Abstract
Human cytomegalovirus (HCMV) lytic DNA replication is initiated at the complex cis-acting oriLyt region, which spans nearly 3 kb. DNA synthesis requires six core proteins together with UL84 and IE2. Previously, two essential regions were identified within oriLyt. Essential region I (nucleotides [nt] 92209 to 92573) can be replaced with the constitutively active simian virus 40 promoter, which in turn eliminates the requirement for IE2 in the origin-dependent transient-replication assay. Essential region II (nt 92979 to 93513) contains two elements of interest: an RNA/DNA hybrid domain and an inverted repeat sequence capable of forming a stem-loop structure. Our studies now reveal for the first time that UL84 interacts with a stem-loop RNA oligonucleotide in vitro, and although UL84 interacted with other nucleic acid substrates, a specific interaction occurred only with the RNA stem-loop. Increasing concentrations of purified UL84 produced a remarkable downward-staircase pattern, which is not due to a nuclease activity but is dependent upon the presence of secondary structures, suggesting that UL84 modifies the conformation of the RNA substrate. Cross-linking experiments show that UL84 possibly changes the conformation of the RNA substrate. The addition of purified IE2 to the in vitro binding reaction did not affect binding to the stem-loop structure. Chromatin immunoprecipitation assays performed using infected cells and purified virus show that UL84 is bound to oriLyt in a region adjacent to the RNA/DNA hybrid and the stem-loop structure. These results solidify UL84 as the potential initiator of HCMV DNA replication through a unique interaction with a conserved RNA stem-loop structure within oriLyt.
The mechanism of initiation of human cytomegalovirus (HCMV) lytic DNA replication is largely undefined. The process of HCMV lytic DNA replication requires six core replication proteins: UL54 (polymerase), UL44 (polymerase accessory protein), UL57 (single-stranded DNA binding protein), UL70 (primase), UL102 (primase-associated factor), and UL105 (helicase) (13, 30). These six core proteins make up the generic replication machinery responsible for lytic DNA replication of the HCMV genome (27). Although the proposed activities of these components are postulated through homology to other herpesvirus systems, the exact mechanism of their assembly at the site of the origin is unknown. Our laboratory seeks to understand the early events in HCMV origin-dependent DNA replication that lead to the assembly and function of the HCMV DNA replication machinery.
Most herpesviruses, as well as many other DNA viruses, encode an initiation factor, or origin binding protein (OBP), responsible for recognizing the lytic origin and creating a local environment where DNA is separated and the replication machinery can assemble. Many initiation proteins such as those for simian virus 40 (SV40) T antigen, papillomavirus E1, and herpes simplex virus type 1 (HSV-1) UL9 have enzymatic activities such as DNA-dependent nucleoside triphosphatase activity and helicase activity (10, 11, 29, 44). In addition, a few of the initiation proteins such as Epstein-Barr virus Zta and human herpesvirus 8 K-bZIP also play a dual role as transcriptional activators/repressors as well as their role in origin recognition (12, 18, 19, 21, 31). Other viral mechanisms for initiation of DNA synthesis involve the creation of a site of initiation and are exemplified by adeno-associated virus (AAV), where the AAV Rep protein recognizes and nicks a stem-loop structure within the origin of replication (6). All of these proteins exhibit specific activity for nucleic acid binding to a sequence or structure within the lytic origin, and this activity consequently targets the protein to the site of DNA synthesis (20, 21, 24, 35, 40, 42, 45). The initiation of HCMV DNA replication appears to be more complex than that of other herpesviruses in that oriLyt spans nearly 3 kb and contains many cis-acting features such as transcription factor binding sites, RNA/DNA hybrid structures, and proposed stem-loop structures (2, 25, 32).
It was shown previously that the only noncore protein required for HCMV lytic DNA replication is UL84 (34, 46, 48). In addition, the major immediate early protein IE2 (IE86) is also necessary for the process of initiation of HCMV lytic DNA replication; however, IE2 appears to supply a transactivator function, as demonstrated by the successful functional replacement of an IE2-responsive element within oriLyt essential region I (ERI) with the constitutive SV40 promoter (47). This indicated that UL84 performs a function that is essential for the initiation of HCMV origin-dependent DNA replication, and the function is imparted within a specific region, ERII, of oriLyt. ERII contains an RNA/DNA hybrid structure as well as two proposed stem-loop structures, one of which is variably reiterated, adding up to an additional 300 bp within laboratory strains (2, 32).
UL84 is a unique protein, exhibiting little to no homology to any other protein in nature with the exception of homologues present in closely related cytomegaloviruses (CMVs), such as chimpanzee CMV. Previously, our laboratory and others have demonstrated that UL84 is an essential multifunctional phosphoprotein that displays early kinetics (8, 17). The protein is an enzyme that exhibits UTPase activity that is not stimulated by nucleic acids and that represses IE2-mediated transactivation of the UL112-113 promoter, indicating that UL84 can both positively and negatively affect IE2 transactivation function (8, 16). It is assumed that the latter activity is dependent upon the ability of UL84 to form a stable interaction with IE2 (38). Our previous studies also demonstrated that UL84 is able to self-associate, and this interaction is essential for oriLyt-dependent DNA replication (7). A recent report by Lischka and colleagues demonstrated that UL84 acts as a nucleocytoplasmic shuttling protein, using a CRM-1-dependent pathway facilitated by two leucine-rich nuclear export signals within the protein (23).
In an effort to understand the function of UL84 and to characterize possible nucleic acid substrates for the protein we now show for the first time that UL84 interacts with a specific region of oriLyt in vitro and in vivo. Although purified UL84 interacts with a variety of nucleic acid substrates nonspecifically, it binds with high affinity and specificity to the stem-loop configuration that is part of the RNA/DNA hybrid structure within the defined lytic origin of DNA replication, oriLyt. This binding displays a unique downward-staircase pattern, suggesting that UL84 acts to change the conformation of the stem-loop RNA oligonucleotide. This evidence suggests that UL84 recognizes and binds to this structure within the origin of replication in the early events of initiation of HCMV DNA replication. Analysis of the interaction of UL84 with oriLyt using the chromatin immunoprecipitation (ChIP) assay showed that binding was observed in a region of oriLyt known to contain an RNA/DNA hybrid.
MATERIALS AND METHODS
Cells and viruses.
