Abstract
Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus associated with many malignant and nonmalignant human diseases. Life-long latent EBV persistence occurs in blood-borne B lymphocytes, while EBV intermittently productively replicates in mucosal epithelia. Although several models have previously been proposed, the mechanism of EBV transition between these two reservoirs of infection has not been determined. In this study, we present the first evidence demonstrating that EBV latently infects a unique subset of blood-borne mononuclear cells that are direct precursors to Langerhans cells and that EBV both latently and productively infects oral epithelium-resident cells that are likely Langerhans cells. These data form the basis of a proposed new model of EBV transition from blood to oral epithelium in which EBV-infected Langerhans cell precursors serve to transport EBV to the oral epithelium as they migrate and differentiate into oral Langerhans cells. This new model contributes fresh insight into the natural history of EBV infection and the pathogenesis of EBV-associated epithelial disease.
Epstein-Barr virus (EBV) is a human herpesvirus that ultimately infects 95% of adults worldwide. EBV is transmitted via mucosal fluids and establishes life-long persistent infection. Although persistent infection is asymptomatic in most people, EBV is associated with benign syndromes, including infectious mononucleosis and oral hairy leukoplakia, and a wide variety of both lymphoid and mucosal epithelial malignancies. Persistent latent EBV infection occurs in memory B lymphocytes that circulate in blood (22). While latent EBV infection also occurs in some epithelial malignancies (29), EBV infection of epithelial cells typically results in productive replication, as demonstrated in oral hairy leukoplakia (9) and in normal oral epithelium (8, 10, 48).
Three different models have previously been proposed to explain the transition of EBV from the latent reservoir of infection in blood-borne B lymphocytes to sites of productive replication in oral epithelium. Model 1 proposes that B lymphocytes carrying latent EBV infection migrate from the blood to the epithelium, where the EBV reactivates and infects adjacent epithelial cells (12), but evidence of intraepithelial B lymphocytes in normal oral epithelium or in oral hairy leukoplakia is lacking (30, 33). Model 2 proposes that EBV virions produced by B lymphocytes in the oral submucosa bind submucosal EBV-specific dimeric immunoglobulin A (IgA) and enter basal oral epithelial cells by endocytosis via the polymeric Ig receptor (37), but the polymeric Ig receptor is not expressed in oral epithelium (23). Model 3 proposes that EBV virions produced by B lymphocytes in oral lymphoid tissues (26) gain access to and infect middle- and upper-layer oral epithelial cells as a result of microscopic traumatic epithelial injury, such as that which occurs during mastication. However, this model is contradicted by evidence that EBV transitions into oral epithelium as cell-associated latent infection and that EBV reactivates from a persistent latent oral mucosal source to productively replicate in the oral epithelium (44, 47, 50).
We now propose a fourth model of EBV transition from blood into oral epithelium. Langerhans cells (LC) are dendritic antigen-presenting cells that reside in the basal and suprabasal layers of cutaneous and mucosal epithelia. Bone marrow-derived LC precursor cells (pre-LC) circulate in the blood before they migrate into epithelia and differentiate into LC (4). We hypothesized that EBV latently infects blood-borne pre-LC and that these EBV-infected pre-LC serve as transporters of the EBV as they migrate and differentiate into epithelium-resident LC. EBV reactivation in oral LC could infect adjacent epithelial cells that could result in productive EBV replication in the epithelial cells. In this study, we present evidence demonstrating that blood-borne pre-LC are latently infected with EBV and that oral epithelium cells likely to be LC harbor EBV infection that can reactivate into productive EBV replication.
MATERIALS AND METHODS
Human research subjects and tissue specimens.
Human experimentation research was approved by the University of Texas Medical Branch at Galveston Institutional Review Board and the University of Texas Health Science Center at Houston Committee for the Protection of Human Subjects. Informed consent was obtained from each research subject. Subjects were either healthy individuals or patients infected with human immunodeficiency virus (HIV), with or without AIDS. Tissue specimens obtained included 100 ml of venous blood, tongue and buccal mucosal epithelium cells collected by nonsurgical oral epithelial brush biopsy (45), and tongue and buccal mucosal tissue collected by surgical excisional biopsy (44, 46-48, 50). Some subjects contributed specimens more than once, and the numerical identity of each subject was maintained throughout the study.
Isolation of pre-LC from blood.
Mononuclear cells were isolated from anticoagulated whole blood by density gradient centrifugation with Ficoll-Hypaque and washed and suspended in phosphate-buffered saline. The major cell subsets were then removed from the mononuclear cell population with mouse monoclonal antibodies against defining human cell surface molecules, followed by magnetic separation with sheep monoclonal antibody against mouse IgG conjugated to magnetic microbeads (Dynabeads; Dynal Biotech, Oslo, Norway) in the following sequence: T lymphocytes (Table 1, antibody 1), B lymphocytes (Table 1, antibody 2), and monocytes (Table 1, antibody 3). Finally, the CD1a+ cells were positively selected and isolated from the remaining mononuclear cells by using anti-CD1a antibody (Table 1, antibody 4) and magnetic separation with magnetic microbeads (CELLection Pan Mouse IgG Kit; Dynal Biotech, Oslo, Norway) and then released from the microbeads with DNase I. Cells were maintained at 4°C throughout the procedure.
TABLE 1.
Antibodies used in this study
| Antibody | Antibody origin | Antigen target | Conjugate | Clone | Source |
|---|---|---|---|---|---|
| 1 | Mouse | CD3 | None | HIT3a | BD/Pharmingena |
| 2 | Mouse | CD19 | None | HIB19 | BD/Pharmingen |
| 3 | Mouse | CD14 | None | M5E2 | BD/Pharmingen |
| 4 | Mouse | CD1a | None | HI149 | BD/Pharmingen |
| 5 | Mouse | CD1a | FITC | WM35 | Research Diagnosticsb |
| 6 | Mouse | CD1a | PEf | SK9 | BD/Pharmingen |
| 7 | Mouse | CD11c | PE | B-ly6 | BD/Pharmingen |
| 8 | Mouse | CCR6 | PE | 11A9 | BD/Pharmingen |
| 9 | Mouse | E-cadherin | FITC | 36 | BD/Pharmingen |
| 10 | Mouse | CD207/Langerin | PE | DCGM4 | Beckman Coulter Immunotechc |
| 11 | Mouse | CD21 | PE | B-ly4 | BD/Pharmingen |
| 12 | Mouse | CD3 | PerCPg | SK7 | BD/Pharmingen |
| 13 | Mouse | CD19 | PE | HIB19 | BD/Pharmingen |
| 14 | Mouse | CD14 | FITC | MΦP9 | BD/Pharmingen |
| 15 | Goat | FITC | Biotin | Polyclonal | Vector Laboratoriesd |
| 16 | Goat | Rhodamine | Biotin | Polyclonal | Vector Laboratories |
| 17 | Goat | CD207/Langerin | Biotin | Polyclonal | R&D Systemse |
BD Biosciences/Pharmingen, San Jose, CA.
Research Diagnostics, Inc., Flanders, NJ.
Beckman Coulter Immunotech, Fullerton, CA.
Vector Laboratories, Inc., Burlingame, CA.
R&D Systems, Inc., Minneapolis, MN.
PE, phycoerythrin.
PerCP, peridinin-chlorophyll-protein complex.
Isolation of LC from oral epithelium.
Cells obtained with the oral epithelial biopsy brush were suspended and washed in phosphate-buffered saline. CD1a+ cells were positively selected and isolated from the total cell population by using anti-CD1a antibody (Table 1, antibody 4) and magnetic separation with magnetic microbeads (CELLection Pan Mouse IgG Kit; Dynal Biotech, Oslo, Norway) and then released from the microbeads with DNase I and 0.25% trypsin. Cells were maintained at 4°C throughout the procedure.
Histologic and molecular characterization of oral surgical biopsy tissues.
One half of each surgical biopsy specimen was formalin fixed and paraffin embedded. Tissue sections were placed on positively charged glass slides, stained with hematoxylin and eosin, and examined by an oral and maxillofacial pathologist. Specimens were classified as oral hairy leukoplakia if they demonstrated hyperparakeratosis, acanthosis, “koilocyte”-like cells, nuclear chromatin margination, and molecular evidence of productive EBV replication. The other half of each surgical biopsy specimen was frozen, processed for nucleic acid extraction, and analyzed by reverse transcription (RT)-PCR for CD19, CD45, and EBV gene expression by using a previously validated molecular definition of productive EBV replication (45-48, 50).
Culture of LC and chemical induction of EBV.
CD1a+ cells were cultured in RPMI 1640 medium with 15% fetal bovine serum, penicillin, streptomycin, and amphotericin B at 37°C in a 5% CO2 atmosphere. Induction of EBV replication was achieved by adding 30 ng/ml phorbol 12-myristate 13-acetate (MP Biochemicals, Irvine, CA), sodium n-butyrate (Sigma-Aldrich, St. Louis, MO) to a 3 mM final concentration, and 50 μg/ml 5-iodo-2′-deoxyuridine (Sigma-Aldrich, St. Louis, MO) to the medium and incubating it for 48 to 72 h.
Fluorescent immunostaining of cell surface markers.
CD1a+ cells were stained with fluorochrome-conjugated monoclonal antibodies against cell surface molecules (Table 1, antibody 5 to 14). Appropriate fluorochrome-conjugated isotype control antibodies were used. After staining, cells were fixed in 4% paraformaldehyde and cell staining was analyzed with a FACSort flow cytometer with CellQuest software (Becton Dickinson, San Jose, CA) or by imaging on a Zeiss LSM 510 UV META laser scanning confocal microscope.
