To analyze virus vector replication and spread in vivo, subcutaneous U87-MG tumors were established in 4-week-old male Hsd:athymic Nude-Foxn1nu mice (Harlan). All animal experiments were discussed and approved by the institutional ethics committee (GZ 68.205/53-BrGt/2004). Briefly, mice were anesthetized by intraperitoneal injection of ketamine (1 mg/10 g body weight) and xylazine (39 μg/10 g body weight) and 5 × 105 U87-MG glioma cells were injected into the right dorsal flank. Identification of the individual mouse was ensured by subcutaneous implantation of a microchip transponder (Virbac) into the left flank. Tumor growth was monitored twice a week by measurement with a caliper, and tumor size was calculated according to the formula length × width × width/2. At a tumor size of 60 to 100 mm3, normally 3 to 4 weeks after cell injection, 1 × 105 infectious viral particles, as previously determined by titration of each vector on U87-MG cells in vitro, were suspended in 20 μl DMEM and injected directly into the tumor center. Five mice per vector and time point were sacrificed by cervical dislocation 5, 10, and 20 days postinfection. Tumors were excised and divided into pieces. One tumor piece was fixed in 4% formaldehyde solution and used for immunohistochemical analysis. A second piece was used for preparation of a single-cell suspension for FACS analysis. Briefly, tumor pieces were minced in phosphate-buffered saline (PBS) and digested in 5 ml PBS-2 mg collagenase type 3 (Worthington Biochemical Corporation)/ml at 37°C for 1 to 2 h with continuous shaking at 320 cycles per min. Digestion was stopped by addition of an equal amount of DMEM supplemented with 10% fetal bovine serum. The resulting single-cell suspension was centrifuged at 180 × g for 5 min, and the recovered cells were fixed with 4% formaldehyde solution for 1 h, washed twice with PBS, and subjected to FACS analysis.
For immunohistochemical analysis, tumor pieces were fixed in 4% formaldehyde for 4 h, embedded in paraffin, and sectioned. Sections were mounted onto poly-l-lysine-coated slides, deparaffinized, rehydrated, and immersed in 3% hydrogen peroxide in methanol for 15 min to inactivate endogenous peroxidases. Nonspecific binding was blocked with 1.5% goat serum for 30 min. Then, the slides were incubated overnight with a 1:6,000 dilution of a rabbit anti-eGFP antibody (Molecular Probes) and a horseradish peroxidase-conjugated goat anti-rabbit antibody (Immunovision) for 30 min. Immunoreactivity was visualized using diaminobenzidine tetrahydrochloride as chromogen and Mayer's hemalum for counterstaining, Sections were analyzed with a Zeiss light microscope, and images were captured (Axiovision Systems; Zeiss).
RESULTS
Generation of MLV-based RCR vectors.
The accurate comparison of the effects of several parameters such as virus strain, transgene cassette position, and target cell on MLV-based RCR vector characteristics such as replication kinetics, genomic stability, and transgene expression is possible only if the individual genetic differences are compared independently of each other on an otherwise homologous background.
To this end, we generated a series of vectors which could be used to delineate the effects of these variables (Fig. 1A). The effect of insertion of a transgene cassette either contiguously to the 3′ end of the env gene or within the U3 region of the 3′ LTR was analyzed by comparing vector ACE-GFP (36) with vector ACEU3-GFP. Vector ACE-GFP is based on Mo-MLV and contains the 4070A amphotropic env gene and a 1.3-kb IRES-eGFP transgene cassette fused directly to the 3′ end of the env gene. To generate ACEU3-GFP, the IRES-eGFP cassette was excised from ACE-GFP and placed in the LTR, 30 bp downstream from the 5′ end of the U3 region.
The effect of the strain of MLV on which the vector is based was analyzed by comparing vectors ACE-GFP and Akv4070A-eGFP, the latter of which was generated by replacing the ecotropic env gene of Akv-MLV with the 4070A amphotropic env gene and the 1.3-kb IRES-eGFP transgene cassette originating from vector ACE-GFP, such that the resulting vector has sequence identity with ACE-GFP from the 5′ end of the env gene to the 3′ end of the eGFP gene. The remaining sequences of vectors Akv4070A-eGFP and ACE-GFP are wild-type Akv-MLV and wild-type Mo-MLV, respectively.
To analyze the effects which the type of host cell can exert upon vector characteristics, the behaviors of the different vectors were compared in both murine (NIH 3T3) and human (HEK293 and U87-MG) cell lines. In order to circumvent TRIM5-alpha restriction factor-mediated resistance of human cell lines to infection by N-tropic Akv-based vectors (24, 37), overlap extension PCR mutagenesis was employed to introduce a glutamine-to-arginine transition at position 110 in the capsid protein of vector Akv4070A-eGFP, generating the B-tropic vector AkvB4070A-eGFP, which is able to infect human cells.
Multiple serial infection cycles of RCR vectors.
