Abstract
A 1.25-kbp DNA fragment from the right side of the genome containing the lytic origin of replication (oriLyt) of murine gammaherpesvirus 68 (MHV-68) has been identified by a plasmid replication assay. Here we show that a mutant MHV-68 with a deletion of an essential part of this oriLyt, generated by using an MHV-68 bacterial artificial chromosome, was only slightly attenuated and still able to replicate but that a mutant containing an additional deletion on the left side of the genome was replication deficient. The newly identified region was sufficient to support plasmid replication, thus providing evidence for a second oriLyt.
Murine gammaherpesvirus 68 (MHV-68) is a member of the gammaherpesvirus subfamily and is closely related to Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus. Since there are no good animal models for KSHV and Epstein-Barr virus, MHV-68 serves as a model to investigate gammaherpesvirus pathogenesis in vivo (8, 14-16, 18). Since MHV-68 replicates well in tissue culture, it is a good model to study early stages of gammaherpesvirus infection as well as mechanisms controlling viral replication during the lytic cycle. Using in vitro plasmid replication assays which were carried out in the presence of viral replication proteins, Deng and colleagues mapped one lytic origin of replication (oriLyt) to a 1.25-kbp region in the right side of the MHV-68 genome (7). The 1.25-kbp sequence between nucleotides 100,723 and 101,974 was shown to be responsible for replication, and nucleotides 101,248 to 101,974 were identified as being absolutely essential.
To evaluate the impact of the reported oriLyt, we generated a mutant genome with a deletion in its essential part, using an MHV-68 bacterial artificial chromosome (BAC) (1). We generated this BAC mutant (mutant A) (Table 1) lacking nucleotide positions 101,530 to 101,731 by a two-step replacement procedure which does not leave behind any foreign sequence (3, 13). For this purpose, the HindIII D fragment of MHV-68 (9) was cloned into the shuttle plasmid pST76K-SR. Subsequently, nucleotides 101,530 to 101,731 were removed from this fragment by digestion with SfiI and BspEI, followed by blunting of the ends and religation. Surprisingly, we were able to reconstitute a mutant virus which was only slightly attenuated. This finding suggested the presence of a second oriLyt in MHV-68.
TABLE 1.
MHV-68 BACs used in this study
| Mutant | BAC | Deletion(s) (nucleotide positions) |
|---|---|---|
| A | Delta101,530-101,731 | 101,530-101,731 |
| B | Delta26,778-28,191 | 26,778-28,191 |
| C | Delta26,059-28,191 | 26,059-28,191 |
| D | Delta101,530-101,731 plus Delta26,778-28,191 | 101,530-101,731 and 26,778-28,191 |
| E | Delta101,530-101,731 plus Delta26,059-28,191 | 101,530-101,731 and 26,059-28,191 |
| P | Parent |
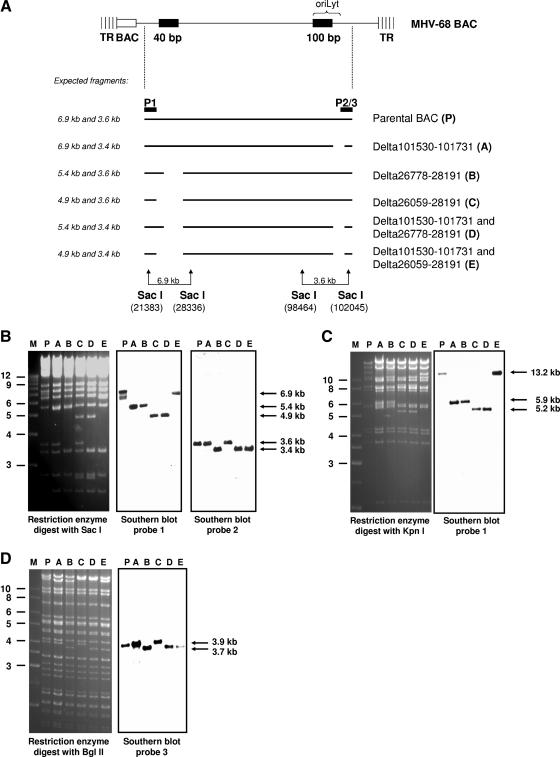
The structure of the MHV-68 oriLyt is similar to that in KSHV, which contains two oriLyts, with one on either side of the genome, sharing an almost identical 1,153-bp sequence and a 600-bp GC-rich downstream repeat sequence (2). In contrast to the case for KSHV, such a duplicated oriLyt sequence has not been identified in the left side of the MHV-68 genome (7). However, MHV-68 contains a GC-rich 40-bp repeat sequence in the left side of the genome (nucleotides 26,778 to 28,191) (17). In addition, the genomic coordinates 101,765 to 101,624 (belonging to the essential part of the oriLyt) have been found to be >70% identical to coordinates 26,232 to 26,374 (5). Thus, we hypothesized that the 40-bp repeat and the adjacent sequences at the left side of the genome may contain a putative second oriLyt of MHV-68. To identify this second oriLyt, we generated additional mutants with deletions both at the left and at the right side of the genome (Table 1). In mutant B, we deleted the 40-bp repeat, and in mutant C, we deleted the 40-bp repeat and the adjacent sequences at the left side. Mutant B was created by “ET cloning” as described previously (19). To this end, a linear PCR fragment