Limiting-dilution PCR analysis of EBV infection.
For each subject, DNA purified from a known number (x) of CD1a+ cells was serially diluted into four sets of 9 or 10 tubes, each tube with the following quantity of DNA: set 1, 0.075 × x cells; set 2, 0.025 × x cells; set 3, 0.0075 × x cells; set 4, 0.0025 × x cells. The DNA in each tube was then amplified by nested PCR to detect a single-copy sequence present in the EBV gene for BALF1 (Table 2). Forty cycles each were performed for the initial reaction with Pfu Turbo Hotstart DNA polymerase (Stratagene, La Jolla, CA) and for the nested reaction with Vent DNA polymerase (New England BioLabs, Beverly, MA). Amplified products were identified by agarose gel electrophoresis and Southern blot hybridization to an internal 32P-labeled oligonucleotide probe as those products consistent with the predicted size. The sensitivity of this PCR assay was one copy of the target EBV DNA sequence. Standard techniques were used to prevent and detect in vitro contamination of the amplification reaction mixtures. The fraction of negative tubes from each set was plotted on a semilog graph versus the number of cells represented by the DNA added to each tube of the set. The number of cells represented by the line crossing the 37% EBV-negative point was taken as the number of CD1a+ cells harboring a single EBV genome in vivo for each subject. Pooled data from all 14 subjects were analyzed by plotting the total fraction of negative tubes per dilution set versus the total number of cells represented by the DNA added to the tubes of each dilution set to estimate the population's mean number of CD1a+ cells harboring a single EBV genome in vivo.
TABLE 2.
EBV PCR target gene, primer sequences, and product size
| EBV gene | Initial primers (5′-3′) | Nested primers (5′-3′) | Final product size (bp) |
|---|---|---|---|
| BALF1 | ATGAGGCCAGCCAAGTCTACA | ATGAGGCCAGCCAAGTCTACA | |
| GAACTGACGTCTCAGCGATCT | GTCATCCAGGTAGTTTCGCAC | 387 |
Multiplex RT-PCR (M-RT-PCR) analysis of EBV gene expression.
RNA purified from CD1a+ cells was synthesized into cDNA with an oligo(dT) primer and an EBER-1 gene-specific primer and then amplified in a multiplexed nested PCR assay that simultaneously detects seven different EBV gene sequences (Table 3). Forty cycles each were performed for the initial reaction with Pfu Turbo Hotstart DNA polymerase (Stratagene, La Jolla, CA) and the nested reaction with Vent DNA polymerase (New England BioLabs, Beverly, MA). Amplified products were identified by agarose gel electrophoresis and Southern blot hybridization to an internal 32P-labeled oligonucleotide probe as those products consistent with the predicted size. Standard techniques were used to prevent and detect in vitro contamination of the amplification reaction mixtures.
TABLE 3.
EBV M-RT-PCR target genes, primer sequences, and product sizes
| EBV gene | Initial primers (5′-3′) | Nested primers (5′-3′) | Spliced transcript final product size (bp) |
|---|---|---|---|
| EBER-1 | TACGCTGCCCTAGAGGTTTT | TACGCTGCCCTAGAGGTTTT | 144a |
| GGACCACCAGCTGGTACTT | CTTGACCGAAGACGGCAGA | ||
| BZLF1 | ACAGCTAGCAGACATTGGTGT | ACAGCTAGCAGACATTGGTGT | 223 |
| GCACATCTGCTTCAACAGGAG | CCTGTCATTTTCAGATGATTTGG | ||
| LMP-2A | ACCGTCACTCGGACTATCAA | ACCGTCACTCGGACTATCAA | 256 |
| AGCTGGCCACTGCTGCCAA | GCGGTCACAACGGTACTAA | ||
| EBNA-1-Fp/Qpb | ATAGCGTGCGCTACCGGATG | ATAGCGTGCGCTACCGGATG | 333 |
| TATGTCTTGGCCCTGATCCTG | CCGTCCTCGTCCATGGTTAT | ||
| EBNA-1-Cp/Wpc | ACGTGGTGTAAAGTTTTGCCTG | ACGTGGTGTAAAGTTTTGCCTG | 348 |
| TATGTCTTGGCCCTGATCCTG | CCGTCCTCGTCCATGGTTAT | ||
| LMP-1 | GAGACCTTCTCTGTCCACTTG | GAGACCTTCTCTGTCCACTTG | 387 |
| GGTAGCTTGTTGAGGGTGCG | GTCATCGTGGTGGTGTTCATC | ||
| gp220 | CAACCTCACCGCACCTGCAA | CAACCTCACCGCACCTGCAA | 418 |
| GCCGTAATCTGTGGTGGGCT | TCCATGTCCTGTGGTGTGCT |
The EBER-1 transcript is not spliced. Control amplification of RNA treated with DNase I but not reverse transcriptase is performed in parallel to exclude the amplification of possible persisting genomic DNA sequences in the RNA sample.
EBNA-1 expressed from either the F or the Q promoter.
EBNA-1 expressed from either the C or the W promoter.
Fluorescence in situ hybridization (FISH) assay for detection of EBV gene expression.
Formalin-fixed tissue sections and paraformaldehyde-fixed CD1a+ cells were placed on positively charged glass slides, deparaffinized with xylene, rehydrated with decreasing concentrations of ethanol mixed with water, and permeabilized with Triton X-100, NP-40, and proteinase K. Slides were treated with 0.25% acetic anhydride in 0.1 M triethanolamine (pH 8.0) and then hybridized at 55°C for 90 min with four fluorescein isothiocyanate (FITC)-conjugated peptide nucleic acid oligonucleotides complementary to EBER-1 and EBER-2 transcripts (EBV EBER PNA Probe/FITC; DakoCytomation, Carpinteria, CA) in a hybridization solution as supplied with the probe, with or without 4 pmol each of three rhodamine-conjugated locked nucleic acid oligonucleotides complementary to BHLF1 transcripts (Table 4). Slides were washed at 55°C in a stringent wash solution (PNA ISH Detection Kit; DakoCytomation, Carpinteria, CA) for 30 min and then blocked with 20% goat serum. Hybridization was detected with biotin-conjugated goat antibodies against FITC and rhodamine (Table 1, antibodies 15 to 16) and binding to avidin conjugated to FITC or rhodamine (Vector Laboratories, Burlingame, CA). For tissue sections, additional EBER signal amplification was accomplished by a repeat of the biotin anti-FITC antibody and FITC-avidin. Slides were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and analyzed on a Zeiss LSM 510 UV META laser scanning confocal microscope. Appropriate controls were included with each batch performance of the protocol.
TABLE 4.
BHLF1 transcript probe oligonucleotides for EBV EBER/BHLF1-FISH
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| IR2-A | CGGTGGGTCCGCTGG |
| IR2-B | CGCTGGGCACCGCTG |
| IR2-C | GGTTCCTGGCGCTCC |
Underlining indicates the bases synthesized by locked nucleic acid chemistry.
Fluorescent immunostaining for detection of LC in tissue sections.
Formalin-fixed tissue sections were placed on positively charged glass slides, deparaffinized with xylene, and rehydrated with decreasing concentrations of ethanol mixed with water. Antigen unmasking was accomplished by boiling the slides in 10 mM citric acid (pH 6.0) with 0.05% Tween 20 for 20 min. Tissue sections were permeabilized with Triton X-100, NP-40, proteinase K, and trypsin. Slides were blocked with 20% goat serum. LC were detected by using biotin-conjugated goat antibody against CD207/Langerin (Table 1, antibody 17) and binding to avidin conjugated to rhodamine (Vector Laboratories, Burlingame, CA). Signal amplification was accomplished with a biotin-conjugated goat anti-rhodamine antibody (Table 1, antibody 16), followed again by rhodamine-avidin. Slides were stained with DAPI and analyzed on a Zeiss LSM 510 UV META laser scanning confocal microscope. Appropriate controls were included with each batch performance of the protocol.
RESULTS
Isolation and characterization of pre-LC from blood.
Blood-borne CD1a+ CD11c+ cells are direct precursors of LC, making up 0.1 to 0.5% of the total blood mononuclear cell population (14). In this study, CD1a+ mononuclear cells were isolated from the blood of eight healthy subjects and six with AIDS to a mean purity of 99% (Table 5). The cumulative mean frequency of CD3+, CD19+, and CD14+ staining of these purified cells was 0.7% (range, 0 to 2.5%), with a cumulative mean frequency for isotype control antibodies of 0.9% (range, 0 to 2.9%). These results demonstrate that CD1a+ cells were isolated to near absolute purity without significant contamination with T lymphocytes, B lymphocytes, or monocytes.
TABLE 5.