A prerequisite for accurate and unbiased comparison of vector replication kinetics and genomic stability is that each vector has a constant supply of fresh cells into which it can spread, since otherwise differences in these parameters may be underestimated. Cells were therefore initially infected with vectors at a low MOI, and after the vector was allowed to spread, filtered supernatant from the infected cells was serially diluted and used to inoculate fresh cells. Preliminary analysis of the vectors in infection experiments clearly demonstrated that those based on Akv-MLV replicated with much slower kinetics than those based on Mo-MLV, and vectors containing the transgene cassette in the U3 region replicated with slower kinetics than those where the transgene cassette was inserted contiguously to the env gene (data not shown). To circumvent this problem, an experimental design was chosen which allowed each vector to replicate at its maximum rate throughout the course of the experiment, whereby viral supernatants transferred from one infection cycle to the next were diluted by a factor large enough to permit the inoculation of fresh cells at an MOI sufficiently low enough to ensure that enough uninfected cells were always available for infection by exponentially replicating vectors, at least until transfer of viral supernatant for inoculation of cells of the following infection cycle. In NIH 3T3 cells, the dilution factors required to achieve this were generally 10,000- to 100,000-fold for ACE-GFP, 100- to 1,000-fold for ACEU3-GFP, and 10- to 1,000-fold for Akv4070A-eGFP. In HEK293 cells, the dilution factors required to achieve this were generally 1,000-fold for ACE-GFP, 10- to 100-fold for ACEU3-GFP, and only 1- to 100-fold for AkvB4070A-eGFP. In U87-MG cells, the dilution factors required to achieve this were generally 1,000-fold for ACE-GFP, 100-fold for ACEU3-GFP, and only 1- to 10-fold for AkvB4070A-eGFP (data not shown). These results indicate not only that HEK293 and U87-MG cells are much more difficult to infect with MLV-based vectors expressing the 4070A envelope than are NIH 3T3 cells but also that vector ACE-GFP replicates with much faster kinetics than vector ACEU3-GFP, which in turn replicates with faster kinetics than Akv4070A-eGFP or AkvB4070A-eGFP.
Genomic stability of vectors in NIH 3T3, HEK293 and U87-MG cells.
Once a cell has been productively infected and begins to express nascent Env proteins, it cannot be subsequently superinfected by a second virus expressing the same Env protein, due to the phenomenon of retroviral superinfection resistance (23). Hence, if a cell is infected with a vector which has lost the ability to express eGFP, i.e., by deletion of the eGFP gene, this cell will remain non-eGFP-expressing even when cultured in the presence of other cells productively infected with eGFP-expressing vectors. FACS analyses of cells infected with MLV-based RCR vectors expressing eGFP can hence serve as an accurate indication of the genomic stability of these vectors (18, 20).
Infection of NIH 3T3 cells generated eGFP expression detectable by FACS in more than 90% of infected cells up to the third serial infection cycle, regardless of the vector used (Fig. 2A). From the fourth cycle onwards, however, vector ACEU3-GFP failed to reach these levels, and less than 50% of infected cells expressed eGFP by the fifth infection cycle. Both vectors, ACE-GFP and Akv4070A-eGFP, on the other hand, generated detectable levels of eGFP expression in more than 90% of infected cells up to the fifth serial infection cycle, and this did not decrease to below 50% until cycles 12 and 11, respectively (Fig. 2A).
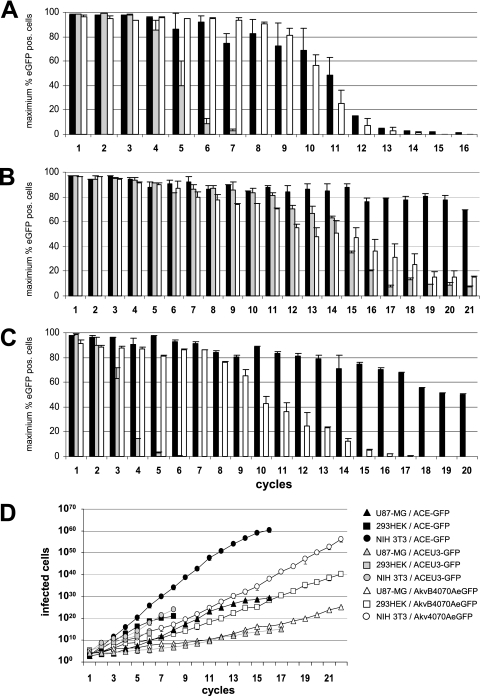
FIG. 2.
Genomic stability and in vitro replication kinetics of vectors over multiple infection cycles. (A to C) NIH 3T3 cells (A), HEK293 cells (B), and U87-MG cells (C) were infected with vectors ACE-GFP (black bars), ACEU3-GFP (gray bars), and Akv4070A-eGFP or AkvB4070A-eGFP (white bars), respectively, using an MOI of 0.001 and passaged until the percentage of eGFP-expressing cells no longer increased from one passage to the next. Cell-free virus-containing supernatant harvested from infected cells at the second passage was used to initiate a new infection cycle using a dilution factor sufficient to allow exponential spread for at least two passages. This process was repeated for multiple serial infection cycles. The bars shown indicate the maximum percentage of eGFP-expressing cells detected during each infection cycle. (D) Spread kinetics of the vectors were calculated using a mathematical model where each vector has an unlimited supply of fresh cells to infect and taking into account the dilution factors used to inoculate cells at each infection cycle and the number of eGFP-expressing cells detected by FACS following two passages postinfection.
In HEK293 cells, all vectors generated detectable levels of eGFP expression in over 90% of infected cells up to the fourth serial infection cycle (Fig. 2B). In contrast to the infection of NIH 3T3 cells, vector ACEU3-GFP did not become so rapidly unstable, proving to be roughly as stable as vector AkvB4070A-eGFP up until the 14th serial infection cycle (Fig. 2B). From cycle 15 onwards, however, vector ACEU3-GFP became unstable more rapidly than vector AkvB4070A-eGFP. As in NIH 3T3 cells, vector ACE-GFP was also clearly the most stable in HEK293 cells and still generated detectable levels of eGFP expression in 70% of infected cells up until the 21st serial infection cycle (Fig. 2B), at which point the experiment was terminated.
In U87-MG cells, as in NIH 3T3 and HEK293 cells, vector ACEU3-GFP was the least stable, with eGFP expression detectable in more than 90% of the infected cells only during the first infection cycle and in less than 50% of infected cells by the fourth infection cycle (Fig. 2C). In cells infected with vector AkvB4070A-eGFP, eGFP expression could be detected in more than 90% of infected cells only in the first serial infection cycle but could also still be detected in more than 50% of infected cells up until the ninth infection cycle. Vector ACE-GFP, however, was clearly the most stable in U87-MG cells, and eGFP could be still be detected in more than 90% of infected cells up until the seventh infection cycle and in more than 50% of infected cells up until the 20th infection cycle (Fig. 2C), at which point the experiment was terminated.