containing an FLP recombination target-flanked tetracycline (Tet) resistance cassette was generated from vector pCP16 (4), using a primer pair that contained 24 nucleotides for amplification of the Tet resistance gene and an additional 50 nucleotides homologous to sequences flanking the region to be deleted. This PCR product was transferred into the MHV-68 BAC by homologous recombination in Escherichia coli strain DH10B already containing the MHV-68 BAC and the plasmid pKD46, which expresses Red recombinase under the control of a regulated (l-arabinose-inducible) promoter (6). Successful recombination led to the replacement of the 40-bp repeat of MHV-68 with a Tet resistance gene. Subsequently, the Tet resistance cassette was removed, resulting in a deletion between nucleotides 26,778 to 28,191 and leaving a small residual insert consisting of an FLP recombination target site and short vector sequences in the disrupted region. Mutant C was also generated by “ET cloning,” resulting in a deletion between nucleotides 26,059 and 28,191. To further evaluate the impact of the deletions in mutants B and C, respectively, we constructed the double deletion mutants D and E by combining both of the single deletions at each side of the genome in one BAC plasmid. All mutant genomes were verified by restriction enzyme analysis with different enzymes, Southern blot analysis, and sequencing (Fig. 1A to D).
FIG. 1.
Generation of MHV-68 oriLyt mutants. (A) Scheme of the mutants generated. “P” indicates the probes used for Southern blot analysis, corresponding to nucleotides 25,097 to 25,833 (P1), 101,141 to 102,045 (P2), and 101,443 to 102,699 (P3). (B) Restriction enzyme and Southern blot analyses of BAC DNAs digested with the restriction enzyme SacI and hybridized with probes P1 and P2. (C) Restriction enzyme and Southern blot analyses of BAC DNAs digested with the restriction enzyme KpnI and hybridized with probe P1. (D) Restriction enzyme and Southern blot analyses of BAC DNAs digested with the restriction enzyme BglII and hybridized with probe P3. Lanes P and A to E, parental and mutant BACs, as indicated in panel A and Table 1. The expected fragments are indicated by arrows on the right. Marker (M) sizes (in kilobase pairs) are indicated on the left.
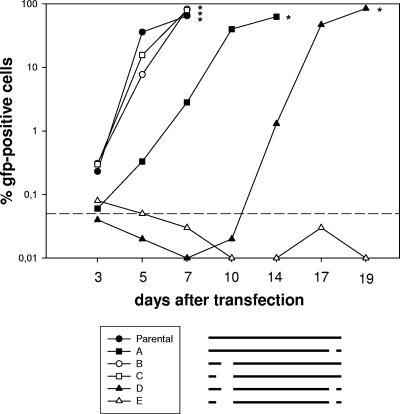
To investigate whether the mutations affected the reconstitution and replication of the appropriate viruses, we used the fact that all of the constructed BAC plasmids contained the gene for green fluorescent protein (GFP) expressed by a cytomegalovirus promoter (1). Thus, the spread of infectious virus within the culture could be followed by monitoring the number of GFP-positive cells. To this end, we transfected permissive BHK-21 cells with 2 μg of BAC DNA of each BAC to be investigated (Table 1) by using Superfect (QIAGEN, Hilden, Germany). Subsequently, we determined the number of GFP-positive cells over time. Recombinant viruses could be reconstituted, as determined by the spread of infectious virus in the cultures from all but one of the BAC-cloned genomes (Fig. 2). After transfection of the BAC DNAs into BHK-21 cells, the single deletion mutants A, B, and C could be reconstituted into infectious viruses, similar to the parental genome, indicating that the mutated regions are not essential for the reconstitution of infectious virus in cell culture. The double deletion mutant D did spread very slowly compared to the single mutants, indicating that the double deletion did heavily impair but not completely prevent virus reconstitution and replication. After transfection of BAC DNA of double deletion mutant E, a similar number of GFP-positive cells was observed initially. However, no further increase was observed and the GFP-positive cells were lost after a few passages, indicating that no infectious virus could be reconstituted. Since we failed to reconstitute this virus from BAC DNA in numerous attempts, we concluded that the deletion of both regions in the same BAC genome resulted in a complete replication deficiency.