Cell surface marker expression in pre-LC newly isolated from blood
| Subject or parameter (status)a | Mononuclear-cell yield (108) | CD1a+ cell yield (105) | % CD1a+ cells | % CD1a+ CD11c+ cells | % CD1a+ E-cad+ cells | % CD1a+ CCR6+ cells | % CD1a+ CD207+ cells | % CD1a+ CD21+ cells |
|---|---|---|---|---|---|---|---|---|
| 3 (H) | 1.72 | 1.33 | 98 | 99 | 4 | 0 | 0 | 0 |
| 4 (H) | 1.48 | 0.89 | 100 | 100 | 3 | 5 | 1 | 0 |
| 5 (H) | 0.96 | 0.80 | 99 | 100 | 13 | 1 | 3 | 1 |
| 6 (H) | 1.27 | 0.66 | 98 | 99 | 10 | 8 | 2 | 2 |
| 7 (H) | 1.24 | 1.25 | 100 | 98 | 16 | 7 | 1 | 1 |
| 8 (H) | 1.67 | 1.67 | 99 | 99 | 19 | 8 | 1 | 1 |
| 9 (H) | 1.47 | 1.76 | 99 | 94 | 3 | 2 | 0 | 1 |
| 10 (H) | 1.25 | 1.87 | 100 | 80 | 1 | 1 | 0 | 1 |
| 11 (A) | 1.47 | 2.08 | 94 | 100 | 64 | 2 | 1 | 1 |
| 12 (A) | 1.81 | 2.17 | 100 | 100 | 76 | 1 | 1 | 0 |
| 13 (A) | 1.25 | 1.50 | 100 | 94 | 5 | 1 | 1 | 4 |
| 14 (A) | 1.11 | 1.62 | 97 | 98 | 5 | 1 | 1 | 1 |
| 15 (A) | 1.85 | 2.52 | 98 | 98 | 0 | 2 | 0 | 0 |
| 16 (A) | 1.21 | 1.81 | 100 | 100 | 1 | 12 | 0 | 0 |
| Mean | 1.41 | 1.56 | 99 | 97 | 16 | 4 | 1 | 1 |
H, healthy; A, AIDS.
Consistent with the pre-LC phenotype, 97% (mean) of the CD1a+ cells coexpressed CD11c, while LC differentiation markers (E-cadherin, CCR6, CD207/Langerin) and the EBV receptor (CD21) were not significantly coexpressed (Table 5). Cultured CD1a+ cells markedly up-regulated E-cadherin and CD207/Langerin expression (Table 6), consistent with a previous report of culture-induced up-regulation of these LC differentiation markers in pre-LC (14).
TABLE 6.
Up-regulation of LC differentiation markers in cultured pre-LC
| Subject or parameter and cell status | % CD1a+ CD11c+ cells | % CD1a+ E-cad+ cells | % CD1a+ CCR6+ cells | % CD1a+ CD207+ cells | % CD1a+ CD21+ cells |
|---|---|---|---|---|---|
| 3 | |||||
| Newly isolated | 98 | 4 | 0 | 0 | 0 |
| Cultured for 3 days | 95 | 76 | 0 | 88 | 0 |
| 4 | |||||
| Newly isolated | 94 | 3 | 5 | 1 | 0 |
| Cultured for 3 days | 96 | 67 | 0 | 95 | 0 |
| 10 | |||||
| Newly isolated | 80 | 1 | 1 | 0 | 1 |
| Cultured for 3 days | 100 | 42 | 2 | 7 | 3 |
| Mean | |||||
| Newly isolated | 91 | 3 | 2 | 0 | 0 |
| Cultured for 3 days | 97 | 62 | 1 | 63 | 1 |
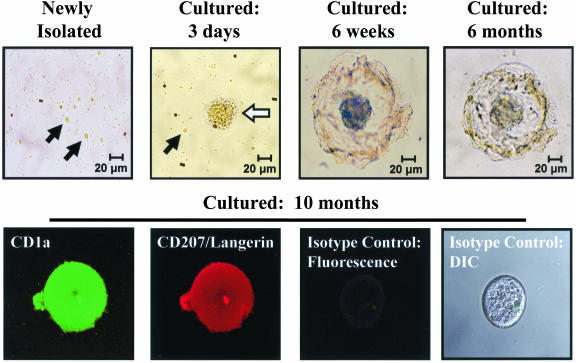
Cultured CD1a+ pre-LC evolved from individual round cells to small cell clusters with short dendritic projections and eventually to larger cluster formations with long dendritic projections that remained stable for more than 10 months (Fig. 1). The cell clusters likely formed as a result of homotypic interaction of newly expressed E-cadherin (31). Immunostaining confirmed CD1a and CD207/Langerin expression in these cell clusters (Fig. 1). These culture-induced expression and morphological changes demonstrate that CD1a+ CD11c+ cells purified from blood are, in fact, pre-LC capable of completing their differentiation into an LC phenotype.
FIG. 1.
Differentiation of blood-derived pre-LC into LC during prolonged culture. Examination by light microscopy revealed that most of the CD1a+ pre-LC newly isolated from blood were small round cells (black arrows). After 3 days, the number of small round cells decreased and larger cell clusters with short dendrite-like projections (white arrow) appeared. By 6 weeks, most cell clusters elaborated a large spherical structure of interlocking dendritic projections surrounding the central core. These structures were stable for more than 10 months of culture and did not increase in number over time, suggesting a lack of cell division. Fluorescent immunostaining and imaging with a laser scanning confocal microscope demonstrated expression of both CD1a and CD207/Langerin in these cell clusters, confirming their differentiation into the LC phenotype. Staining with isotype control fluorescent antibodies was negative. DIC, Nomarski differential interference contrast.
EBV infection in blood-borne pre-LC.
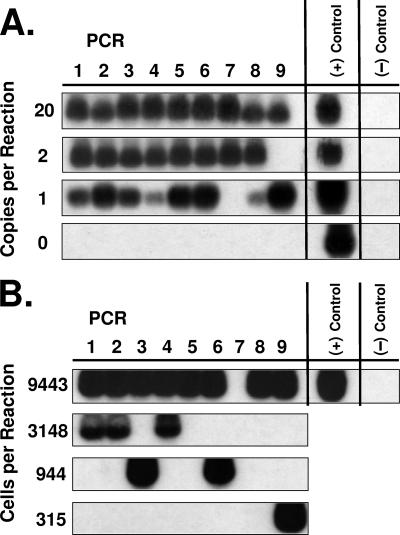
To detect EBV infection in pre-LC, a PCR assay was developed to detect DNA of the gene for EBV BALF1 with a sensitivity of a single target copy (Fig. 2). Similar to the approach used to determine the frequency of EBV infection in B lymphocytes in vivo (22), this BALF1 PCR assay was applied to limiting dilutions of known numbers of purified CD1a+ cells from eight healthy subjects and six with AIDS to detect EBV genomes (Fig. 2) and to estimate their frequency in the CD1a+ cell population. EBV genomes were detectable in the CD1a+ cells for 12 of the 14 subjects and in sufficient quantity to estimate their frequency for 7 of the 14 subjects (Table 7). These seven subjects likely represent the higher end of the frequency range (highest frequency, 2,200 CD1a+ cells per EBV genome), whereas insufficient CD1a+ cells were available for those subjects in which EBV infection occurred at the lower end of the frequency range. To overcome this quantity limitation, pooled data from all 14 subjects were analyzed collectively to estimate a population mean frequency of 12,250 CD1a+ cells per EBV genome (Table 7). There was no significant difference in the mean frequency of CD1a+ cells per EBV genome between healthy subjects and those with AIDS (Table 7). Assuming latent EBV infection with approximately 10 EBV genomes per infected cell (22), it is estimated that EBV infects approximately 1 cell in 105 CD1a+ cells in vivo.
FIG. 2.
Limiting-dilution PCR of the EBV BALF1 gene in pre-LC isolated from blood. (A) For control cells, variable numbers of cells of the Namalwa Burkitt's lymphoma cell line were diluted into a constant number of cells of the Raji Burkitt's lymphoma cell line. Each Namalwa cell carries 2 integrated copies of the EBV genome (two copies of the BALF1 target sequence), whereas each Raji cell carries approximately 50 copies of an EBV genome with the BALF1 gene target sequence naturally deleted. After extraction of the DNA from the admixed cells, PCR amplification was performed in nine identical reaction tubes at each level of dilution containing the indicated number of copies of the BALF1 gene target present along with the DNA of 105 Raji cell equivalents. These results demonstrate that the PCR amplification assay is both sensitive and specific to a single copy of target sequence. (+) Control = B958 lymphoblastoid cell line DNA; (−) Control = no DNA. (B) Newly isolated pre-LC (representative subject example). The DNA from a total of 125,900 CD1a+ cells (Table 7, AIDS subject 15) was extracted, distributed among nine identical reaction tubes at each level of dilution containing the indicated number of cell equivalents, and amplified by PCR for the BALF1 gene target sequence. (+) Control = B958 lymphoblastoid cell line DNA; (−) Control = no DNA.
TABLE 7.
EBV genome frequency in pre-LC isolated from blood
| Subject or parameter (status)a | No. of CD1a+ cells available for PCR assay | Estimated no. of CD1a+ cells/EBV genome in vivo |
|---|---|---|
| 3 (H) | 66,600 | IDEc (>4,955) |
| 4 (H) | 44,500 | 2,200 |
| 5 (H) | 40,000 | 10,750 |
| 6 (H) | 33,000 | IDE (>2,475) |
| 7 (H) | 62,500 | 6,300 |
| 8 (H) | 83,400 | 3,200 |
| 9 (H) | 88,200 | IDE (>6,615) |
| 10 (H) | 93,700 | EBV NDd |
| 11 (A) | 104,200 | IDE (>7,815) |
| 12 (A) | 108,600 | IDE (>8,145) |
| 13 (A) | 75,000 | EBV ND |
| 14 (A) | 81,000 | 13,200 |
| 15 (A) | 125,900 | 4,700 |
| 16 (A) | 90,600 | 13,150 |
| Mean | ||
| All subjects | NAb | 12,250 |
| Healthy subjects | NA | 8,450 |
| AIDS subjects | NA | 15,900 |
H, healthy; A, AIDS.
NA, not applicable (pooled data).
IDE, insufficient data to estimate.
ND, not detectable.