Taken together, these results indicate that positioning of the transgene cassette immediately adjacent to the env gene generates vectors which are genomically more stable than when the transgene was positioned in the U3 region, that the strain of MLV used to generate the RCR vector can also influence the genomic stability of the respective vector, and that this appears to be in turn influenced by the source of the host cell line. While in the mouse-derived NIH 3T3 cells equivalent Mo-MLV- and Akv-MLV-based vectors were equally stable, in the human-derived HEK293 and U87-MG cell lines the Mo-MLV-based vector was more stable than the Akv-MLV-based vector.
To detect deletions and mutations in the transgene cassette which lead to creation of mutants no longer capable of expressing eGFP in infected cells, PCR was performed on genomic DNA extracted from NIH 3T3, HEK293, and U87-MG cells at the end of each infection cycle, using primers annealing to the MLV genome flanking the IRES-eGFP cassette, followed by analysis of the PCR products by agarose gel electrophoresis (data not shown). Generally, the reduction in the percentage of infected cells expressing eGFP observed in later infection cycles corresponded with the emergence of a series of mutants containing defined deletions in the transgene cassette, accompanied by the concomitant gradual disappearance of undeleted vector genomes (M. Paar, submitted for publication).
Replication kinetics and transgene expression levels in NIH 3T3, HEK293, and U87-MG cells.
The experimental design employed in this study simulates a situation in which viruses have an unlimited amount of fresh cells to infect and thus allows the calculation of a theoretical cumulative number of eGFP-expressing cells after each infection cycle, using the formula Nx = Nx−1Dx + Nx−1, where Nx = the cumulative amount of eGFP-expressing cells after two passages of cycle x, Nx−1 = the cumulative amount of eGFP-expressing cells after two passages of the previous cycle, and D = the dilution factor used for inoculation of cycle x.
Application of this formula to the data obtained from the serial infection cycles performed with NIH 3T3, HEK293, and U87-MG cells clearly demonstrates that Mo-MLV-based RCR vectors are able to propagate a functional transgene cassette through cell cultures much more rapidly than Akv-MLV-based RCR vectors and that positioning of the transgene cassette immediately adjacent to the 3′ end of the env gene permits more rapid transgene propagation than positioning the cassette in the U3 region of the 3′ LTR (Fig. 2D). Interestingly, although the Mo-MLV-based vectors spread much more slowly in HEK293 than in NIH 3T3 cells, their greater genomic stability in HEK293 cells than in NIH 3T3 cells implies that they are actually able to propagate functional transgene cassettes to a far greater number of cells over time. Vector ACEU3-GFP, for example, can propagate a functional IRES-eGFP cassette to 109-fold-more HEK293 cells than NIH 3T3 cells before the maximum level of eGFP-expressing cells drops below 50%, whereas for the Akv-MLV-based vectors, the very slow replication kinetics combined with only a moderate genomic stability result in 107-fold-fewer HEK293 cells than NIH 3T3 cells being infected before the maximum level of eGFP-expressing cells drops below 50% (Table 1). In U87-MG cells, the vectors also spread much more slowly than in NIH 3T3 cells and are even less genomically stable than in NIH 3T3 cells (Fig. 2C and D; Table 1). The slower replication kinetics of all vectors in HEK293 and U87-MG cells than in NIH 3T3 cells can be at least in part ascribed to differences in infectivity of these cell types, since approximately 10-fold- and 4-fold-more virus particles are required to infect the same numbers of HEK293 and U87-MG cells, respectively, than NIH 3T3 cells (data not shown).
TABLE 1.
Calculated number of cells which can be infected before the majority of each vector becomes unstable
| Vector | HEK293
|
NIH 3T3
|
U87-MG
|
|||
|---|---|---|---|---|---|---|
| Cycle no.a | No. of eGFP-positive cellsb | Cycle no. | No. of eGFP-positive cells | Cycle no. | No. of eGFP-positive cells | |
| ACE-GFP | ≥21 | ≥5.5 × 1052 | 11 | 1.2 × 1046 | 20 | 6.4 × 1023 |
| ACEU3-GFP | 15 | 9.9 × 1024 | 5 | 1.1 × 1014 | 4 | 2.2 × 1011 |
| Akv(B)4070A-eGFP | 15 | 1.7 × 1016 | 11 | 2.9 × 1021 | 10 | 3.3 × 108 |
Serial infection cycle number at which more than 50% of infected cells no longer express detectable levels of eGFP.
Theoretical maximum number of cells to which a functional eGFP-expressing vector has been propagated by this time point (cycle).
The levels of eGFP expression mediated by the different vectors were analyzed by determining the mean fluorescence intensity (MFI) of cells infected with each vector at different time points during the course of the experiment. Generally, all of the vectors mediated the highest levels of eGFP expression in NIH 3T3 cells and the lowest levels in HEK293 cells. The Mo-MLV-based vectors mediated higher eGFP expression levels than the Akv-MLV-based vectors (data not shown), which was in turn generally reflected by the differences in replication kinetics of the vectors in the respective cell lines.