FIG. 2.
Kinetics of GFP expression after transfection of permissive BHK-21 cells with BAC DNAs of different mutants. Cells were transfected with 2 μg of BAC DNA. At the indicated time points, cells were harvested and passaged. A part of the harvested cells was used to determine the number of GFP-positive cells by fluorescence-activated cell sorting analysis. Since there was a dilution effect due to passaging of the cells, the numbers of GFP-positive cells in some samples dropped temporarily below the detection limit, followed by their reappearance after virus replication reached an appropriate extent. Recombinant viruses could be reconstituted, as determined by increasing numbers of GFP-positive cells in the cultures, from all BAC-cloned genomes except for the double deletion mutant E. The asterisks indicate the time points when the particular cell cultures were stopped due to massive viral cytopathic effects. The dashed line indicates the detection limit, which was defined on the basis of the background fluorescence of untransfected BHK-21 cells. Below this limit, it is not possible to differentiate between specific and unspecific fluorescence. The pictogram schematically illustrates the mutants investigated. Data shown are from a representative experiment which was repeated twice with similar results.
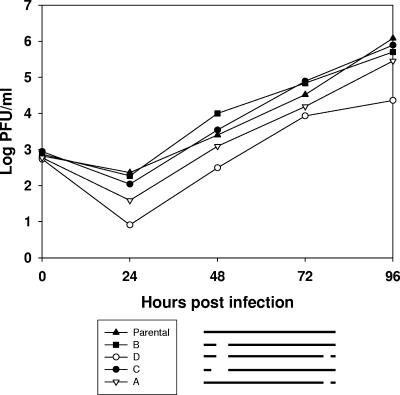
To further investigate the in vitro growth properties of the different mutants, virus stocks were generated from all reconstituted viruses. Multistep growth experiments were conducted by infecting NIH 3T3 cells at a multiplicity of infection of 0.01. Infected cultures were harvested at the indicated time points after infection, and infectious virus in each culture was titrated by plaque assays on BHK-21 cells. The single deletion mutants A, B, and C showed a similar growth pattern to that of the parental virus, although mutant A did so with slightly delayed kinetics. In contrast, the double deletion mutant D showed a replication deficit of approximately 2 orders of magnitude (Fig. 3).
FIG. 3.
Multistep growth properties of MHV-68 mutants. NIH 3T3 cells were infected at a multiplicity of infection of 0.01 at 37°C. After 1 h of adsorption at 37°C, the inoculum was removed and fresh medium was added. Cells and supernatants were harvested at the indicated time points, and viral titers were determined by plaque assay. The pictogram schematically illustrates the mutants investigated. Data shown are mean values compiled from up to three independent experiments.