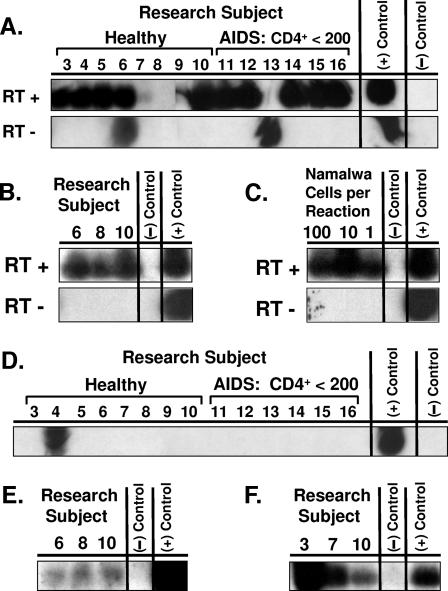
To identify the state of EBV infection in pre-LC, an M-RT-PCR assay was developed to simultaneously detect the transcription of seven different EBV genes in a single reaction (EBV M-RT-PCR) such that the EBV gene expression profile could differentiate among latent infection, nonproductive infection, and productive replication (47, 50). This EBV M-RT-PCR assay was used to analyze purified CD1a+ cells from the blood of the eight healthy subjects and six with AIDS. Expression of EBER-1 was detected in newly isolated CD1a+ cells from 11 subjects and in cultured CD1a+ cells from 3 of 3 subjects (Fig. 3). Expression of replication-associated BZLF1 was detected in newly isolated CD1a+ cells from one subject (Fig. 3). BZLF1 expression was weakly detected in cultured CD1a+ cells from three of three subjects and strongly detected in chemically induced CD1a+ cells from three of three subjects (Fig. 3). Expression of LMP-1, LMP-2A, EBNA-1-Fp/Qp, EBNA-1-Cp/Wp, and gp220 was not detected in the newly isolated or cultured CD1a+ cells from any subject. Two subjects showed weak expression of LMP-1 and EBNA-1-Fp/Qp after chemical induction. These results suggest that EBV infects blood-borne pre-LC as a latency type 0 infection (EBER only), consistent with latent EBV persistence in a nondividing cell. The inducible expression of BZLF1, LMP-1, and EBNA-1-Fp/Qp suggests a potential for EBV in pre-LC to enter a nonproductive infection state (47, 50).
FIG. 3.
EBV M-RT-PCR of pre-LC isolated from blood. CD1a+ pre-LC were isolated from the blood of eight healthy subjects and six with AIDS, each with fewer than 200 CD4+ cells/ml of blood (mean, 70; range, 4 to 164). Cells were studied as newly isolated, after culture for 3 days, or after culture for 3 days and treatment with chemical inducers of EBV replication for 2 more days. RNA extracted from the cells was studied by EBV M-RT-PCR amplification and specific probe hybridization. (+) Control = B958 lymphoblastoid cell line DNA and Akata Burkitt's lymphoma cell line DNA; (−) Control = no DNA. (A) Newly isolated pre-LC. EBER-1 expression was demonstrated in healthy subjects 3, 4, 5, 6, 7, and 10 and in AIDS subjects 11, 12, 14, 15, and 16. With a band in the reverse transcriptase-negative (RT −) reaction mixture, it is uncertain if the result for subject 6 represents true EBER-1 expression or detection of viral genomic DNA sequences in the RNA preparation. Despite a band in the reverse transcriptase-negative reaction mixture for subject 13, the absence of a band in the corresponding reverse transcriptase-positive (RT +) reaction mixture was interpreted as an absence of EBER expression for this subject. (B) Cultured pre-LC. EBER-1 expression was demonstrated in all three subjects, including subject 8, in whom EBER-1 expression was not detected in newly isolated pre-LC. (C) Control cells. The EBV-positive Namalwa Burkitt's lymphoma cell line expresses the least EBER-1 of the known EBV-positive cell lines. The sensitivity of detection of the EBV M-RT-PCR assay for EBER-1 expression was demonstrated to be a single Namalwa cell diluted into 105 cells of the EBV-negative RHEK-1 cell line. (D) Newly isolated pre-LC. BZLF1 expression was demonstrated in healthy subject 4. (E) Cultured pre-LC. New weak BZLF1 expression was demonstrated in all three healthy subjects in whom BZLF1 expression was not detected in newly isolated pre-LC. (F) Cultured and induced pre-LC. Newly induced strong BZLF1 expression was demonstrated in all three healthy subjects in whom BZLF1 expression was not detected in newly isolated pre-LC.
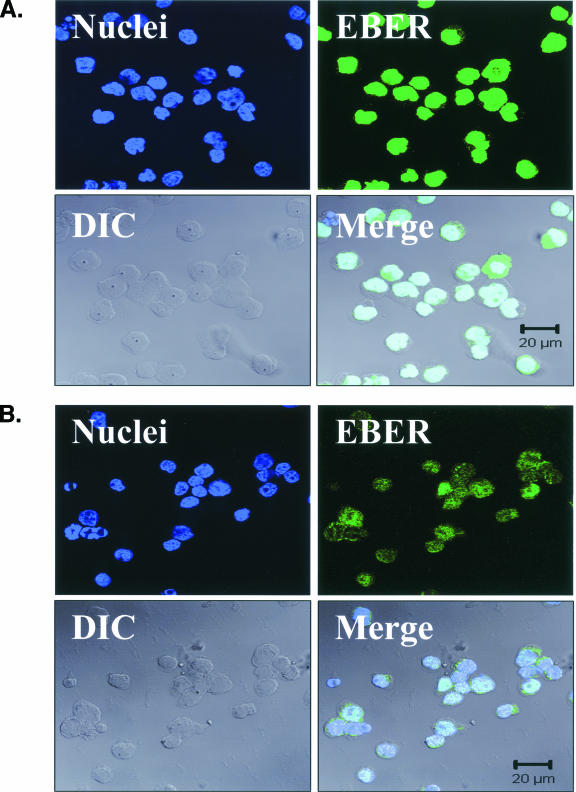
To confirm the presence of EBER expression in individual EBV-infected pre-LC, a highly sensitive FISH assay was developed that is capable of detecting even the lowest level of EBER transcription found in the Namalwa cell line (2, 38) (EBER-FISH; Fig. 4) and used to examine purified CD1a+ cells from the blood of multiple healthy subjects. In total, two unequivocally EBER-positive cells were identified (Fig. 4), each with low-level EBER expression similar to that in the Namalwa cell line. The rarity of these EBER-FISH-positive CD1a+ cells was consistent with the estimated 1-in-105 frequency of EBV pre-LC infection in vivo and confirmed the EBV M-RT-PCR EBER-1 expression results.
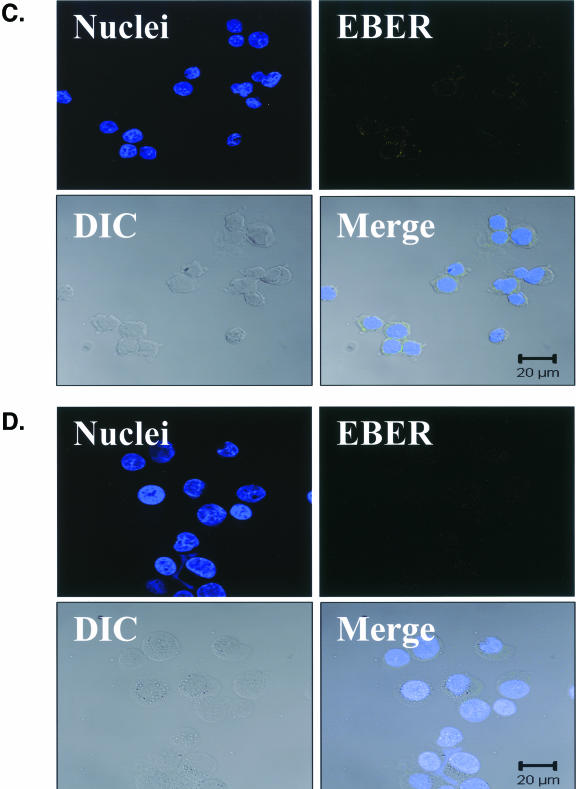
FIG. 4.
EBV EBER-FISH of pre-LC isolated from blood. Control cells and purified CD1a+ pre-LC isolated from the blood of healthy subjects were examined by in situ hybridization for latency-associated EBER transcription. Cells were imaged by fluorescent laser scanning confocal microscopy. DIC, Nomarski differential interference contrast. (A) EBV-positive Raji Burkitt's lymphoma cells express high levels of EBER (up to 105 or 106 transcripts per cell). Nuclear saturation of fluorescence was consistently detected. (B) EBV-positive Namalwa Burkitt's lymphoma cells express much lower levels of EBER (at least 100-fold lower than Raji cells). Variable levels of nuclear EBER expression were easily detected in most Namalwa cells. (C) EBV-negative BJAB Burkitt's lymphoma cells did not show EBER hybridization. (D) EBV-negative RHEK-1 epithelial cells did not show EBER hybridization. (E) Nuclear EBER expression was detected in a newly isolated CD1a+ pre-LC. The numerous black circles represent antibody-conjugated magnetic microbeads that are still attached to most of the CD1a+ cells, including the EBER-positive cell. (F) Nuclear EBER expression was detected in a CD1a+ pre-LC that was cultured in vitro for 3 days before EBER in situ hybridization.
EBV infection in oral epithelial LC.