Vero and COS-1 cells used for production and purification of recombinant UL84 and IE2 protein were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) bovine growth serum (HyClone) (7). Ad293 cells (Stratagene) used to produce the UL84 recombinant adenovirus were propagated in DMEM supplemented with 10% (vol/vol) bovine growth serum (HyClone). Primary human foreskin fibroblasts (HF) were cultured in DMEM supplemented with 10% (vol/vol) fetal bovine serum. HCMV strain AD169 (ATCC VR-538) was maintained as frozen stocks and propagated as described previously (36).
Antibodies.
The UL84-specific antibody (Virusys; CA144) was used for electrophoretic mobility shift assays (EMSA), the ChIP assay, and coimmunoprecipitation assays. The IE2-specific antibody G13-12E2 (Vancouver Biotech) was used for Western blotting identification of IE2 coimmunoprecipitated protein.
Virion preparation.
For the ChIP assay, 10 roller bottles totaling 1 × 108 HF were infected with AD169 at a multiplicity of infection of 1 to 5, and supernatant was harvested 2 weeks postinfection. Cells were subjected to three freeze-thaw cycles and centrifuged at 3,000 rpm for 10 min to pellet cell debris. Supernatant virus was pelleted at 25,000 rpm for 1 h using an SW28 rotor. The virus pellet was resuspended in 2 ml 1× phosphate-buffered saline (PBS), layered on top of a discontinuous sucrose gradient, and centrifuged at 25,000 rpm for 1 h using an SW28 rotor. The virion layer was removed, suspended in 2 ml 1× PBS, and centrifuged at 25,000 rpm for 1 h using an SW28 rotor. The pelleted virus was resuspended in 2 ml 1× PBS and treated with 20 U micrococcal nuclease at 37°C for 10 min and then again purified using a discontinuous sucrose gradient. The resulting purified virions were stored in 2 ml 1× PBS at −80°C.
Oligonucleotides.
All oligonucleotides were purchased from Integrated DNA Techonologies. DNA oligonucleotides were purified by polyacrylamide gel electrophoresis (PAGE), and RNA oligonucleotides were purified by RNase-free high-performance liquid chromatography for use in the EMSA. SL-RNA and SL-DNA sequences are derived from nucleotides 93127 to 93181. This sequence contains part of the stem-loop from the larger stem-loop mapping from nucleotide (nt) 93096 to 93211. EMSA oligonucleotides include SL-RNA, 5′-CGGGCCCGUCCCACCGCCCUGGAGCACCAUCCGGGGCCGUGGGCCGGGC A-3′, SL-RNA PS, 5′-C*PS*GGGCCCGUCCCACCGCCCUGGAGCACCAUCCGGGGCCGUGGGCCGGGC*PS*A-3′, and SL-DNA, 5′-CGGGCCCGTCCCACCGCCCTGGAGCACCATCCGGGGCCGTGGGCCGGGCA-3′ (*PS* indicates phosphorothioate linkages). ChIP assay primers include UL144 F, 5′-GCGGGTGGTGTTGCGCGGCGA-3′, UL144 R, 5′-GCCGACCGCCGCCGCCCTCCC-3′, oriLyt F, 5′-ACGGCGCACATCTAGTGGAATTTTACCG-3′, and oriLyt R, 5′-CTCCGGAACCGGGGGGGGCAAATTTTTA-3′.
Protein purification.
Purified UL84 from mammalian (Vero) cells was produced using a recombinant adenovirus containing the UL84 open reading frame in frame with a 3′ FLAG tag as described previously by our laboratory (7, 8). All protein was evaluated for concentration and purity by sodium dodecyl sulfate (SDS)-PAGE of gel stained with Coomassie blue. Only protein samples deemed to be greater than 90% purity were used in binding assays. All protein quantities were approximated by comparison to bovine serum albumin standards. Similarly, IE2 protein was purified from transfection of 8- to 10-cm dishes of COS-1 cells with a phCMV1-Xi-IE2 FLAG plasmid that expresses the IE2 cDNA in frame with a 3′ FLAG tag (47). IE2 FLAG protein was purified and evaluated in the same manner as described previously for UL84 (7, 8).
EMSA.
All binding reactions were carried out at room temperature for 30 min in a 20-μl reaction volume. The components of the reaction mixture include binding buffer (10 mM Tris-HCl, pH 7.5, 0.5 mM MgCl2, 100 mM NaCl, and 10% glycerol), between 0 and 0.5 μg purified UL84 protein, in some cases with the addition of IE2 protein (see figure legends), and 100 nM 32P-abeled probe. All samples containing RNA-based substrates also included 1 μl of RNasin (Promega). Elution buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol) was used to fill reaction mixtures when unequal volumes of protein were used in order to maintain buffer and salt concentrations throughout the experiments. Twenty microliters of loading buffer (40% glycerol, 10 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanole) was added to each reaction mixture, and 20 μl of the mixture was subsequently separated through a 0.6% agarose gel at 4.5 V/cm for 4.5 h. The gel was dried and imaged using a phosphorimager (GE Healthcare). For cross-linking experiments, reactions were carried out as described above except samples were UV irradiated for 30 min on ice in a UV Stratalinker 2400 (Stratagene).
ChIP.
For infected-cell samples, 1 × 107 HF cells were infected with AD169 (multiplicity of infection, 10) for 3 days. Samples were fixed for 10 min with 1% formaldehyde, washed twice with 1× PBS, and lysed for 15 min in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.1% Tween 20, 1 mM EDTA). Samples were sonicated to shear DNA (∼500 bp), and fragments were diluted sixfold with ChIP buffer (12.5 mM Tris, pH 8, 200 mM NaCl, 1% Triton X-100) and precleared with mouse immunoglobulin G-AC (Santa Cruz Biotechnology) at 4°C for 30 min. For each immunoprecipitation, 2.64 μg of antibody (Mab84) was incubated with the lysate at 4°C overnight. No-antibody and antibody isotype control immunoprecipitations were also performed. Protein G-Plus-agarose beads (Santa Cruz Biotechnology) were blocked with various amounts of sheared salmon sperm DNA and bovine serum albumin at 4°C overnight and then washed with ChIP buffer. The blocked and washed protein G-Plus beads were incubated with the lysate at 4°C for 1 h. The beads were washed once with low-salt buffer (0.1% SDS, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 150 mM NaCl), once with high-salt buffer (0.1% SDS, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 500 mM NaCl), once with LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris pH 8), and twice with Tris-EDTA (TE). Beads were resuspended in TE, incubated with RNase A at 37°C for 30 min, and then incubated with proteinase K and 10% SDS at 37°C for 4 h, followed by incubation at 65°C overnight. For input control samples, NaCl was added to the sonicated lysate to a final concentration of 0.3 M and incubated at 65°C overnight. After the antibody, no-antibody, and isotype control immunoprecipitated samples were filtered through a 0.45-μm filter to remove the agarose beads, they were extracted with phenol-chloroform and ethanol precipitated. DNA was then used as a template for PCR with primers that spanned the oriLyt region, or, in the case of a control sample, PCR primers from the HCMV UL144 locus were used.