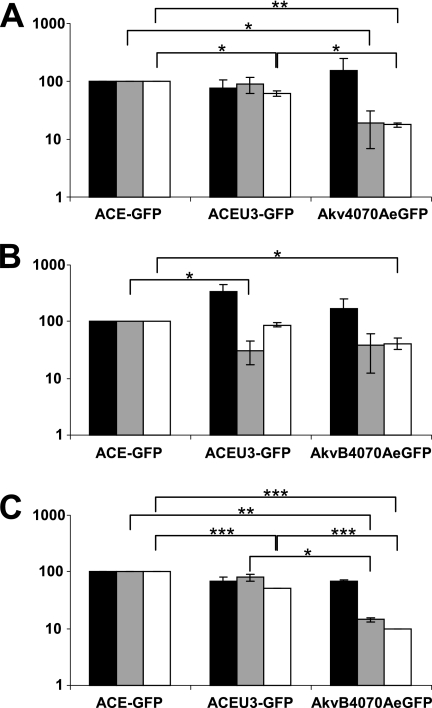
In order to further investigate the effects which differences in virus strain and transgene cassette position of vectors ACE-GFP, ACEU3-GFP, and Akv4070A-eGFP exert on virus replication, we investigated the levels of virus particle production and the infectivity of these particles on the respective cell lines. By comparing the amount of nascent virus particles released into the culture medium to the amount of integrated proviruses from which they were transcribed, by real-time RT-PCR of virus RNA and real-time PCR of proviral DNA and cellular rRNA gene sequences, it could be shown that the levels of nascent particle production were comparable for the vectors regardless of the host cell line (Fig. 3A to C, black bars). Using a similar MOI to infect fresh cells and measuring the amount of eGFP-expressing cells detectable by FACS analysis 48 h postinfection, however, significant differences were observed in the relative infectivities of the nascent viral particles (Fig. 3A to C, gray bars), indicating that the lower infectivity of Akv-MLV-based vectors is the main reason for their slower replication kinetics. When both nascent particle production and infectivity are taken into account by including the two values in the same calculation (Fig. 3A to C, white bars), the cumulative differences demonstrate a statistically significant disadvantage for Akv-MLV-based vectors compared to either of the Mo-MLV-based vectors in each cell line which also reflects the observed differences in the replication kinetics of each vector (Fig. 2D).
FIG. 3.
Virus-containing supernatant harvested from NIH 3T3 (A), HEK293 (B), and U87-MG (C) cells in infection cycle 1 was used to infect fresh cells of the corresponding cell line, and 50 μM zidovudine was applied 24 h postinfection to prevent secondary infection events. The relative quantity of integrated proviral copies per infected producer cell and the relative quantity of nascent virus released into the supernatant from each infected producer cell were determined by real-time PCR and real-time RT-PCR, respectively. Values shown indicate the numbers of nascent virions released per proviral copy in infected producer cells (black bars), the quantity of eGFP-positive cells generated per virus particle (gray bars), and the cumulative effect of the differences in nascent virus particle production and infectivity (white bars), in each case relative to the value obtained for ACE-GFP. Statistical significance of differences between vectors is indicated by asterisks (t test; P < 0.0001 is indicated by three asterisks, P < 0.001 is indicated by two asterisks, and P < 0.02 is indicated by one asterisk).
Replication kinetics, genomic stability, and transgene expression of vectors in preformed U87-MG tumors in vivo.
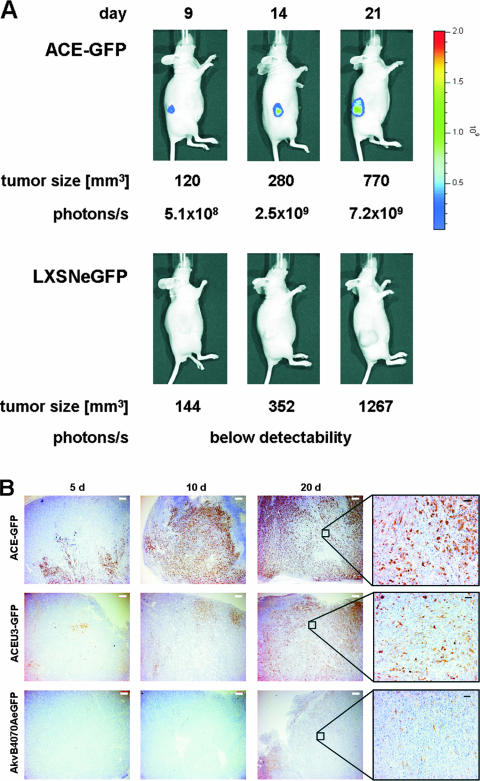
To evaluate whether the effects on vector spread and stability observed in vitro with vectors differing in transgene location and virus strain also apply for the situation in vivo, preformed subcutaneous U87-MG tumors growing in nude mice were infected with 1 × 105 infectious virus particles, with titers determined on U87-MG cells in vitro, of each of the MLV-based RCR vectors and the MLV-based replication-deficient retroviral vector LXSNeGFP (15). Virus spread was monitored in living mice using a charge-coupled device camera (IVIS50; Xenogen) to detect eGFP expression. Infection of the preformed tumors with vector ACE-GFP, for example, led to increasing levels of eGFP fluorescence over time and at a rate which was over and above that which could be explained by tumor growth alone (Fig. 4A). In comparison, tumors infected with the RDR vector LXSNeGFP did not reveal increasing eGFP fluorescence over time (Fig. 4A).
FIG. 4.
Intratumoral delivery of 1 × 105 viral particles to nude mice bearing subcutaneous U87-MG tumors. (A) Spread of the replication-competent retrovirus vector ACE-GFP and the replication-deficient vector LXSNeGFP was followed in a living mouse 9, 14, and 21 days after virus administration by using the IVIS50 device (Xenogen). Signals were measured by photon counts (p/s/cm2), and tumor size was determined. (B) Immunohistological analysis of infected tumor cells. Tumor sections from 5, 10, and 20 days postinfection were probed with an eGFP-specific antibody. White and black scale bars represent 500 μm and 100 μm, respectively.