If EBV latently infects blood-borne pre-LC that will eventually migrate into epithelia and differentiate into LC, then EBV-infected LC should be detectable in oral epithelium. To identify EBV infection in oral LC, an oral epithelial biopsy brush was used to collect cells from the normal oral epithelium of seven healthy subjects. This brush obtains cells down to the basal layer, minimizing cells from the submucosa (45). LC were isolated from the oral cells by CD1a selection, enriching them to a mean purity of 93% as defined by coexpression of CD1a and CD207/Langerin. In newly isolated oral LC, patterns of EBV expression were identified by EBV M-RT-PCR in two subjects consistent with latent/nonproductive (EBER-1 + LMP-1 + BZLF1) and superimposed productive (EBNA-1-Fp/Qp + EBNA-1-Cp/Wp) infections (Fig. 5), as previously described for oral EBV infection (47, 50). Chemically induced oral LC up-regulated EBV genes associated with oral productive EBV replication (BZLF1 + EBNA-1-Cp/Wp + gp220) (Fig. 5) (47, 48, 50). These results suggest that EBV infects oral LC primarily as a latent/nonproductive infection and that EBV in oral LC can reactivate to productive replication.
FIG. 5.
EBV M-RT-PCR of LC isolated from oral epithelium. CD1a+ LC were isolated from cells obtained by brush biopsy of grossly normal oral mucosal epithelia of seven healthy subjects. Half of each oral LC specimen was studied as newly isolated cells, and the other half was studied after culture of the cells in the presence of chemical inducers of EBV replication for 3 days. RNA extracted from the cells was studied by EBV M-RT-PCR amplification and specific probe hybridization. (+) Control = B958 lymphoblastoid cell line DNA and Akata Burkitt's lymphoma cell line DNA; (−) Control = no DNA. (A) Newly isolated oral LC. EBER-1, BZLF1, EBNA-1-Fp/Qp, and EBNA-1-Cp/Wp expression was demonstrated in subjects 10 and 20. LMP-1 expression was demonstrated in subject 10. LMP-2A and gp220 expression was not detected. RT +, with reverse transcriptase; RT −, without reverse transcriptase. (B) Cultured and induced oral LC. Newly induced BZLF1 expression was demonstrated in subjects 4, 5, 17, 18, and 21 and was especially strong in subject 5. (Note that the PCR product of subject 10 leaked from the well of the gel, explaining the apparent lack of BZLF1 hybridization for the induced cells of that subject, and that BZLF1 expression was also previously detected in subject 20 prior to induction.) Newly induced LMP-2A and EBNA-1-Cp/Wp expression was demonstrated in subjects 5 and 18. Newly induced strong gp220 expression was demonstrated in subject 5. LMP-1 and EBNA-1-Fp/Qp expression was not detected.
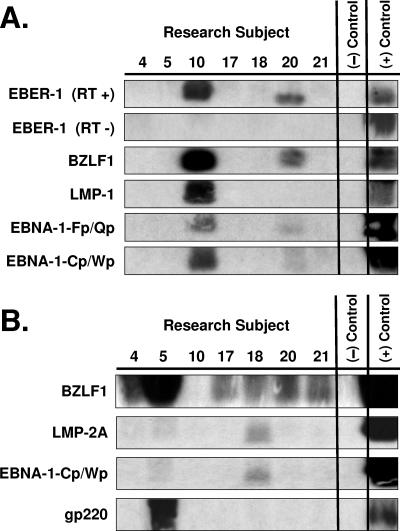
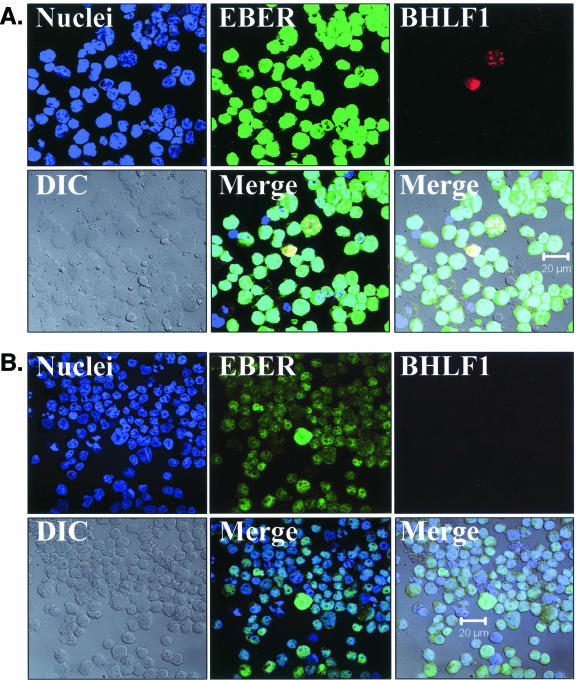
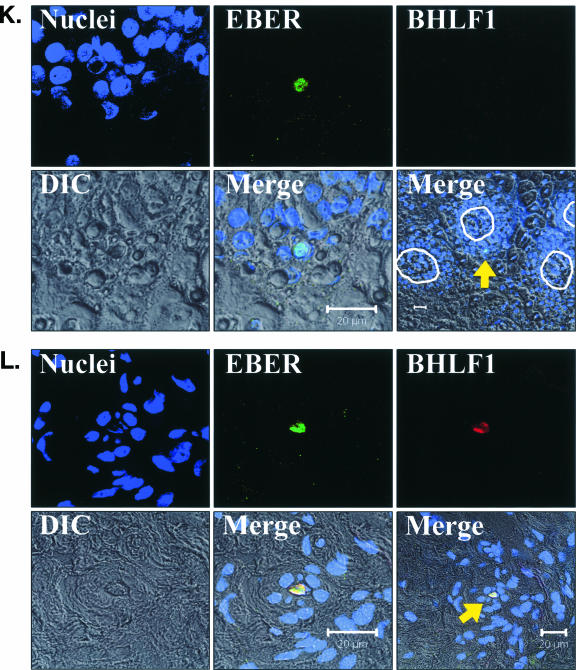
To detect both latent and replicative EBV infections in oral epithelium tissues, a highly sensitive FISH assay was developed to simultaneously detect both EBER and BHLF1 transcription (EBER/BHLF1-FISH) (Fig. 6) and used to examine 248 tissue sections from 62 surgical biopsy specimens (29 oral hairy leukoplakia, 4 normal epithelium with EBV replication, and 29 normal epithelium without EBV replication) obtained from 28 HIV-positive subjects. Tissues harboring EBV replication hybridized with both EBER and BHLF1 (Fig. 6).
FIG. 6.
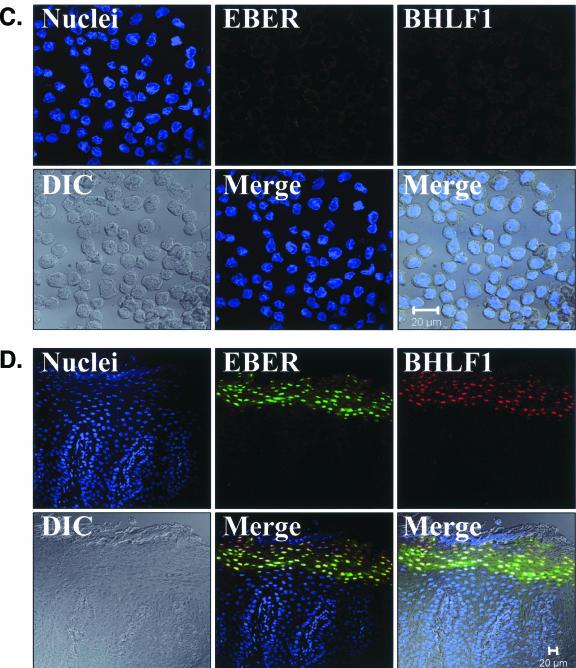
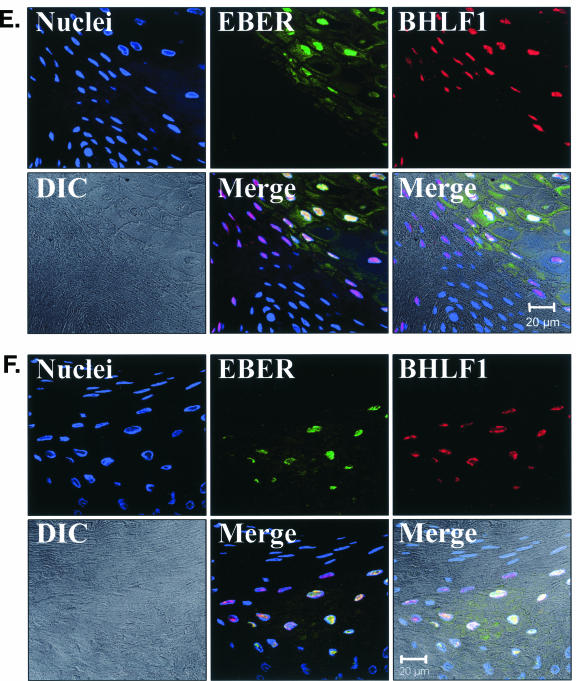
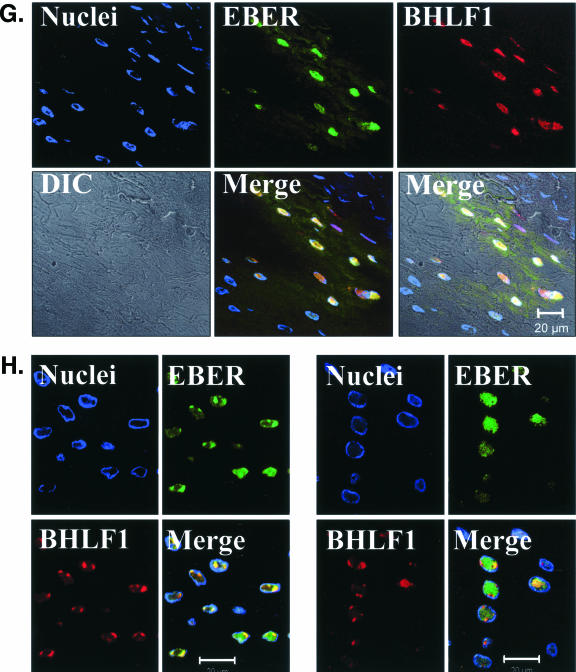
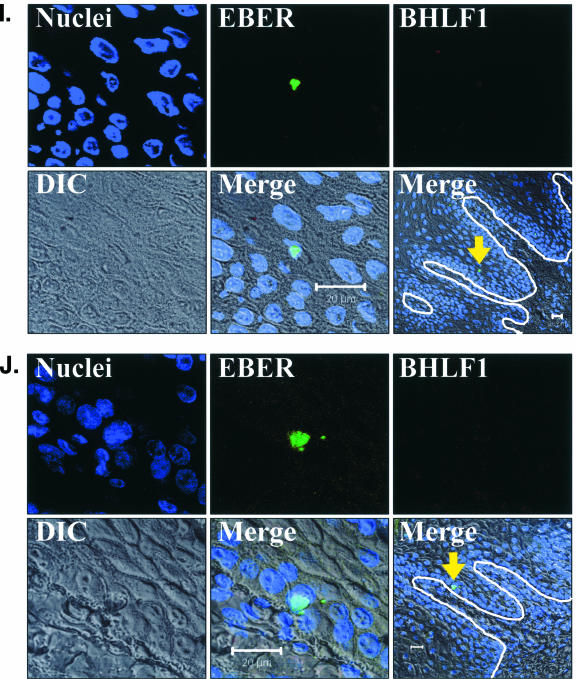
EBV EBER/BHLF1-FISH of oral epithelial tissue. Control cells and oral surgical biopsy tissue sections from HIV-positive subjects were examined by in situ hybridization for latency-associated EBER transcription and replication-associated BHLF1 transcription. Tissues were imaged by fluorescence laser scanning confocal microscopy. Tissue section panels are oriented with the mucosal surface to the top. DIC, Nomarski differential interference contrast. (A) EBV-positive B958 lymphoblastoid cells express high levels of EBER in most cells, and nuclear saturation of fluorescence was consistently detected. Approximately 1 to 5% of B958 cells also express replicative genes, including the early replicative gene BHLF1. Two different patterns of nuclear fluorescence were seen with BHLF1 expression, diffuse and punctate. (B) EBV-positive Namalwa Burkitt's lymphoma cells express much lower levels of EBER (at least 100-fold lower than B958 cells). Variable levels of nuclear EBER expression were easily detected in most Namalwa cells, but BHLF1 expression was not detected in these cells, which harbor only latent EBV infection. (C) EBV-negative RHEK-1 epithelial cells did not hybridize to either EBER or BHLF1. (D) Hybridization with both the EBER and BHLF1 probes was seen in a band-like pattern in the upper spinous layer of oral hairy leukoplakia, consistent with the known localization of productive EBV replication in the oral epithelium. (E, F, G, and H) Nuclear EBER probe hybridization was always associated with nuclear cohybridization of the BHLF1 probe in the upper spinous layer of oral hairy leukoplakia. Nuclear chromatin margination was present, and the most intense EBER probe hybridization strongly colocalized with the BHLF1 probe in the punctate hybridization pattern. This phenomenon of EBER probe hybridization in the upper spinous layer of oral hairy leukoplakia does not represent latent EBV infection but instead is consistent with EBER probe cross-hybridization to EBER gene sequences present in single-stranded EBV DNA synthesized in the nuclei of these cells during productive EBV replication, as previously described for EBER in situ hybridization in oral hairy leukoplakia (28). In the cells with the strongest nuclear EBER hybridization, additional weaker EBER hybridization was often seen in the cytoplasm and likely represents