RESULTS
UL84 binds to a synthetic oligonucleotide stem-loop structure complementary to a region within the HCMV lytic origin of replication.
One defining prerequisite for a protein that initiates DNA synthesis is that it must bind to nucleic acid either directly or indirectly. Since our hypothesis is that UL84 initiates lytic DNA replication, we assume that this protein must interact with a specific region(s) within oriLyt. Based on previous studies, it is known that a region upstream of nt 92888 (NotI site) in oriLyt was able to be replaced with the SV40 promoter and still retain function (Fig. 1) (47). Although we acknowledge that UL84 could interact with this region, we were interested in potential interaction sites of UL84 outside of this promoter domain and within ERII. Therefore we decided to focus on the region downstream of the NotI site where a previously reported RNA/DNA hybrid structure was identified (Fig. 1). Within this region there is a series of reiterated repeat sequences that have the capacity to form a stem-loop structure. This stem-loop is also part of the essential variably reiterated region of HCMV oriLyt, spanning nt 92887 to 93513 (2, 32). This structure seemed an attractive substrate for an origin recognition protein such as UL84. Consequently, we chose to evaluate the ability of purified UL84 to bind to synthetic oligonucleotides complementary to the proposed stem-loop sequence within oriLyt.
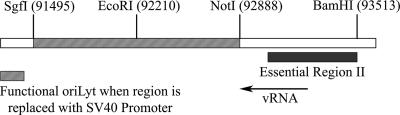
FIG. 1.
Schematic of the HCMV origin of DNA replication, oriLyt. The relative positions and nucleotide numbers of notable restriction enzyme recognition sequences are shown. The area between SgfI and NotI sites denotes the region that can be replaced with the SV40 promoter. Also shown are the relative positions of ERII and the viral RNA (vRNA) in comparison to oriLyt.
To test this potential structural target, we generated a synthetic RNA oligonucleotide (since the region is part of the RNA/DNA hybrid) that had the same sequence present within HCMV oriLyt. We initially used this oligonucleotide (SL-RNA) as a substrate in the EMSA as described in Materials and Methods. Our attempt to resolve a UL84-nucleic acid complex using a PAGE system failed due to the large size of the protein-nucleic acid complex; hence, we switched to an agarose gel resolution system similar to one used previously for the HSV-1 replication protein ICP8 (4).
SL-RNA was able to interact with UL84, and this interaction was confirmed by the presence of a supershifted species upon incubation with the UL84-specific antibody Mab84 (Fig. 2A, lanes 2 and 3, respectively). The shifted band was efficiently eliminated by competition with unlabeled SL-RNA oligonucleotides added to the binding mixture at concentrations of 50- or 100-fold molar excess (Fig. 2A, lanes 4 and 5, respectively).
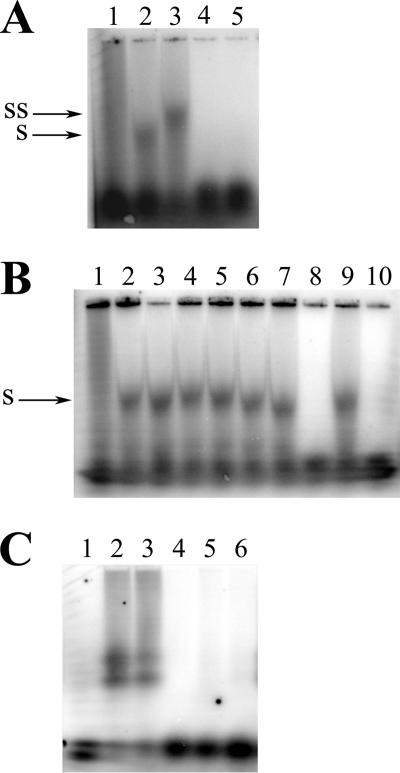
FIG. 2.
UL84 specifically recognizes an RNA stem-loop oligonucleotide in vitro. Autoradiographs of dried agarose gels show purified UL84 incubated with SL-RNA, a stem-loop oligonucleotide with an RNA backbone complementary to a sequence found within the essential RNA/DNA hybrid region of oriLyt. (A) Lanes: 1, SL-RNA alone; 2, 3 μl of purified UL84 plus SL-RNA; 3, 3 μl of purified UL84 plus SL-RNA incubated with Mab84; 4, 3 μl of purified UL84 plus SL-RNA plus 50× unlabeled SL-RNA; 5, 3 μl of purified UL84 plus SL-RNA plus 100× unlabeled SL-RNA. (B) Interaction of UL84 with SL-RNA is specific. Lanes: 1, SL-RNA alone; 2, 3 μl of purified UL84 plus SL-RNA; 3, 3 μl of purified UL84 plus SL-RNA incubated with 100× yeast tRNA; 4, 3 μl of purified UL84 incubated with SL-RNA incubated 100× poly-r(A); 5, 3 μl of purified UL84 plus SL-RNA incubated with 100× salmon sperm DNA; 6, 3 μl of purified UL84 plus SL-RNA incubated with 100× activated calf thymus DNA; 7, 3 μl of purified UL84 plus SL-RNA incubated with100× poly-r(U); 8, 3 μl of purified UL84 plus SL-RNA incubated with 100× SL-RNA PS; 9, 3 μl of purified UL84 plus SL-RNA incubated with 100× SL-DNA; 10, 3 μl of purified UL84 plus SL-RNA incubated with 100× SL-RNA. (C) The DNA version of the stem-loop oligonucleotide does not interact with UL84. Lanes: 1, SL-DNA alone; 2, 10 μl of purified UL84 plus SL-DNA; 3, 10 μl of purified UL84 plus SL-DNA-UL84 plus Mab84; 4, 10 μl of purified UL84 plus SL-DNA incubated with 100× unlabeled SL-DNA; 5, 10 μl of purified UL84 plus SL-DNA incubated with 100× salmon sperm DNA; 6, 10 μl of purified UL84 plus SL-DNA incubated with 100× activated calf thymus DNA. SS, supershifted band upon incubation with anti-UL84 antibody. S, shifted UL84-SL-RNA species.