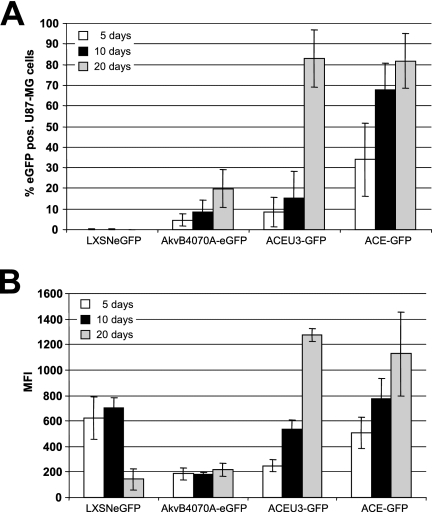
In addition, cohorts of five mice each were sacrificed at 5, 10, and 20 days after virus injection to analyze vector spread in the tumor tissue by FACS of single-cell suspensions of the excised tumor mass and immunohistochemistry using an eGFP-specific antibody. FACS analysis of infected tumors revealed that the RCR vectors were indeed spreading throughout the tumor mass but, however, that there were large differences in the performances of the different vectors. In general, the behavior of the RCR vectors in vivo reflected the situation as observed in vitro, namely, that vectors based on Mo-MLV spread more efficiently and led to greater levels of transgene expression in infected cells than did those based on Akv-MLV and that vectors containing the transgene cassette immediately adjacent to the 3′ end of the env gene spread more efficiently than those in which it was inserted in the U3 region of the 3′ LTR (Fig. 5A). Vector AkvB4070A-eGFP reached only about 20% of the tumor cells at day 20 postinfection, by which time point both of the Mo-MLV-based vectors had infected over 80% of the tumor mass (Fig. 5A). Unfortunately, it was not possible to follow up virus spread over a longer time period, since due to the rapid tumor growth all mice had to be euthanized for ethical reasons after the 20-day time point. Clearly, however, vector ACE-GFP also spreads with kinetics superior to those of vector ACEU3-GFP in vivo, since after only 10 days of infection with ACE-GFP almost 70% of the tumor cells expressed eGFP, whereas in the case of vector ACEU3-GFP less than 20% of tumor cells expressed eGFP by this time point. The more rapid replication kinetics of the Mo-MLV-based vectors also led to a large increase in eGFP expression levels in infected tumor cells over time (Fig. 5B), likely due to multiple simultaneous infection events of single cells, such that the MFI of eGFP-expressing tumor cells was about sixfold higher 20 days postinfection using the Mo-MLV-based vectors in comparison to the Akv-MLV-based vector (Fig. 5B).
FIG. 5.
Propagation and transgene expression of vectors in vivo. Subcutaneous established U87-MG tumors in nude mice were infected with 1 × 105 viral particles. On days 5, 10, and 20 after injection mice were sacrificed, the tumor was excised, and tumor cell suspensions were analyzed by FACS. The percentages (A) and the MFIs (B) of infected and eGFP-expressing cells are shown.
Immunohistochemical analyses of tumor sections from infected mice sacrificed 5, 10, and 20 days postinfection indicated that eGFP expression is stronger as well as being more widespread in tumors infected with the Mo-MLV-based vectors than in those infected with the Akv-MLV-based vector at any given time point postinfection (Fig. 4B), strongly supporting the data obtained in FACS analyses. Application of this technique also provides information regarding the pattern of vector spread throughout the tumor. It was observed, for example, that eGFP expression is restricted to the tumor mass, revealing a defined border representing the interface between tumor (eGFP expressing) and eGFP expression-negative healthy mouse cells (Fig. 4B). Within the tumor mass itself, virus spread appears not to occur in a regular and concentric fashion, as might be expected following a single application into the center of the preformed tumor, but seems rather to advance on several separate fronts at different speeds. In addition, eGFP expression is not detected in some tumor cells even when they are surrounded by other eGFP-expressing cells (Fig. 4B). Higher-power magnification of representative areas of the tumor slices clearly reveals that eGFP-negative cells are also present in viable nonnecrotic regions (Fig. 4B), suggesting either that they are infected but express undetectable levels of eGFP, that they have been infected by vectors which have already lost the transgene cassette, or that they may be refractory to infection.
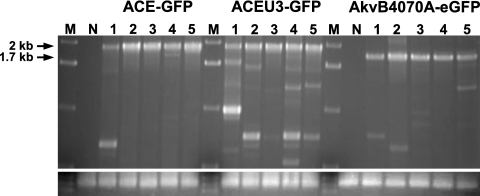
In order to determine the genomic stability of the vectors following replication in vivo, PCR was performed on genomic DNA extracted from tumors from mice sacrificed 20 days postinfection using primers annealing to the MLV genome flanking the IRES-eGFP cassette, followed by analysis of the PCR products by agarose gel electrophoresis. Vector ACEU3-GFP proved to be the least stable in vivo, as several PCR products representing deletion mutants were readily detectable from DNA of tumors removed from each of the five mice analyzed (Fig. 6). Vector AkvB4070A-eGFP appears to be more stable than vector ACEU3-GFP in vivo, since only one or two faintly detectable PCR products representing deletion mutants were amplified from tumor cell DNA (Fig. 6). Vector ACE-GFP, however, was clearly the most stable vector in vivo, since PCR products representing deletion mutants were readily detectable in DNA of only one out of the five tumors infected with this vector (Fig. 6).
FIG. 6.
Genomic stability of vectors in vivo. Genomic DNA was extracted from U87-MG tumor tissue of mice infected with vectors ACE-GFP, ACEU3-GFP, and AkvB4070A-eGFP 20 days after virus injection. PCR was performed on DNA of five mice each, with the use of primers annealing to the vector genome flanking the IRES-eGFP cassette. Lane M, DNA size markers; lane N, DNA extracted from noninfected tumors. Signals at 2 kb (in the case of vectors ACE-GFP and ACEU3-GFP) and 1.7 kb (in the case of vector AkvB4070A-eGFP) indicate the full-length vector fragment. Signals revealing smaller DNA fragments indicate deletions within the amplified IRES-eGFP region of the respective vectors. The lower panel shows amplification of a glyceraldehyde-3-phosphate dehydrogenase-specific fragment to verify similar amounts of DNA used for PCR.