EBER probe cross-hybridization to EBER gene sequences present in artifactually denatured double-stranded EBV DNA in maturing virions being prepared for release from the cells. Furthermore, the cells immediately below and immediately above the EBER-BHLF1 cohybridizing cells often showed nuclear hybridization with only the BHLF1 probe. This result is consistent with early gene expression both preceding and persisting after viral DNA synthesis in the differentiation-dependent cascade of replicative EBV gene expression in oral epithelium, as previously described in oral hairy leukoplakia (40, 52). (I, J, and K) Three tissue sections (panel I, normal tongue epithelium without EBV replication; panels J and K, tongue epithelium with oral hairy leukoplakia) each demonstrated a solitary EBER-expressing cell located in or immediately above the basal layer. The locations of the epithelial basement membrane and basal layer are illustrated by the white lines and circles. The tissue section in panel K represents a cut through the mucosal rete ridges (white circles) in a plane that is perpendicular to the plane represented by the tissue sections in panels I and J. In all three cases, the EBER probe localization was confirmed to be intranuclear in a single cell by computer-generated three-dimensional reconstruction of the cell with a sequential series of 0.6-μm-deep confocal microscopy images. This expression of EBER in the absence of BHLF1 indicates the presence of latent EBV infection in each of these three solitary cells, similar to that demonstrated in the Namalwa cell line (panel B). (L) A tissue section of tongue epithelium with oral hairy leukoplakia demonstrated a solitary EBER- and BHLF1-coexpressing cell in the basal or lower spinous epithelial layer. The location of the epithelial basement membrane is not evident in this photomicrograph, but the cell appears to be located at the top of a rete ridge and was distinctly distant from the EBV replication in the upper spinous epithelial layer. The punctate nuclear colocalization of BHLF1 with the more diffuse EBER and the absence of EBER in the cell cytoplasm together suggest that this cell represents EBV reactivation of early replicative gene expression in a previously latently infected cell, similar to that demonstrated in the B958 cell line (panel A).
Three examples of solitary intraepithelial cells unequivocally expressing only EBER were identified (Fig. 6), consistent with latent EBV infection. The level of EBER expression in each cell was low, similar to that in the Namalwa cell line. One example of a solitary intraepithelial cell unequivocally expressing both EBER and BHLF1 was identified (Fig. 6), consistent with EBV reactivation from latency in that cell. No EBV-positive cells were found in the submucosa of any tissue section, and transcription of CD19 and CD45 was not detected in the tissues with these four EBV-positive cells. All four EBV-positive cells localized to the basal or lower spinous layer of the epithelium, the same locations where LC reside (Fig. 7). Attempts to combine the EBER/BHLF1-FISH and LC immunostaining assays were technically unsuccessful. Nevertheless, these data demonstrate the presence of rare individual cells in the lowest layers of oral epithelium that harbor latent EBV infection and support EBV reactivation.
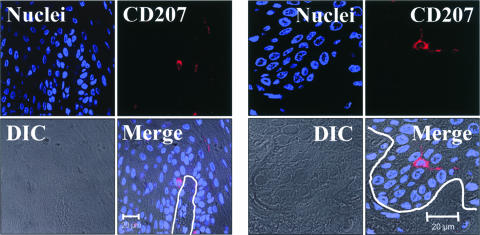
FIG. 7.
CD207/Langerin immunostaining of LC in oral epithelial tissue. Oral surgical biopsy tissue sections of normal tongue epithelium were immunostained for CD207/Langerin. Tissues were imaged by fluorescent laser scanning confocal microscopy. Tissue section panels are oriented with the mucosal surface to the top. DIC, Nomarski differential interference contrast. LC were identified in the basal layer and the immediate suprabasal region of the lower spinous layer of the oral epithelium. The location of the epithelial basement membrane and basal layer is illustrated by the white lines. These results demonstrate that oral LC localize to the same lower epithelial layers as the solitary EBV-positive cells identified in Fig. 6, suggesting a possible LC identity for these EBV-positive cells.
DISCUSSION
The results of this study demonstrate EBV infection of blood-borne pre-LC and of oral epithelium cells likely to be LC. Approximately 1 in 105 pre-LC harbors latent EBV infection in vivo, a frequency similar to EBV infection of B lymphocytes in healthy individuals (6, 22). Blood-borne CD1a+ CD11c+ cells were shown to be pre-LC by their ability to differentiate into the LC phenotype. Remarkably, the EBV gene expression patterns, from latency to nonproductive infection and then productive replication, escalated in parallel with progression of cell differentiation from blood-borne pre-LC into oral LC. The nonproductive EBV expression pattern demonstrated in the purified oral LC matched that originally described for EBV infection in normal oral epithelium without productive replication (47, 48, 50).
Importantly, latent EBV infection and EBV reactivation were identified in rare individual cells in the lower layers of oral epithelium. These data confirm the suspected presence of a cellular reservoir of latent EBV infection in the oral epithelium that serves as a reactivation source for productive EBV replication (44, 47, 50). Although multiple previous studies have failed to identify EBV in the lower layers of oral epithelium (21, 24, 25, 28, 32, 40, 48, 50-52), the singular success of this study is likely a result of two important methodological advantages. First, the uniquely high sensitivity of the EBER/BHLF1-FISH assay made it possible to detect the lowest levels of EBER expression. Second, examination of an unprecedented number of tissue sections, 248, greatly increased the odds of finding the rare (1 in 105) oral LC expected to harbor EBV infection.