We next investigated if the binding of UL84 with SL-RNA could be eliminated in a competition assay using a variety of nucleic acid substrates. No other nonspecific substrate, including unlabeled Saccharomyces cerevisiae tRNA, poly-r(A), poly-r(U), salmon sperm DNA, and activated calf thymus DNA, was able to competitively inhibit the specific UL84-SL-RNA interaction (Fig. 2B, lanes 2 to 7). Interestingly, even an oligonucleotide with the same sequence but composed of a DNA backbone did not compete with the specific UL84-SL-RNA interaction (Fig. 2B, lane 9). However, an SL-RNA oligonucleotide capped at the 3′ and 5′ ends with thioester linkages efficiently competed with UL84-SL-RNA binding (Fig. 2B, lane 10).
To further define the affinity of UL84 with RNA, we investigated the interaction of UL84 with a DNA stem-loop substrate. We performed an EMSA using an oligonucleotide with the same sequence as SL-RNA but having a DNA backbone instead of RNA. This oligonucleotide, SL-DNA, did bind weakly with the protein preparation containing UL84 but required almost threefold more purified protein in the binding reaction mixture. In addition, the interaction between UL84 and SL-DNA was eliminated in cold competition assays when either specific or nonspecific unlabeled substrates were added to the binding reaction mixture (Fig. 2C). Also, the apparently shifted band could not be supershifted by the addition of the UL84-specific antibody to the binding reaction mixture (Fig. 2C, lane 3). Based on these results, we conclude that the interaction with the DNA version of the oligonucleotide is nonspecific and that the band observed on the EMSA may be the result of a low-abundance contaminating protein due to the increased amount of protein preparation used. When the same binding conditions used for SL-RNA (i.e., 3 μl of protein) were used for SL-DNA, no band was detected in the EMSA (data not shown).
These experiments indicated that UL84 interacts with and favors RNA and may specifically bind to oligonucleotides forming a stem-loop whose primary sequence is found within the oriLyt region.
The secondary structure of SL-RNA is required for interaction with UL84.
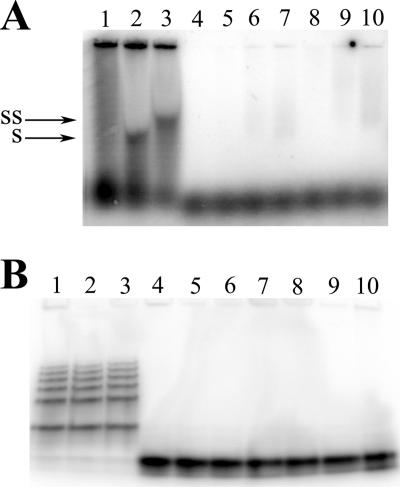
Once we determined that UL84 interacted with SL-RNA, we wanted to establish if the presence of secondary structure, primarily the stem-loop configuration, contributed to the binding. We eliminated the secondary structure of SL-RNA by heating the oligonucleotide to 95°C for 3 min followed by incubation at 4°C for 5 min before performing the binding reaction. This treatment completely reduced SL-RNA to a single-stranded configuration (Fig. 3B, compare lanes 3 and 4). The pattern of the native SL-RNA shows the presence of several “high-order” complexes. These complexes may be the result of intramolecular interactions. If UL84 has a binding preference for the stem-loop secondary structure, then this denatured oligonucleotide should not act as an efficient substrate in vitro. We then used this denatured SL-RNA (dSL-RNA) in the EMSA and compared the interaction of UL84 to that of the native stem-loop version, SL-RNA. No apparent binding was observed when using the dSL-RNA under the same conditions where a UL84-SL-RNA interaction was present (Fig. 3B, compare lane 2 to 4, 5, and 6). This result strongly suggests that a stem-loop substrate was required for efficient binding and recognition by UL84.
FIG. 3.
Binding of UL84 to SL-RNA is dependent on oligonucleotide secondary structure. (A) Autoradiograph of dried agarose gel showing heat denaturation of SL-RNA. Lanes: 1, no protein; 2, 3 μl UL84 plus nondenatured SL-RNA; 3, 3 μl UL84 plus nondenatured SL-RNA and UL84 MAb; 4, 1 μl UL84 plus denatured SL-RNA; 5, 5 μl UL84 plus denatured SL-RNA; 6, 10 μl UL84 plus denatured SL-RNA; 7, 1 μl UL84 plus denatured SL-RNA and UL84 MAb; 8, 5 μl UL84 plus denatured SL-RNA and UL84 MAb; 9, 10 μl UL84 plus denatured SL-RNA and UL84 MAb. (B) Autoradiograph of dried polyacrylamide gel showing the same samples deproteinized. SS, supershifted band upon incubation with anti-UL84 antibody; S, shifted UL84-SL-RNA species.
Increasing amounts of UL84 elicit a downward-staircase binding pattern with SL-RNA.
Initial EMSA experiments used one concentration of UL84 and demonstrated a specific interaction with SL-RNA. We next investigated the effects on complex mobility of using increasing concentrations of UL84 purified protein. EMSA experiments were performed as before except increasing concentrations of UL84 purified protein were incubated with SL-RNA. The UL84-SL-RNA interaction now resulted in an observed increased mobility within the gel resembling a downward staircase (Fig. 4A). Since one possible explanation of this downward-staircase observation could be a progressive degradation of the input oligonucleotide, we examined the integrity of the SL-RNA oligonucleotide after incubation with purified UL84 under the same conditions used for the EMSA. Three shifted bands (Fig. 4A, lanes 5, 6, and 7) corresponding to SL-RNA oligonucleotides from lanes with increasing concentrations of UL84 were excised from the EMSA agarose gel and resolved through a denaturing 10% PAGE urea gel. If input SL-RNA was degraded by UL84, we would then see the resulting degradation products in the denaturing gel. Figure 4B clearly shows that the length of the probe did not change (from the untreated control lane) and that no apparently less-than-full-length oligonucleotide was detected. This result suggested that SL-RNA remained intact in the presence of UL84.
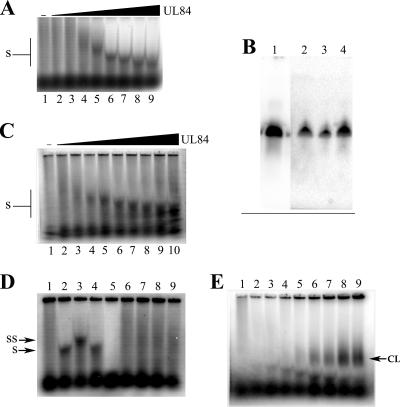
FIG. 4.