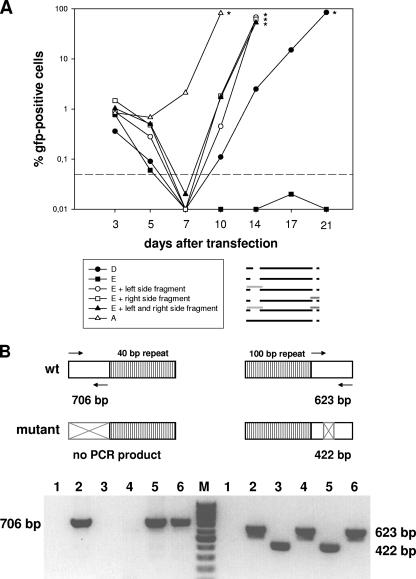
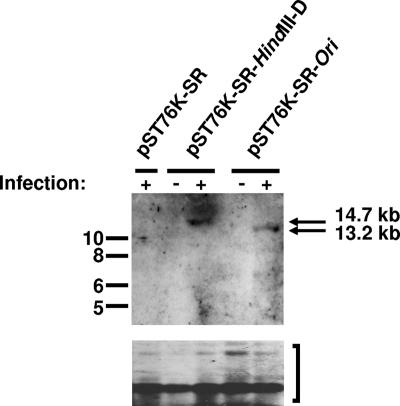
To prove that the lack of infectious virus after reconstitution of double deletion mutant E was due to the two deletions and not to rearrangements outside the mutated regions, cotransfection experiments were performed. Permissive BHK-21 cells were cotransfected with 2 μg of BAC DNA of the double deletion mutant E and 4 μg of pST76K-SR-derived plasmid DNA carrying unmutated genomic fragments of the mutated regions (1). Cotransfection with either one or both of the plasmids led to the reconstitution of infectious virus, as determined by an increase in the number of GFP-positive cells over time after transfection (Fig. 4A). Thus, the unmutated fragments were able to rescue the phenotype of double deletion mutant E. Since origins of replication can act only in cis, the reconstitution of infectious virus after cotransfection indicated that the ori-containing unmutated genomic fragments recombined into the viral genome. To formally prove that these recombination events actually occurred, we employed a PCR strategy, as illustrated in Fig. 4B. In all cases, cotransfection of the unmutated genomic fragments resulted in an output virus which was revertant, as demonstrated by the reappearance of PCR products corresponding to wild-type (wt) virus (Fig. 4B). The reconstitution of infectious virus (revertants) after cotransfection with the unmutated genomic fragments additionally clearly indicated that the observed phenotype of mutant E was not due to rearrangements outside the mutated regions. Next, we tested whether the newly identified region was sufficient to support the replication of a plasmid by performing a plasmid-based replication experiment as described by others (7). A 6.9-kb fragment of MHV-68 (nucleotide positions 21,383 to 28,336) containing the region of interest was cloned into the vector pST76K-SR (6.3 kb) (1) to generate the plasmid pST76K-SR-Ori. To test this construct, 1.5 μg of plasmid DNA was transfected into 293 cells in six-well plates, and 24 h after transfection, the cells were either left uninfected or infected with wt MHV-68 at a multiplicity of infection of 0.1 to provide the trans factors required for viral DNA replication (7). Seventy-two hours after infection, extrachromosomal DNAs were prepared according to the Hirt method (12). The DNAs were digested with DpnI (which cuts only the unreplicated input plasmid DNA) and KpnI or AflII. The latter two enzymes cut at unique sites in both the input and replicated plasmids. The digested DNAs were analyzed by Southern blotting with a digoxigenin-labeled probe hybridizing to vector pST76K-SR (Fig. 5). In contrast to the empty vector, both vector pST76K-SR-Ori and the positive control vector replicated successfully in infected cells, as demonstrated by the presence of 13.2-kb and 14.7-kb fragments, respectively, that were resistant to digestion with DpnI. There was no replication in uninfected cells, indicating that replication was dependent on factors expressed in trans during lytic infection. Thus, the plasmid-based replication assay complements our mutagenesis data and provides additional evidence that the newly identified region contains a second oriLyt of MHV-68.
FIG. 4.