Although the present data cannot prove that the four solitary EBV-positive cells identified in this study are LC, they are likely to be LC for the following reasons. EBV latently infects blood-borne pre-LC that are known to migrate to epithelia and differentiate into LC. The lower-layer epithelium locations of the four EBV-positive cells are consistent with the known localization of oral LC. The four EBV-positive cells are solitary, suggesting that they are nondividing cells, consistent with known LC biology. If EBV latently infected a basal epithelial stem or transit amplifying cell, the EBV would be passed to all progeny cells and EBER in situ hybridization would identify a cluster of EBV-positive cells instead. Finally, LC are the most abundant immune cells found in the oral epithelium. Based upon the absence of CD19 and CD45 expression in these tissues and upon current knowledge of EBV and epithelial biology, it is unlikely that a rare oral epithelium-resident B-lymphocyte, T-lymphocyte, melanocyte, or Merkel cell would be harboring the EBV identified in these four cells, but this possibility cannot be absolutely excluded by present data.
While the results of this study do not directly discredit any of the three previously proposed models, they are most consistent with a new model of EBV oral epithelial entry, persistence, and reactivation based upon EBV infection of blood-borne pre-LC (Fig. 8). Although only 1% of pre-LC expressed the CD21 EBV receptor (Table 5), this frequency is 1,000-fold higher than the 1 in 105 pre-LC that apparently becomes infected with EBV in vivo. Given that productive EBV replication in blood-borne, EBV-infected B lymphocytes is rare (3, 6), the source of the EBV that infects blood-borne pre-LC is uncertain. CD21 is expressed on upper spinous epithelial cells of nonkeratinized and parakeratinized oral epithelium but not on lower spinous or basal epithelial cells (5, 40). EBV produced in a basal LC could access the CD21-positive epithelial cells if virions were released from dendritic projections extending into the upper spinous layer. Alternatively, EBV could infect epithelial cells through a CD21-independent mechanism, such as virion binding to cellular integrins (41) or direct cell-to-cell transfer (12), with subsequent lateral spread of EBV infection to adjacent epithelial cells (41). The resulting EBV replication in the upper spinous layer could then occur as either a localized focus in normal oral epithelium (8, 10, 48) or a wider band-like distribution in oral hairy leukoplakia (9).
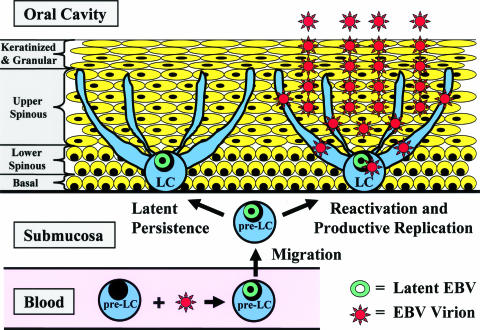
FIG. 8.
Proposed model of EBV oral epithelial entry, persistence, and reactivation. EBV latently infects pre-LC in the blood. The pre-LC migrate from the blood, through the submucosa, and into the oral epithelium, transporting latent EBV infection. In the epithelium, the pre-LC differentiate into LC that reside in the lower epithelial layers and extend dendrites into the upper spinous layer. EBV may persist in LC as a latent infection or may reactivate in LC to productive replication. Infection of adjacent epithelial cells results in productive EBV replication in the upper spinous layer, sometimes causing the pathological changes of oral hairy leukoplakia and ultimately releasing infectious virions into the oral cavity.
The results of this study are consonant with the hypothesis that productive EBV replication in oral epithelium is the major source of virus in oral secretions (15). Although EBV replication has been detected in occasional tonsillar crypt B lymphocytes (1, 11, 26, 27), it is uncertain if so few cells can account for the high levels of virus often detected in saliva (19, 42). EBV transition into oral epithelium via pre-LC and subsequent replication at multiple oral epithelial foci would serve as a viral production amplifier, achieving greater quantities in saliva and more efficient transmission.
The results of this study are also consonant with the observed transition of multiple EBV strains from blood to oral epithelium (44) and the continuously evolving populations of multiple EBV strains seen in the oral secretions of healthy persons (34, 42) and mononucleosis patients (7, 35) and in oral hairy leukoplakia tissues (43, 44, 47). Given that pre-LC appear to be inhibited from entering the oral hairy leukoplakia lesion (46), the EBV-positive cells found in the lower layers of oral hairy leukoplakia tissue (Fig. 6J to L) likely entered prior to development of the lesion. Thus, EBV reactivation in oral LC could not only give rise to new epithelial EBV replication but also potentially contribute new EBV strains to preexisting epithelial EBV replication (Fig. 6L).
Interestingly, the frequency of pre-LC EBV infection did not significantly differ between healthy subjects and those with AIDS (Table 7), despite a large difference in oral hairy leukoplakia prevalence between healthy and immunocompromised persons (9, 17, 20). This result suggests that the conversion of a focus of EBV replication in normal oral epithelium into an oral hairy leukoplakia lesion depends more upon local epithelial immune deficiencies (18), EBV mechanisms of immune evasion (46), and specific EBV gene expression patterns (48, 50) than upon the frequency with which one or more EBV strains enter and reactivate within the oral epithelium (44, 47).
Finally, the results of this study have implications for understanding the pathogenesis of other EBV-associated epithelial diseases. EBV-infected pre-LC likely migrate to all types of epithelial tissues with equal affinity, but the probability of EBV reactivation in an epithelial LC and the outcome of subsequent EBV infection of an epithelial cell may be determined by the local environment unique to each type of epithelium. It makes biological sense that EBV is produced and shed from those mucosal epithelial surfaces with the greatest potential for transmission of EBV to new hosts, such as oropharyngeal (19, 42), genital (13, 36), and lactating mammary (16) epithelia. Other types of mucosal epithelia may be less permissive of EBV replication and more susceptible to latent transforming EBV infection, such as nasopharyngeal (29) and gastric (39) epithelia. The rarity of finding EBV directly in cutaneous epithelial disease (49) suggests that epidermal cells may be resistant to both latent and productive EBV infections.
Acknowledgments
This work was supported in part by a grant from the John Sealy Memorial Endowment Fund for Biomedical Research to Dennis M. Walling.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Anagnostopoulos, I., M. Hummel, C. Kreschel, and H. Stein. 1995. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood 85:744-750. [PubMed] [Google Scholar]
- 2.Arrand, J. R. 2000. Expressed but enigmatic RNAs. EBV Rep. 7:145-149. [Google Scholar]
- 3.Babcock, G. J., L. L. Decker, R. B. Freeman, and D. A. Thorley-Lawson. 1999. Epstein-Barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J. Exp. Med. 190:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 5.Corso, B., L. R. Eversole, and L. Hutt-Fletcher. 1989. Hairy leukoplakia: Epstein-Barr virus receptors on oral keratinocyte plasma membranes. Oral Surg. Oral Med. Oral Pathol. 67:416-421. [DOI] [PubMed] [Google Scholar]
- 6.Decker, L. L., L. D. Klaman, and D. A. Thorley-Lawson. 1996. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J. Virol. 70:3286-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fafi-Kremer, S., P. Morand, J. P. Brion, P. Pavese, M. Baccard, R. Germi, O. Genoulaz, S. Nicod, M. Jolivet, R. W. Ruigrok, J. P. Stahl, and J. M. Seigneurin. 2005. Long-term shedding of infectious Epstein Barr virus after infectious mononucleosis. J. Infect. Dis. 191:985-989. [DOI] [PubMed] [Google Scholar]
- 8.Frangou, P., M. Buettner, and G. Niedobitek. 2005. Epstein-Barr virus (EBV) infection in epithelial cells in vivo: rare detection of EBV replication in tongue mucosa but not in salivary glands. J. Infect. Dis. 191:238-242. [DOI] [PubMed] [Google Scholar]
- 9.Greenspan, J. S., D. Greenspan, E. T. Lennette, D. I. Abrams, M. A. Conant, V. Petersen, and U. K. Freese. 1985. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N. Engl. J. Med. 313:1564-1571. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann, K., P. Frangou, J. Middeldorp, and G. Niedobitek. 2002. Epstein-Barr virus replication in tongue epithelial cells. J. Gen. Virol. 83:2995-2998. [DOI] [PubMed] [Google Scholar]
- 11.Hudnall, S. D., Y. Ge, L. Wei, N.-P. Yang, H.-Q. Wang, and T. Chen. 2005. Distribution and phenotype of Epstein-Barr virus-infected cells in human pharyngeal tonsils. Mod. Pathol. 18:519-527. [DOI] [PubMed] [Google Scholar]
- 12.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israele, V., P. Shirley, and J. W. Sixbey. 1991. Excretion of the Epstein-Barr virus from the genital tract of men. J. Infect. Dis. 163:1341-1343. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., M. Inaba, K. Inaba, J. Toki, S. Sogo, T. Iguchi, Y. Adachi, K. Yamaguchi, R. Amakawa, J. Valladeau, S. Saeland, S. Fukuhara, and S. Ikehara. 1999. A CD1a+/CD11c+ subset of human blood dendritic cells is a direct precursor of Langerhans cells. J. Immunol. 163:1409-1419. [PubMed] [Google Scholar]