Increasing concentrations of purified UL84 produce a downward-staircase pattern of binding with SL-RNA. EMSA were performed as described in Materials and Methods except increasing amounts of purified protein were used in the binding reactions. (A) Autoradiograph of dried agarose gel showing EMSA using SL-RNA with increasing concentrations of purified UL84. Lanes: 1, UL84 SL-RNA alone; 2, SL-RNA plus 0.5 μl of UL84; 3, SL-RNA plus 1 μl of purified UL84; 4, SL-RNA plus 2 μl of purified UL84; 5, SL-RNA plus 3 μl of purified UL84; 6, SL-RNA plus 5 μl of purified UL84; 7, SL-RNA plus 7 μl of purified UL84; 8, SL-RNA plus 10 μl of purified UL84; 9, SL-RNA plus 13 μl of purified UL84. (B) The downward-staircase pattern of binding to SL-RNA is not due to a shortening or degradation of the oligonucleotide. SL-RNA protein bands corresponding to lanes 5, 6, and 7 from the EMSA shown in panel A were extracted from the gel, deproteinized, and resolved through a denaturing urea gel. Shown is the autoradiograph of the dried urea gel. Lanes: 1, SL-RNA alone; 2, SL-RNA extracted from lane 5 from panel A; 3, SL-RNA extracted from lane 6 from panel A; 4, SL-RNA extracted from lane 6 from panel A. (C) The phosphorothioate-capped SL-RNA displays the same downward-staircase pattern as the unmodified version. EMSA experiments were performed using the same conditions as described for panel A except that SL-RNA PS was used instead of the unmodified SL-RNA. (D) Control experiments demonstrating that the UL84-SL-RNA interaction is genuine. Lanes: 1, SL-RNA alone; 2, SL-RNA plus UL84; 3, SL-RNA plus UL84 plus Mab84; 4, SL-RNA plus UL84 plus the K8 MAb; 5, SL-RNA plus heat-treated UL84; 6, SL-RNA plus IE2 (1 μl); 7, SL-RNA plus IE2 (3 μl); 8, SL-RNA plus IE2 (5 μl); 9, SL-RNA plus IE2 (10 μl). (E) UV cross-linking gives rise to a slower-migrating species. Lanes are the same as in panel A except samples were exposed to 30 min of UV light in a Stratalinker 2400. SS, supershifted band upon incubation with anti-UL84 antibody; S, shifted UL84-SL-RNA species; CL, shift resulting from cross-linking.
As one more test for degradation of the SL-RNA, we used an oligonucleotide that contained phosphorothioate linkages at both the 5′ and 3′ termini (between the first and second and penultimate and last nucleotides). Phosphothioate modifications were added such that the oligonucleotide would be more resistant to any UL84-associated exonuclease activity. This oligonucleotide displayed the same downward-staircase pattern upon incubation with increasing concentrations of UL84 that was observed with SL-RNA (Fig. 4C). Hence, these data indicate that UL84 can change the mobility of an RNA stem-loop oligonucleotide, but this change in mobility is not due to a shortening or degradation of the input oligonucleotide.
To confirm that the UL84-SL-RNA interaction is genuine, we performed EMSA and supershift experiments using an unrelated antibody. Samples were incubated with SL-RNA as before and reacted with either a UL84-specific antibody or an antibody specific for human herpesvirus 8 K8 (K-bZIP). No supershifted band was observed when using the K-bZIP-specific antibody (Fig. 4D, lane 4). To show that binding was due to native UL84 protein, we performed a reaction with a protein sample that was heat denatured (95°C for 5 min). The protein sample was heat denatured and then cooled on ice for 10 min, followed by the addition of probe. No shifted band was detected using the denatured UL84 sample (Fig. 4D, lane 5). Lastly, we also used purified IE2 protein alone in a EMSA experiment. This was done to show that the bands generated by a UL84-SL-RNA mixture are authentic and not due to some contaminant within our purified protein preparation. No banding pattern was observed when increasing concentrations of IE2 purified protein were incubated with SL-RNA (Fig. 4D, lanes 6 to 10). These experiments show that UL84 interacts with SL-RNA in a highly specific manner and that the downward-staircase pattern is due to increasing concentrations of UL84 and not a protein contaminant. These results also indicate that the banding pattern observed is genuine and due to UL84 and that denatured protein no longer interacts with SL-RNA.
We also performed the EMSA under conditions where UL84 protein was cross-linked to SL-RNA using UV light. Cross-linking of samples containing UL84-SL-RNA resulted in the presence of a slower-migrating band in the gel, which intensified upon incubation with increasing concentration of UL84 (Fig. 4E). This suggests that UL84 resolves or reduces some higher-order structure and that cross-linking the samples inhibited the ability of UL84 to act on the RNA substrate.
The SL-RNA-UL84 complex is not affected by the addition of IE2.
It is known that UL84 binds to IE2 in infected cells, and this interaction apparently is responsible for both a decrease in IE2-mediated transactivation of UL112-113 promoter and autoregulation of IE2 itself. This suggests that UL84 diminishes the affinity for IE2 for its DNA substrate, whether it is the UL112-113 promoter or the CRS element. Additionally, IE2 and UL84 are required for the efficient transactivation of the promoter region within oriLyt, which is required for DNA synthesis. We were interested in whether the same would be true for UL84 and the interaction with its substrate. We investigated the effects of increasing amounts of purified IE2 on the ability of UL84 to interact with SL-RNA in vitro. EMSA experiments were performed as before with the addition of increasing amounts of purified IE2 or increasing amounts of UL84 in the presence of a stable concentration of IE2 within the binding reaction mixture. The addition of IE2 to the binding reaction mixture did not change the mobility or intensity of the UL84 and SL-RNA complexes in either scenario (Fig. 5A, lanes 2 to 5 and 7 to 10, respectively). In order to confirm UL84-IE2 complex formation within the binding reaction mixture, we successfully coimmunoprecipitated the complex using the UL84-specific monoclonal antibody (MAb) (Fig. 5B). This result indicated that IE2 did not enhance or interfere with the interaction of UL84 with this nucleic acid substrate and is consistent with the replication data showing that UL84 alone is required for efficient replication of oriLyt with a substituted SV40 promoter.
FIG. 5.