Thus, in summary, the data obtained in this study unravel the effects of basic vector design features such as virus strain and transgene insertion position upon vector performance in respect to spread kinetics and stability in vivo. Our data unequivocally demonstrate that a Mo-MLV-based vector containing a transgene cassette positioned immediately adjacent to the 3′ end of the env gene spreads with faster kinetics and exhibits greater genomic stability in vitro as well as in vivo in solid tumors than a Mo-MLV vector carrying the transgene cassette in the U3 region of the 3′ LTR or when the vector is based on Akv-MLV.
DISCUSSION
Following the recent flurry of publications documenting the generation and application of a variety of MLV-based RCR vectors, it has become abundantly clear that vectors of this type will play an increasingly important role in the development of efficacious gene therapies for solid tumors. Not least because the use of MLV-based RCR vectors for this purpose is, nevertheless, still at a comparatively early stage, it is of great interest to analyze the effects which certain aspects of basic vector design can exert on vector characteristics likely to be of chief importance for successful therapy.
We have therefore undertaken a comprehensive analysis of the extent to which the choice of virus strain and insertion position of the transgene cassette in the viral genome can influence the replication kinetics, genomic stability, and transgene expression of MLV-based RCR vectors in both murine and human cells in cell culture, by following the vectors over multiple infection cycles for up to 100 days of constant propagation. Furthermore, we have demonstrated for the first time that such differences in MLV-based RCR vector design can have dramatic effects on their ability to propagate a functional transgene cassette in a more relevant in vivo setting, by comparing the performance of different vector designs in preformed human tumors in nude mice.
The data obtained clearly demonstrate that insertion of the transgene cassette immediately downstream of the 3′ end of the env gene generates vectors with better replication kinetics and genomic stability than when an identical transgene cassette is placed in the 5′ end of the 3′-LTR U3 region and that Mo-MLV-based vectors have better replication kinetics, genomic stability, and transgene expression levels than equivalent Akv-MLV-based vectors. The data also reveal that the source of the host cell line can have a profound effect upon the genomic stability of MLV-based RCR vectors, with propagation in certain cell lines leading to the more rapid emergence of deletion mutants regardless of viral strain or transgene cassette position. Our results are in broad agreement with those previously published in this field (7, 13, 18, 19), inasmuch as they show that both Akv-MLV and Mo-MLV can stably propagate a transgene cassette over several infection cycles and that this can be achieved with a transgene cassette placed either directly downstream of the env gene or in the U3 region of the 3′ LTR. However, only a direct comparative evaluation of these vectors, as performed in this study, unravels such major effects on vector stability and spread kinetics due to basic vector design differences.
The clear superiority of vectors containing the transgene expression cassette immediately adjacent to the 3′ end of the env gene (ACE-GFP) rather than in the 5′ end of the 3′-LTR U3 region of an otherwise identical vector (ACEU3-GFP) is due to the presumably more rapid emergence of deletion mutants in cells infected with ACEU3-GFP and reflects the greater fitness of these mutants in terms of replication kinetics compared to the parental vector (Fig. 2A to D). However, it is not immediately clear why ACEU3-eGFP should replicate more slowly than ACE-GFP. The 3′ UTR of Mo-MLV contains sequences important for efficient virus replication (27), but fusion of the IRES-GFP cassette directly to the 3′ end of the env gene does not interrupt these critical sequences. Similarly, the U3 region of Mo-MLV has multiple functions, including reverse transcription, transcription, and possibly also processing of nascent viral RNA following integration into the host cell genome (17), insertion of the transgene cassette into the 5′ end of the 3′-LTR U3 region ensures that none of the known transcription factor binding sites or sequences known to be involved in RNA processing are disrupted, neither individually nor in their spatial relationships to one another. This is also indicated by the fact that we did not observe any significant differences in transgene expression levels in infected cells mediated by vectors ACE-GFP and ACEU3-GFP (data not shown). Although it has not been investigated, it is conceivable, however, that the presence of two copies of the transgene cassette in the proviral DNA of vector ACEU3-GFP in infected cells may lead to higher rates of recombination between these repeats, which may in turn lead to markedly slower replication kinetics as well as to the more rapid emergence of deletion mutants.
The observed lower levels of transgene expression mediated in all cell lines by infection with Akv-MLV-based vectors in comparison to those based on Mo-MLV (data not shown) might be due to different promoter activities of the respective U3 regions, as it has been shown previously that the Mo-MLV LTR directs much higher levels of transcription than the Akv-MLV LTR in cell lines derived from host target cells (3, 28). However, it seems highly unlikely that the roughly twofold difference in transgene expression levels observed could be entirely responsible for the major differences in spreading kinetics (Fig. 2D) between equivalent Mo-MLV-based and Akv-MLV-based vectors. This is also indicated by our findings that, although the relative levels of nascent virus particle production do not differ significantly between the different vectors in infected HEK293, NIH 3T3, or U87-MG cells, Akv-MLV-based vectors are significantly less infectious than Mo-MLV-based vectors on both NIH 3T3 and U87-MG cells (Fig. 3A and C). Moreover, in general, the apparent greater genomic stability of the Mo-MLV-based vectors, at least in human cells, combined with their greatly superior spreading kinetics (Fig. 2A to D; Table 1), indicates that Mo-MLV-based vectors are preferable to Akv-MLV-based vectors when it is of paramount importance that as many target cells as possible are transduced with a functional transgene cassette, as is the case in cancer gene therapy.