- 15.Jiang, R., R. S. Scott, and L. M. Hutt-Fletcher. 2006. Epstein-Barr virus shed in saliva is high in B-cell-tropic glycoprotein gp42. J. Virol. 80:7281-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junker, A. K., E. E. Thomas, A. Radcliffe, R. B. Forsyth, A. G. Davidson, and L. Rymo. 1991. Epstein-Barr virus shedding in breast milk. Am. J. Med. Sci. 302:220-223. [DOI] [PubMed] [Google Scholar]
- 17.King, G. N., C. M. Healy, M. T. Glover, J. T. Kwan, D. M. Williams, I. M. Leigh, and M. H. Thornhill. 1994. Prevalence and risk factors associated with leukoplakia, hairy leukoplakia, erythematous candidiasis, and gingival hyperplasia in renal transplant recipients. Oral Surg. Oral Med. Oral Pathol. 78:718-726. [DOI] [PubMed] [Google Scholar]
- 18.Lilly, E. A., J. E. Cameron, K. V. Shetty, J. E. Leigh, S. Hager, K. M. McNulty, C. Cheeks, M. E. Hagensee, and P. L. Fidel, Jr. 2005. Lack of evidence for local immune activity in oral hairy leukoplakia and oral wart lesions. Oral Microbiol. Immunol. 20:154-162. [DOI] [PubMed] [Google Scholar]
- 19.Ling, P. D., J. A. Lednicky, W. A. Keitel, D. G. Poston, Z. S. White, R. Peng, Z. Liu, S. K. Mehta, D. L. Pierson, C. M. Rooney, R. A. Vilchez, E. O. Smith, and J. S. Butel. 2003. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J. Infect. Dis. 187:1571-1580. [DOI] [PubMed] [Google Scholar]
- 20.Lozada-Nur, F., J. Robinson, and J. A. Regezi. 1994. Oral hairy leukoplakia in nonimmunosuppressed patients. Report of four cases. Oral Surg. Oral Med. Oral Pathol. 78:599-602. [DOI] [PubMed] [Google Scholar]
- 21.Miller, D. R., P. L. Heard, M. P. Cagle, D. DiMaio, Y. Ench, D. G. Morrison, P. A. Eagan, M. L. Gulley, H. B. Jenson, and M. P. Moyer. 1994. Absence of a reservoir of Epstein-Barr virus (EBV) in normal tongue epithelium. J. Oral Pathol. Med. 23:156-160. [DOI] [PubMed] [Google Scholar]
- 22.Miyashita, E. M., B. Yang, K. M. Lam, D. H. Crawford, and D. A. Thorley-Lawson. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80:593-601. [DOI] [PubMed] [Google Scholar]
- 23.Mogi, G. 1975. Secretory immunoglobulin A in oral and respiratory passages in man. Ann. Otol. Rhinol. Laryngol. 84:1-23. [DOI] [PubMed] [Google Scholar]
- 24.Murray, P. G., G. Niedobitek, E. Kremmer, F. Grasser, G. M. Reynolds, A. Cruchley, D. M. Williams, N. Muller-Lantzsch, and L. S. Young. 1996. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J. Pathol. 178:44-47. [DOI] [PubMed] [Google Scholar]
- 25.Murray, P. G., L. J. Swinnen, C. M. Constandinou, J. M. Pyle, T. J. Carr, J. M. Hardwick, and R. F. Ambinder. 1996. BCL-2 but not its Epstein-Barr virus-encoded homologue, BHRF1, is commonly expressed in posttransplantation lymphoproliferative disorders. Blood 87:706-711. [PubMed] [Google Scholar]
- 26.Niedobitek, G., A. Agathanggelou, N. Steven, and L. S. Young. 2000. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol. Pathol. 53:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niedobitek, G., A. Agathanggelou, H. Herbst, L. Whitehead, D. H. Wright, and L. S. Young. 1997. Epstein-Barr virus (EBV) infection in infectious mononucleosis: viral latency, replication and phenotype of EBV-infected cells. J. Pathol. 182:151-159. [DOI] [PubMed] [Google Scholar]
- 28.Niedobitek, G., L. S. Young, R. Lau, L. Brooks, D. Greenspan, J. S. Greenspan, and A. B. Rickinson. 1991. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J. Gen. Virol. 72:3035-3046. [DOI] [PubMed] [Google Scholar]
- 29.Pathmanathan, R., U. Prasad, R. Sadler, K. Flynn, and N. Raab-Traub. 1995. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 333:693-698. [DOI] [PubMed] [Google Scholar]
- 30.Riccardi, R., N. Pimpinelli, G. Ficarra, L. Borgognoni, D. Gaglioti, D. Milo, and P. Romagnoli. 1990. Morphology and membrane antigens of nonlymphoid accessory cells in oral hairy leukoplakia. Hum. Pathol. 21:897-904. [DOI] [PubMed] [Google Scholar]
- 31.Riedl, E., J. Stockl, O. Majdic, C. Scheinecker, K. Rappersberger, W. Knapp, and H. Strobl. 2000. Functional involvement of E-cadherin in TGF-β1-induced cell cluster formation of in vitro developing human Langerhans-type dendritic cells. J. Immunol. 165:1381-1386. [DOI] [PubMed] [Google Scholar]
- 32.Sandvej, K., L. Krenacs, S. J. Hamilton-Dutoit, J. L. Rindum, J. J. Pindborg, and G. Pallesen. 1992. Epstein-Barr virus latent and replicative gene expression in oral hairy leukoplakia. Histopathology 20:387-395. [DOI] [PubMed] [Google Scholar]
- 33.Séguier, S., G. Godeau, and N. Brousse. 2000. Immunohistological and morphometric analysis of intra-epithelial lymphocytes and Langerhans cells in healthy and diseased human gingival tissues. Arch. Oral Biol. 45:441-452. [DOI] [PubMed] [Google Scholar]
- 34.Sitki-Green, D., M. Covington, and N. Raab-Traub. 2003. Compartmentalization and transmission of multiple Epstein Barr virus strains in asymptomatic carriers. J. Virol. 77:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sitki-Green, D. L., R. H. Edwards, M. M. Covington, and N. Raab-Traub. 2004. Biology of Epstein-Barr virus during infectious mononucleosis. J. Infect. Dis. 189:483-492. [DOI] [PubMed] [Google Scholar]
- 36.Sixbey, J. W., S. M. Lemon, and J. S. Pagano. 1986. A second site for Epstein-Barr virus shedding: the uterine cervix. Lancet 2:1122-1124. [DOI] [PubMed] [Google Scholar]
- 37.Sixbey, J. W., and Q. Y. Yao. 1992. Immunoglobulin A-induced shift of Epstein-Barr virus tissue tropism. Science 255:1578-1580. [DOI] [PubMed] [Google Scholar]
- 38.Stowe, R. P., M. L. Cubbage, C. F. Sams, D. L. Pierson, and A. D. Barrett. 1998. Detection and quantification of Epstein-Barr virus EBER1 in EBV-infected cells by fluorescent in situ hybridization and flow cytometry. J. Virol. Methods 75:83-91. [DOI] [PubMed] [Google Scholar]
- 39.Takada, K. 2000. Epstein-Barr virus and gastric carcinoma. Mol. Pathol. 53:255-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, J. A., D. H. Felix, D. Wray, J. C. Southam, H. A. Cubie, and D. H. Crawford. 1991. Epstein-Barr virus gene expression and epithelial cell differentiation in oral hairy leukoplakia. Am. J. Pathol. 139:1369-1380. [PMC free article] [PubMed] [Google Scholar]
- 41.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 42.Walling, D. M., A. L. Brown, W. Etienne, W. A. Keitel, and P. D. Ling. 2003. Multiple Epstein-Barr virus infections in healthy individuals. J. Virol. 77:6546-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walling, D. M., S. N. Edmiston, J. W. Sixbey, M. Abdel-Hamid, L. Resnick, and N. Raab-Traub. 1992. Coinfection with multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc. Natl. Acad. Sci. USA 89:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walling, D. M., W. Etienne, A. J. Ray, C. M. Flaitz, and C. M. Nichols. 2004. Persistence and transition of Epstein-Barr virus genotypes in the pathogenesis of oral hairy leukoplakia. J. Infect. Dis. 190:387-395. [DOI] [PubMed] [Google Scholar]
- 45.Walling, D. M., C. M. Flaitz, K. Adler-Storthz, and C. M. Nichols. 2003. A non-invasive technique for studying oral epithelial Epstein-Barr virus infection and disease. Oral Oncol. 13:436-444. [DOI] [PubMed] [Google Scholar]
- 46.Walling, D. M., C. M. Flaitz, F. G. Hosein, M. Montes-Walters, and C. M. Nichols. 2004. Effect of Epstein-Barr virus replication on Langerhans cells in pathogenesis of oral hairy leukoplakia. J. Infect. Dis. 189:1656-1663. [DOI] [PubMed] [Google Scholar]
- 47.Walling, D. M., C. M. Flaitz, and C. M. Nichols. 2003. Epstein-Barr virus replication in oral hairy leukoplakia: response, persistence, and resistance to treatment with valacyclovir. J. Infect. Dis. 188:883-890. [DOI] [PubMed] [Google Scholar]
- 48.Walling, D. M., C. M. Flaitz, C. M. Nichols, S. D. Hudnall, and K. Adler-Storthz. 2001. Persistent productive Epstein-Barr virus replication in normal epithelial cells in vivo. J. Infect. Dis. 184:1499-1507. [DOI] [PubMed] [Google Scholar]
- 49.Walling, D. M., S. D. Hudnall, and A. Yen-Moore. 2002. Epstein-Barr virus, p. 145-171. In S. K. Tyring (ed.), Mucocutaneous manifestations of viral diseases. Marcel Dekker, Inc., New York, NY.
- 50.Walling, D. M., P. D. Ling, A. V. Gordadze, M. Montes-Walters, C. M. Flaitz, and C. M. Nichols. 2004. Expression of Epstein-Barr virus latent genes in oral epithelium: determinants of the pathogenesis of oral hairy leukoplakia. J. Infect. Dis. 190:396-399. [DOI] [PubMed] [Google Scholar]
- 51.Webster-Cyriaque, J., J. Middeldorp, and N. Raab-Traub. 2000. Hairy leukoplakia: an unusual combination of transforming and permissive Epstein-Barr virus infections. J. Virol. 74:7610-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, A. B. Rickinson, and P. J. Farrell. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]