Interaction of UL84 with SL-RNA is not affected by the presence of IE2. (A) Binding reactions were carried out as for the experiments described in Fig. 4 except variable amounts of IE2 were added to the reaction mixture. Shown is the autoradiograph of the dried agarose gel. Lanes: 1, no protein; 2, 3 μl UL84 plus SL-RNA; 3, 3 μl UL84 plus SL-RNA and 1 μl IE2; 4, 3 μl UL84 plus SL-RNA and 5 μl IE2; 5, 3 μl UL84 plus SL-RNA and 10 μl IE2; 6, no protein; 7, 1 μl UL84 plus SL-RNA and 3 μl IE2; 8, 3 μl UL84 plus SL-RNA and 3 μl IE2; 9, 5 μl UL84 plus SL-RNA and 3 μl IE2; 10, 10 μl UL84 plus SL-RNA and 3 μl IE2. (B) Western blot of coimmunoprecipitation of IE2 and UL84 from binding reaction with UL84 MAb. Lanes: 1, UL84 immunoprecipitated with UL84 MAb and detected with UL84 MAb with no SL-RNA in binding reaction; 2, UL84 immunoprecipitated with UL84 MAb and detected with UL84 MAb; 3, IE2 immunoprecipitated with UL84 MAb and detected with IE2 MAb with no SL-RNA in binding reaction; 4, IE2 immunoprecipitated with UL84 MAb and detected with IE2 MAb. S, shifted UL84-SL-RNA species.
UL84 interacts with HCMV oriLyt in vivo.
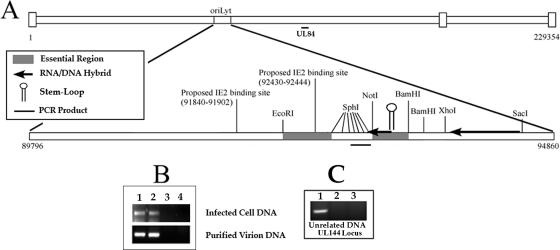
The results of the in vitro assay showed that UL84 interacted in a sequence- and structure-specific manner with one of the proposed stem-loop structures found within oriLyt. In addition, UL84 appeared to have a distinct preference for RNA substrates over DNA counterparts with the same sequence. Based on this observation, we decided to investigate the interaction of UL84 within the context of the cellular and virion environments using the ChIP assay. We designed PCR primers that flanked the oriLyt region known to contain an RNA/DNA hybrid structure (32). These primers and the resulting PCR product were immediately adjacent to and upstream from the proposed stem-loop structure used in the in vitro binding experiments (Fig. 6A). Primer sets just outside of the RNA/DNA hybrid were chosen because PCR amplification of the region flanking the stem-loop consistently resulted in a very weak PCR amplification product. This was probably due to the presence of the RNA/DNA hybrid and the variable length of this region (data not shown).
FIG. 6.
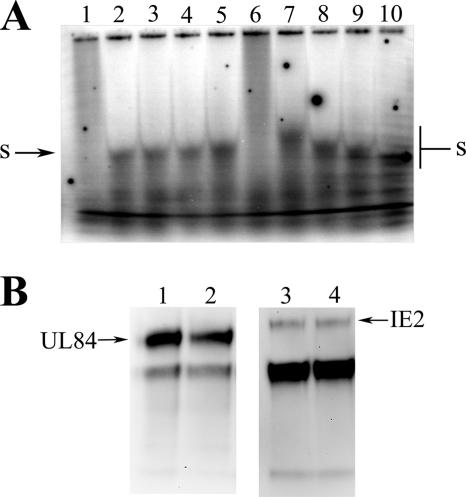
UL84 binds to the HCMV oriLyt within infected cells and purified virions. (A) Schematic of the HCMV origin of DNA replication showing the location within the HCMV genome as well as other distinguishing features including the relative locations of the regions essential for DNA synthesis, the RNA/DNA hybrid region, the SL-RNA stem-loop structure, the region amplified by PCR in the ChIP assay, and the proposed IE2 binding sites. (B) ChIP assay of AD169-infected cells and AD169 purified virion with the UL84-specific MAb (described in Materials and Methods). (Top panel) Lanes: 1, input control sample from infected-cell lysate amplified with oriLyt F and R primers; 2, ChIP sample from infected cell lysate amplified with oriLyt F and R primers; 3, no-antibody control ChIP sample from infected-cell lysate amplified with oriLyt F and R primers. (Bottom panel) Lanes: 1, input control sample from purified virion lysate amplified with oriLyt F and R primers; 2, ChIP sample from purified virion lysate amplified with oriLyt F and R primers; 3, ChIP sample from infected-cell lysate using the isotype control antibody amplified with oriLyt F and R primers; 4, no-antibody control ChIP sample from purified virion lysate amplified with oriLyt F and R primers. (C) ChIP assay of AD169-infected cells with the UL84-specific MAb amplified by primers corresponding to the UL144 locus. Lanes: 1, input control sample from infected-cell lysate amplified with UL144 primers; 2, ChIP sample from infected-cell lysate amplified with UL144 primers; 3, no-antibody control ChIP sample from infected-cell lysate amplified with UL144 primers.
HCMV-infected cells or sucrose gradient-purified virion DNA was used as the starting material for the ChIP assay. Protein-DNA complexes were immunoprecipitated using a UL84 MAb (Virusys; CA144) or no antibody in the case of a control reaction. We also performed two additional controls. The first used an isotype-matched unrelated antibody. The second was a control for the ChIP assay itself, where we amplified a region of the genome outside HCMV oriLyt, the UL144 open reading frame.
Figure 6B shows the result of the ChIP assay showing that positive PCR products were in both the input control lane and the sample immunoprecipitated with the UL84-specific antibody from infected-cell DNA harvested 3 days postinfection (Fig. 6B, top panel, lanes 1 and 2, respectively). No PCR amplification product was observed in the no-antibody control sample (Fig. 6B, lane 4) or the isotype control antibody lane (Fig. 6B, lane 3).
We also investigated the possibility that UL84 was bound to viral DNA within the virion. This seemed plausible since a recently published report using highly sensitive mass spectrometric analysis identified UL84 as a component of the virion (41). Consequently we applied the ChIP assay to DNA isolated from purified virions. Virions were isolated from infected cells and subsequently treated with micrococcal nuclease to ensure that all DNA analyzed in the ChIP assay was from packaged virions. Interestingly, a positive PCR amplification product was observed from ChIP assays performed on DNA isolated from virions (Fig. 6B, lane 2). In order to ensure the accuracy of our results, we used an isotype-matched control antibody in the ChIP assay as well as PCR amplification of an unrelated region of the HCMV genome after immunoprecipitation with the UL84-specific antibody. In either case, no false-positive PCR signal was observed (Fig. 6B, lane 3, and C).