Differences in the genomic stabilities of MLV vectors in different cell types have not been documented previously, and it is difficult to propose a plausible mechanism for the rather striking differences observed in this study. Since NIH 3T3 cells are known to contain many endogenous MLV-related sequences, it is tempting to suppose that recombination of the vectors with such sequences may lead to the more rapid loss of the transgene cassette in these cells, as has previously been reported (18). Alternatively, it could be a reflection of the intrinsic genetic stability of each cell line, whereby a host cell line exhibiting chromosomal instability may lead to recombination of integrated vector proviruses and thus to more rapid emergence of deletion mutants.
In solid tumors, all of the RCR vectors tested showed a clear advantage over an equivalent RDR vector in terms of infection efficiency (Fig. 5A). Between the different RCR vector designs, however, there were also dramatic differences in infection efficiency, transgene expression levels, and genomic stability (Fig. 5A and B and 6). By far the best vector, in every category, was vector ACE-GFP. Over 30% of tumor cells were eGFP positive as early as 5 days postinfection with vector ACE-GFP, a level which was attained only 20 days postinfection with vector ACEU3-GFP and not at all following infection with vector AkvB4070A-eGFP (Fig. 5A). Moreover, eGFP expression levels in tumor cells infected with vector ACE-GFP were much higher than those in cells infected with AkvB4070A-eGFP (Fig. 5B). Although vector ACEU3-GFP had reached a similar amount of tumor cells and expressed similar amounts of eGFP in infected cells as vector ACE-GFP by 20 days postinfection (Fig. 5A and B), PCR analysis of DNA from infected tumor cells demonstrated that considerably more transgene deletion mutants had arisen in ACEU3-GFP-infected tumors than in those infected with ACE-GFP (Fig. 6). When a quantitative PCR assay was used to determine the amount of transgene- and env-specific sequences in DNA isolated from infected tumors in order to calculate the ratio of parental to deleted proviral vectors, however, no statistically significant differences were observed between the signals obtained from tumor DNAs infected with different vectors. The in vivo results obtained with vector ACE-GFP are in agreement with those obtained by the Kasahara group (36), who also showed that this vector and the respective CD-expressing therapeutic vector, ACE-CD, could transduce subcutaneous and intracranial growing tumors with high efficiency. Moreover, PCR and Southern blot analyses also demonstrated that the detection of deletion mutants of vectors ACE-GFP and ACE-CD was very rare (36), further supporting our findings in this respect. Vectors based on Akv-MLV or containing a transgene expression cassette in the 3′-LTR U3 region have, to our knowledge, never been tested in solid tumors in vivo until now.
All in all, this study clearly demonstrates that MLV-based RCR vectors spread with faster kinetics and are genomically far more stable, both in cell culture and in preformed tumors in vivo, when the transgene cassette is inserted immediately downstream of the env gene rather than in the 5′ end of the U3 region of the 3′ LTR. Moreover, this study convincingly shows that Mo-MLV is far more suitable than Akv-MLV for the generation of RCR vectors for cancer gene therapy, in terms of spread kinetics, genomic stability, and transgene expression levels. Thus, these unambiguous findings should prove to be of considerable value in the future design of therapeutic vectors for efficient gene delivery in tumor therapy.
Acknowledgments
We thank Noriyuki Kasahara (UCLA, Los Angeles, CA) for providing plasmid pACE-GFP and Finn Skou Pedersen, University of Aarhus, Aarhus, Denmark, for providing plasmid pAkvEn-GFP. Special thanks go to Elzbieta Knapp, Reinhard Ertl, and Magdalena Pusch for help with FACS; Marielle König-Schuster and Helga Petznek for assistance with the in vivo studies; and Alexander Tichy from the Institute of Medical Physics and Biostatistics, University of Veterinary Medicine, Vienna, Austria, for mathematical and statistical advice. We also specially thank Chris Logg (UCLA, Los Angeles, CA) for his helpful advice and expert opinions during this work.
This study was supported by the Christian-Doppler Forschungsgesellschaft, Austria.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Bachrach, E., M. Duch, M. Pelegrin, H. Dreja, F. S. Pedersen, and M. Piechaczyk. 2003. In vivo infection of mice by replication-competent MLV-based retroviral vectors. Methods Mol. Med. 76:343-352. [DOI] [PubMed] [Google Scholar]
- 2.Bachrach, E., M. Pelegrin, M. Piechaczyk, F. S. Pedersen, and M. Duch. 2002. Efficient gene transfer into spleen cells of newborn mice by a replication-competent retroviral vector. Virology 293:328-334. [DOI] [PubMed] [Google Scholar]
- 3.Celander, D., and W. A. Haseltine. 1984. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature 312:159-162. [DOI] [PubMed] [Google Scholar]
- 4.Coulombe, J., Y. Avis, and D. A. Gray. 1996. A replication-competent promoter-trap retrovirus. J. Virol. 70:6810-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalba, C., D. Klatzmann, C. R. Logg, and N. Kasahara. 2005. Beyond oncolytic virotherapy: replication-competent retrovirus vectors for selective and stable transduction of tumors. Curr. Gene Ther. 5:655-667. [DOI] [PubMed] [Google Scholar]
- 6.Dillon, P. J., J. Lenz, and C. A. Rosen. 1991. Construction of a replication-competent murine retrovirus vector expressing the human immunodeficiency virus type 1 Tat transactivator protein. J. Virol. 65:4490-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duch, M., M. L. Carrasco, T. Jespersen, B. D. Hansen, and F. S. Pedersen. 2004. Transgene stability for three replication-competent murine leukemia virus vectors. Gene 329:61-69. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein, M. L., M. R. Abedi, J. Wixon, and R. M. Edelstein. 2004. Gene therapy clinical trials worldwide 1989-2004—an overview. J. Gene Med. 6:597-602. [DOI] [PubMed] [Google Scholar]