Kinetics of GFP expression after cotransfection of permissive BHK-21 cells. (A) Cells were transfected with 2 μg of BAC DNA of the double deletion mutant E, the double deletion mutant D, or the single deletion mutant A and, in cases of cotransfections, were additionally transfected with 4 μg of plasmid DNA carrying an unmutated genomic fragment spanning the mutated region. Cotransfection with either one or both such plasmids led to the reconstitution of infectious virus, as determined by an increase in the number of GFP-positive cells over timer after transfection. At the indicated times, cells were harvested and passaged. A part of the harvested cells was used to determine the number of GFP-positive cells by fluorescence-activated cell sorting analysis. The asterisks indicate the times when the particular cell cultures were stopped due to massive viral cytopathic effects. The dashed line indicates the detection limit, which was defined on the basis of the background fluorescence of untransfected BHK-21 cells. The pictogram schematically illustrates the mutants investigated, with the unmutated genomic fragments used for cotransfections represented as light and dark gray bars. Data shown are from a representative experiment which was repeated once with similar results. (B) (Top) PCR strategy to determine the presence or absence of either the wt or mutated genomic fragment in the genomes of the reconstituted viruses after cotransfection of BAC DNA of the double deletion mutant E with overlapping unmutated fragments spanning the mutated regions, as illustrated in the pictogram in panel A. The arrows indicate the primer pairs used for PCR amplification. For the left side of the genome, PCR amplification results in a PCR product of 706 bp in the case of the wt genome but in no PCR product (due to the lack of the primer binding sites) in the case of the mutant genome. For the right side of the genome, PCR amplification results in a PCR product of 623 bp in the case of the wt genome and in a PCR product of 422 bp (due to the 201-bp deletion) in the case of the mutant genome. (Bottom) Virion DNAs were isolated from viruses reconstituted after cotransfection with either one or both of the fragments indicated in panel A, and the presence or absence of either the wt or mutated genomic fragment was investigated by PCR. Lanes 1, negative control (water); lanes 2, parental BAC; lanes 3, mutant E; lanes 4, mutant E plus cotransfection with the fragment spanning the deletion at the right side; lanes 5, mutant E plus cotransfection with the fragment spanning the deletion at the left side; lanes 6, mutant E plus cotransfection with both fragments. As expected, the wt pattern for the left side was restored after cotransfection with the left-side fragment or both fragments but not with the right-side fragment, and vice versa, i.e., the wt pattern for the right side was restored after cotransfection with the right-side fragment or both fragments but not with the left-side fragment.
FIG. 5.
Identification of a second MHV-68 oriLyt with a plasmid-based replication assay. 293 cells were transfected with plasmid pST76K-SR (6.3 kb) containing either the 8.4-kb HindIII D fragment including the previously identified oriLyt (pST76K-SR-HindIII-D) (7) or a 6.9-kb fragment of MHV-68 containing the region of interest (pST76K-SR-Ori), followed by MHV-68 infection. Seventy-two hours after infection, extrachromosomal DNAs were prepared and digested with DpnI to digest input plasmid DNA and also with KpnI and AflII, which cut only once in all plasmids. The digested DNAs were analyzed by Southern blotting with a digoxigenin-labeled probe hybridizing to vector pST76K-SR. The arrows indicate the 13.2-kb and 14.7-kb DpnI-resistant replicated DNA fragments, whereas the bracket indicates the DpnI-sensitive input DNA. Marker sizes (in kilobase pairs) are indicated on the left.
The presence of two lytic origins of replication and the functionality with only one oriLyt copy is reminiscent of the case for other gammaherpesviruses (2, 10, 11). To our knowledge, this study is the first that addresses the function of the MHV-68 oriLyt in the viral context. Understanding the replication elements is crucial for the further development of MHV-68 as an important experimental system to study the pathogenesis of gammaherpesviruses.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Ad121/2-1, -2-2, and -2-4 and HA1754-6) and the BMBF (NGFN-2 and FKZ 01GS0405).
We are grateful to B. Adler for a critical reading of the manuscript.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E.-M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, H. M., I. Brierley, and P. G. Stevenson. 2003. An internal ribosome entry site directs translation of the murine gammaherpesvirus 68 MK3 open reading frame. J. Virol. 77:13093-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, H., J. T. Chu, N. H. Park, and R. Sun. 2004. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J. Virol. 78:9123-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 10.Feederle, R., and H. J. Delecluse. 2004. Low level of lytic replication in a recombinant Epstein-Barr virus carrying an origin of replication devoid of BZLF1-binding sites. J. Virol. 78:12082-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine γ-herpesvirus infection. Philos. Trans. R. Soc. Lond. B 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 16.Speck, S. H., and H. W. Virgin IV. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 17.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virgin, H. W., IV, and S. H. Speck. 1999. Unraveling immunity to γ-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y., J. P. Muyrers, G. Testa, and A. F. Stewart. 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18:1314-1317. [DOI] [PubMed] [Google Scholar]