Data from the ChIP assay indicate that UL84 interacts with oriLyt probably within the region of oriLyt that contains RNA/DNA hybrid and stem-loop structures. Also UL84, which was previously shown to be a component of the virion, interacts with oriLyt DNA within the virion. These results, coupled with the data from the in vitro binding assays, strongly suggest that a possible mechanism of initiation of HCMV DNA replication is via a UL84 direct interaction with specific structures within oriLyt.
DISCUSSION
The mechanism for initiation of HCMV lytic DNA synthesis is thought to be unique among the herpesvirus family since no homolog to an OBP was identified from initial studies using the cotransfection replication assay (30). Several reports identified UL84 as the only protein still required after elimination of viral transactivators and ancillary proteins such as IRS1, UL112-113, and UL36-38 (34, 48). Additionally, the immediate early protein IE2, which apparently also has a transactivator function in lytic DNA replication, works in concert with UL84 to positively activate a promoter within oriLyt (47).
Most initiation proteins bind to a specific sequence of DNA within the origin of replication; however, in some cases the initiation protein requires a binding partner to complete this function (1, 39). This is true in the case of papillomavirus E1, where sequence specificity is conferred only in the presence of E2. Although we did observe specific binding of UL84 with a nucleic acid substrate, this interaction or another as yet undiscovered function could be enhanced with the addition of a cellular protein or virally encoded factor. For example, the single-stranded DNA binding protein greatly increases the enzymatic activity of the HSV-1 UL9 OBP (5, 10). Our studies suggest that IE2 does not contribute to the binding of UL84 to the SL-RNA. The interaction of IE2 with UL84 may play another role in the regulation of initiation of DNA replication or the viral life cycle.
The recent finding that UL84 has an intrinsic UTPase activity suggested that this protein could fulfill the characteristics needed to function as a true OBP (8). Many initiation factors possess a helicase activity that is utilized to initially separate the strands of DNA using the energy from the hydrolysis of nucleotide triphosphates. While it was clear that UL84 does not have considerable homology to any other protein in nature and cannot be immediately classified as a helicase, the protein does have limited homology to the DExD/H family of proteins (8). This comparison is based not merely on structural homology but also on functional similarities to this group of enzymes. The fact that UL84 fills a role as a suppresser and an activator is more evidence that the protein fits in the DExD/H family of proteins (43). This diverse family includes proteins with a large array of functions, although they are usually involved with some type of nucleic acid disassociation activity. The group of enzymes can function to separate nucleic acid from RNA, DNA, or protein (reviewed in references 15 and 22). The recent evidence that UL84 is a UTPase, contains several conserved DExD/H sequences, and is both a negative and positive transcriptional effector, paired with the new data that UL84 can bind to RNA, gives further evidence for inclusion within this family. Additionally, many DExD/H proteins shuttle RNA from the nucleus to the cytoplasm, a characteristic recently shown to be shared by UL84 (3, 23, 49). This shuttling could serve to regulate DNA replication in a manner similar to that of HSV-1 ICP27, where UL84 may enhance the accumulation of viral transcripts encoding replication proteins in the cytoplasm (33, 37).
A recent report suggested that UL84 is a dUTPase-related protein based upon computer-generated analysis (9). While it is possible that UL84 does contain a six-stranded beta-barrel structural component at the C terminus, until a crystal structure of the protein is analyzed, this analysis cannot be confirmed. Furthermore this structural fold is found in many classes of enzymes and by no means strictly defines the protein as a dUTPase (14, 28). Our initial structural inquiries indicate that the beta-barrel fold in UL84 more closely resembles that in a tRNA synthetase-like enzyme and may represent an RNA stem-loop binding site (unpublished data). We feel there is overwhelming biochemical and functional evidence that UL84 functions as a DExD/H family member independent of the novel protein structure observed.
As a prerequisite to determining any enzymatic role for UL84, such as helicase or helix destabilization, it was essential to investigate potential nucleic acid substrates or sequences. The most obvious region to search for such a potential substrate within oriLyt was the locus between nt 92887 and 93513. This region was chosen based on several characteristics: (i) the region is part of the ERII of oriLyt, (ii) the region has sequences that form a stem-loop structure that is unique to the HCMV genome, and (iii) the region is part of the RNA/DNA hybrid of oriLyt. In addition, this area of the genome is amplified in laboratory-adapted HCMV strains such that the region containing the stem-loop can be reiterated such that 300 extra nucleotides are present (32). This inverted-repeat structure is also conserved among other CMV oriLyt sequences (26).
The evidence that the RNA stem-loop structure is one of the conformations recognized by UL84 in vitro is based on several lines of data. Although other substrates interacted with UL84 in the EMSA, the amount of protein necessary to shift SL-RNA was almost threefold lower than that required for any other substrate tested. The UL84-SL-RNA interaction was also the only interaction that was not eliminated by nonspecific unlabeled RNA or DNA, whereas UL84 binding to the DNA form of stem-loop oligonucleotide was competed by all nonspecific-DNA-unlabeled substrates tested and could not be supershifted with the UL84-specific antibody. Finally, a supershift of the protein-RNA complex was observed upon incubation with the UL84-specific antibody.
One characteristic that distinguishes the SL-RNA substrate from any other nucleic acid substrate is the unexpected result observed when increasing amounts of UL84 were added to the SL-RNA probe. As more protein is added to the reaction mixture, the complex formed between UL84 and the probe shifts progressively smaller, as judged by determining the complex mobility. This phenomenon was not observed with any DNA substrate that was tested. This downward-staircase observation may indicate that UL84 is able to modify this structure. Since there was no shortening or degradation of this substrate observed, we assume that UL84 may be unwinding the stem-loop or resolving some higher-order structures forming with this oligonucleotide. The binding of UL84 to SL-RNA is absolutely dependent upon the presence of the stem-loop since heat denaturation led to undetectable binding in the in vitro assay.
Several control experiments also show that the observed banding pattern is genuine and due to UL84 binding to SL-RNA: (i) there was no supershifted band when using a nonspecific antibody (anti-human herpesvirus 8 K-bZIP); (ii) no banding pattern was observed when SL-RNA was reacted with heat denatured UL84; and (iii) another purified protein, IE2, alone did not interact with SL-RNA, even when increasing concentrations were used in the reaction mixture. Also, cross-linking experiments show that UL84 formed an additional complex that did not display the typical staircase pattern. This suggests that UL84 changes the conformation of the SL-RNA substrate.