- 9.Erlwein, O., C. J. Buchholz, and B. S. Schnierle. 2003. The proline-rich region of the ecotropic Moloney murine leukaemia virus envelope protein tolerates the insertion of the green fluorescent protein and allows the generation of replication-competent virus. J. Gen. Virol. 84:369-373. [DOI] [PubMed] [Google Scholar]
- 10.Finger, C., Y. Sun, L. Sanz, L. Alvarez-Vallina, C. J. Buchholz, and K. Cichutek. 2005. Replicating retroviral vectors mediating continuous production and secretion of therapeutic gene products from cancer cells. Cancer Gene Ther. 12:464-474. [DOI] [PubMed] [Google Scholar]
- 11.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 12.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Jespersen, T., M. Duch, M. L. Carrasco, S. Warming, and F. S. Pedersen. 1999. Expression of heterologous genes from an IRES translational cassette in replication competent murine leukemia virus vectors. Gene 239:227-235. [DOI] [PubMed] [Google Scholar]
- 14.Klein, D., B. Bugl, W. H. Gunzburg, and B. Salmons. 2000. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 7:458-463. [DOI] [PubMed] [Google Scholar]
- 15.Klein, D., S. Indraccolo, K. von Rombs, A. Amadori, B. Salmons, and W. H. Gunzburg. 1997. Rapid identification of viable retrovirus-transduced cells using the green fluorescent protein as a marker. Gene Ther. 4:1256-1260. [DOI] [PubMed] [Google Scholar]
- 16.Logg, C. R., and N. Kasahara. 2004. Retrovirus-mediated gene transfer to tumors: utilizing the replicative power of viruses to achieve highly efficient tumor transduction in vivo. Methods Mol. Biol. 246:499-525. [DOI] [PubMed] [Google Scholar]
- 17.Logg, C. R., A. Logg, R. J. Matusik, B. H. Bochner, and N. Kasahara. 2002. Tissue-specific transcriptional targeting of a replication-competent retroviral vector. J. Virol. 76:12783-12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logg, C. R., A. Logg, C. K. Tai, P. M. Cannon, and N. Kasahara. 2001. Genomic stability of murine leukemia viruses containing insertions at the Env-3′ untranslated region boundary. J. Virol. 75:6989-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logg, C. R., C. K. Tai, A. Logg, W. F. Anderson, and N. Kasahara. 2001. A uniquely stable replication-competent retrovirus vector achieves efficient gene delivery in vitro and in solid tumors. Hum. Gene Ther. 12:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzl, C., D. Mischek, B. Salmons, W. H. Gunzburg, M. Renner, and D. Portsmouth. 2006. Tissue- and tumor-specific targeting of murine leukemia virus-based replication-competent retroviral vectors. J. Virol. 80:7070-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, D. G., M. A. Adam, and A. D. Miller. 1990. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol. Cell. Biol. 10:4239-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nethe, M., B. Berkhout, and A. C. van der Kuyl. 2005. Retroviral superinfection resistance. Retrovirology 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rainov, N. G., and H. Ren. 2003. Clinical trials with retrovirus mediated gene therapy—what have we learned? J. Neurooncol. 65:227-236. [DOI] [PubMed] [Google Scholar]
- 26.Reik, W., H. Weiher, and R. Jaenisch. 1985. Replication-competent Moloney murine leukemia virus carrying a bacterial suppressor tRNA gene: selective cloning of proviral and flanking host sequences. Proc. Natl. Acad. Sci. USA 82:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robson, N. D., and A. Telesnitsky. 2000. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J. Virol. 74:10293-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Short, M. K., S. A. Okenquist, and J. Lenz. 1987. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J. Virol. 61:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sliva, K., O. Erlwein, A. Bittner, and B. S. Schnierle. 2004. Murine leukemia virus (MLV) replication monitored with fluorescent proteins. Virol. J. 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solly, S. K., S. Trajcevski, C. Frisen, G. W. Holzer, E. Nelson, B. Clerc, E. Abordo-Adesida, M. Castro, P. Lowenstein, and D. Klatzmann. 2003. Replicative retroviral vectors for cancer gene therapy. Cancer Gene Ther. 10:30-39. [DOI] [PubMed] [Google Scholar]
- 31.Stuhlmann, H., R. Jaenisch, and R. C. Mulligan. 1989. Construction and properties of replication-competent murine retroviral vectors encoding methotrexate resistance. Mol. Cell. Biol. 9:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, Y., C. Finger, L. Alvarez-Vallina, K. Cichutek, and C. J. Buchholz. 2005. Chronic gene delivery of interferon-inducible protein 10 through replication-competent retrovirus vectors suppresses tumor growth. Cancer Gene Ther. 12:900-912. [DOI] [PubMed] [Google Scholar]
- 33.Tai, C. K., C. R. Logg, J. M. Park, W. F. Anderson, M. F. Press, and N. Kasahara. 2003. Antibody-mediated targeting of replication-competent retroviral vectors. Hum. Gene Ther. 14:789-802. [DOI] [PubMed] [Google Scholar]
- 34.Tai, C. K., W. J. Wang, T. C. Chen, and N. Kasahara. 2005. Single-shot, multicycle suicide gene therapy by replication-competent retrovirus vectors achieves long-term survival benefit in experimental glioma. Mol. Ther. 12:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., C. K. Tai, A. D. Kershaw, S. K. Solly, D. Klatzmann, N. Kasahara, and T. C. Chen. 2006. Use of replication-competent retroviral vectors in an immunocompetent intracranial glioma model. Neurosurg. Focus 20:E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, W. J., C. K. Tai, N. Kasahara, and T. C. Chen. 2003. Highly efficient and tumor-restricted gene transfer to malignant gliomas by replication-competent retroviral vectors. Hum. Gene Ther. 14:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]