The subtle differences between the affinity of UL84 for DNA and that for RNA or specific sequences could be measured using a more quantitative method. Our current methods limit this measurement because our purified protein is contaminated with 3× FLAG peptide, which was used to efficiently elute the protein from the affinity column. While this poses no problems in the binding assay, it prohibits our quantitation of purified protein to the level necessary to measure an accurate dissociation constant for UL84 binding to RNA and DNA substrates. The only large difference in affinity and specificity was witnessed with the SL-RNA substrate.
With the results from the in vitro assay in hand, we then investigated if UL84 could interact with this substrate in the cellular environment, as well as within packaged virions. Since recent evidence indicated that UL84 was a component of the virion, we performed the ChIP assay using infected cells or purified virus samples (41). The ChIP assay demonstrated that UL84 does interact with the region of oriLyt that contains the stem-loop and RNA/DNA hybrid structures. Because of the nature of the ChIP assay, where the genome is fragmented into approximately 500- to 1,000-bp pieces, coupled with the inherent redundancy of the stem-loop region, PCR amplification was efficiently achieved only using primers complementary to regions adjacent to the RNA/DNA domain. Nevertheless, this is the first evidence that UL84 interacts with the HCMV genome within the oriLyt region in the in vivo environment.
The nucleic acid structure in the stem-loop region may require an initial transcription event prior to the onset of DNA synthesis, and one strand of the RNA/DNA hybrid region is able to form stable RNA stem-loop structures. These structures also remain in the packaged virion, as demonstrated previously, and can then be available for specific binding of UL84 (32). The fact that UL84 was associated with the origin not only in infected cells at 72 h postinfection but also in purified virions is remarkable and further implicates UL84 as a key initiation factor. The binding of UL84 to this region could then allow for efficient initiation of lytic DNA synthesis. It could also be that the binding of UL84 to this region would serve to regulate the onset of DNA synthesis by inhibiting the interaction of factors necessary for DNA replication. Both possibilities will be investigated further.
Acknowledgments
This work was supported by NIH Public Health Service grant AI45096.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Abbate, E. A., J. M. Berger, and M. R. Botchan. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders, D. G., M. A. Kacica, G. Pari, and S. M. Punturieri. 1992. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J. Virol. 66:3373-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askjaer, P., R. Rosendahl, and J. Kjems. 2000. Nuclear export of the DEAD box An3 protein by CRM1 is coupled to An3 helicase activity. J. Biol. Chem. 275:11561-11568. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer, P. E. 2004. RNA binding and R-loop formation by the herpes simplex virus type-1 single-stranded DNA-binding protein (ICP8). Nucleic Acids Res. 32:4576-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehmer, P. E., and I. R. Lehman. 1993. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl. Acad. Sci. USA 90:8444-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brister, J. R., and N. Muzyczka. 2000. Mechanism of Rep-mediated adeno-associated virus origin nicking. J. Virol. 74:7762-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 78:9203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280:11955-11960. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., and N. D. Stow. 2005. New genes from old: redeployment of dUTPase by herpesviruses. J. Virol. 79:12880-12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson, M. S., and I. R. Lehman. 1993. The herpes simplex virus type I origin binding protein. DNA-dependent nucleoside triphosphatase activity. J. Biol. Chem. 268:1213-1219. [PubMed] [Google Scholar]
- 11.Fierer, D. S., and M. D. Challberg. 1992. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J. Virol. 66:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freist, W., J. F. Verhey, A. Ruhlmann, D. H. Gauss, and J. G. Arnez. 1999. Histidyl-tRNA synthetase. Biol Chem. 380:623-646. [DOI] [PubMed] [Google Scholar]
- 15.Fuller-Pace, F. V. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 66:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder, P. 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 34:4168-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lischka, P., C. Rauh, R. Mueller, and T. Stamminger. 2006. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J. Virol. 80:10274-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macao, B., M. Olsson, and P. Elias. 2004. Functional properties of the herpes simplex virus type I origin-binding protein are controlled by precise interactions with the activated form of the origin of DNA replication. J. Biol. Chem. 279:29211-29217. [DOI] [PubMed] [Google Scholar]
- 25.Masse, M. J., S. Karlin, G. A. Schachtel, and E. S. Mocarski. 1992. Human cytomegalovirus origin of DNA replication (oriLyt) resides within a highly complex repetitive region. Proc. Natl. Acad. Sci. USA 89:5246-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masse, M. J., M. Messerle, and E. S. Mocarski. 1997. The location and sequence composition of the murine cytomegalovirus replicator (oriLyt). Virology 230:350-360. [DOI] [PubMed] [Google Scholar]
- 27.McGeoch, D. J., M. A. Dalrymple, A. Dolan, D. McNab, L. J. Perry, P. Taylor, and M. D. Challberg. 1988. Structures of herpes simplex virus type 1 genes required for replication of virus DNA. J. Virol. 62:444-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. James. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 29.Myers, R. M., D. C. Rio, A. K. Robbins, and R. Tjian. 1981. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell 25:373-384. [DOI] [PubMed] [Google Scholar]
- 30.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prichard, M. N., S. Jairath, M. E. Penfold, S. St Jeor, M. C. Bohlman, and G. S. Pari. 1998. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 72:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmons, D. T., D. Gai, R. Parsons, A. Debes, and R. Roy. 2004. Assembly of the replication initiation complex on SV40 origin DNA. Nucleic Acids Res. 32:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, J. A., and G. S. Pari. 1995. Human cytomegalovirus UL102 gene. J. Virol. 69:1734-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Titolo, S., K. Brault, J. Majewski, P. W. White, and J. Archambault. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weir, H. M., J. M. Calder, and N. D. Stow. 1989. Binding of the herpes simplex virus type 1 UL9 gene product to an origin of viral DNA replication. Nucleic Acids Res. 17:1409-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, B. J., G. J. Bates, S. M. Nicol, D. J. Gregory, N. D. Perkins, and F. V. Fuller-Pace. 2004. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol. Biol. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, V. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275-290. [DOI] [PubMed] [Google Scholar]
- 45.Wu, F. Y., S. E. Wang, Q. Q. Tang, M. Fujimuro, C. J. Chiou, Q. Zheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein α and p21CIP-1. J. Virol. 77:8893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Y., S. A. Cei, A. R. Huete, and G. S. Pari. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J. Virol. 78:10360-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, Y., S. A. Cei, A. Rodriguez Huete, K. S. Colletti, and G. S. Pari. 2004. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 78:11664-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Y., K. S. Colletti, and G. S. Pari. 2002. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yedavalli, V. S., C. Neuveut, Y. H. Chi, L. Kleiman, and K. T. Jeang. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381-392. [DOI] [PubMed] [Google Scholar]