Abstract
Mutations in the thymidine kinase gene (tk) of herpes simplex virus type 1 (HSV-1) explain most cases of virus resistance to acyclovir (ACV) treatment. Mucocutaneous lesions of patients with ACV resistance contain mixed populations of tk mutant and wild-type virus. However, it is unknown whether human ganglia also contain mixed populations since the replication of HSV tk mutants in animal neurons is impaired. Here we report the detection of mutated HSV tk sequences in human ganglia. Trigeminal and dorsal root ganglia were obtained at autopsy from an immunocompromised woman with chronic mucocutaneous infection with ACV-resistant HSV-1. The HSV-1 tk open reading frames from ganglia were amplified by PCR, cloned, and sequenced. tk mutations were detected in a seven-G homopolymer region in 11 of 12 ganglia tested, with clonal frequencies ranging from 4.2 to 76% HSV-1 tk mutants per ganglion. In 8 of 11 ganglia, the mutations were heterogeneous, varying from a deletion of one G to an insertion of one to three G residues, with the two-G insertion being the most common. Each ganglion had its own pattern of mutant populations. When individual neurons from one ganglion were analyzed by laser capture microdissection and PCR, 6 of 14 HSV-1-positive neurons were coinfected with HSV tk mutants and wild-type virus, 4 of 14 were infected with wild-type virus alone, and 4 of 14 were infected with tk mutant virus alone. These data suggest that diverse tk mutants arise independently under drug selection and establish latency in human sensory ganglia alone or together with wild-type virus.
The deoxyguanosine homolog acyclovir (ACV) is the most common drug used to treat herpes simplex virus (HSV) infections. After being taken up into cells, ACV is sequentially converted into ACV monophosphate, ACV diphosphate, and finally its active form, ACV triphosphate. The first step of the sequential phosphorylations requires HSV-encoded thymidine kinase (TK); however, cellular enzymes perform the additional phosphorylations. ACV triphosphate is more efficiently incorporated into replicating DNA by HSV DNA polymerase than by the cellular DNA polymerase (8). These characteristics of ACV result in its selectivity for virus-infected cells and its extremely low toxicity to uninfected cells. However, if HSV loses its TK function (including an alteration in substrate specificity or the loss of TK activity) or its DNA polymerase has altered substrate affinity, the virus becomes ACV resistant (Acvr). While viral TK function is crucial for ACV activity, TK is not essential for HSV to replicate in dividing cells (22) such as human epithelial cells, presumably due to the abundance of nucleotides in these cells. Thus, a TK− HSV mutant that is resistant to ACV therapy can still replicate in epithelial cells and cause lesions. In contrast, TK activity is important for virus replication in resting cells or neurons (22, 38). Though Acvr HSV infection rarely has clinical significance in immunocompetent individuals (7, 15, 24, 27, 37), severe disease can occur in immunocompromised persons such as AIDS patients and bone marrow transplant recipients (29, 40).
The majority (>95%) of Acvr clinical isolates from human skin lesions have mutations in the viral tk gene, while the remainder have mutations in the viral DNA polymerase gene (2, 9, 29). A vast array of mutations in the HSV tk gene affect TK activity. Among reported clinical isolates, about 50% of the tk mutants have insertion or deletion mutations in one of two homopolymer regions, either a seven-G string at nucleotides (nt) 430 to 436 or a six-C cord at nt 548 to 553 of the tk open reading frame (ORF) (6, 9, 16, 17, 26, 30, 33, 36). HSV type 1 (HSV-1) TK is encoded by the UL23 gene, and the polypeptide is 376 amino acids in length. The ATP and nucleoside binding domains are essential for TK activity. The main nucleoside binding domain is located from amino acids 161 to 192 (corresponding to nt 483 to 576 of the tk ORF) (1, 3). The insertion and deletion mutations in the homopolymer regions result in frameshifts before or within the nucleoside binding domain and thus drastically reduce TK activity.
HSV-1 tk mutants, including clinical isolates and recombinants engineered in the laboratory, have been studied in animal models. In the mouse model, the replication of tk mutants at the inoculation site is comparable to that of the wild-type (WT) virus (38), but replication terminates earlier than that of the WT virus (16). HSV tk mutants induce cutaneous lesions comparable to those caused by WT virus (17). HSV TK− mutants with losses of functional domains or replacements of critical amino acids that cannot be restored spontaneously show impaired replication and establishment of latency in mouse ganglia and are unable to reactivate (5, 11, 12, 16, 20, 21, 23, 38, 39). In contrast, some of the HSV tk mutants expressing low levels of the TK enzyme have various levels of ability to replicate and reactivate from latency in mouse ganglia (12, 13, 16).
While HSV Acvr tk mutants have been detected in mouse ganglia, it is unknown if similar mutants infect and establish latency in human ganglia. Here we report the presence of HSV-1 tk mutants in multiple ganglia of an immunocompromised patient (patient 708) with a history of chronic Acvr skin lesions, and we discuss the implications of these findings.
MATERIALS AND METHODS
HSV-1 antiviral sensitivity testing.
Swabs collected from the patient's mucocutaneous lesions were used to infect cells which were cultured in various concentrations of ACV or foscarnet, and 2 days later, plaques were counted. The dose of ACV or foscarnet required to inhibit plaque formation by 50%, the 50% inhibitory concentration (IC50), was calculated by comparing the plaque numbers in culture wells with and without antiviral drugs. Resistance to ACV or foscarnet was defined as an IC50 of ≥2 μg/ml or ≥150 μg/ml, respectively.
Ganglion tissue.
Trigeminal ganglia (TG) and thoracic to sacral (T2 to S5) dorsal root ganglia (DRG) were obtained at autopsy. DNA was isolated immediately from the left TG (LTG) and left thoracic (LT3, LT6, LT8, and LT11), left lumbar (LL2 and LL5), and left sacral (LS1 to LS5) ganglia. The remainder of the left ganglia and the right sacral 1 (RS1) ganglion were frozen on dry ice and stored at −80°C. Frozen LT2, LT4, LT5, LT7, LT9, LT10, LT12, LL1, LL3, LL4, and RS1 ganglia were divided into four portions, one each for DNA extraction, explant cocultivation, the preparation of homogenate for the detection of infectious virus, and the preparation of paraffin-embedded tissue sections.
Total ganglionic DNA isolation.
Fresh or frozen ganglion tissue was minced and digested with proteinase K (600 μg/ml) in lysis buffer (PureGene DNA isolation kit; Gentra Systems, Minneapolis, MN) at 100 mg of tissue per 2 ml of buffer and incubated at 55°C overnight. DNA was isolated with the PureGene DNA isolation kit by following the manufacturer's instructions.
LCM of single neurons and DNA extraction from individual cells.
Laser capture microdissection (LCM) was carried out as described previously (41). Briefly, tissue sections were dewaxed in xylenes and air dried. Single neurons were randomly picked up using PixCell II microdissection equipment and CapSure Macro LCM 0211 caps (Arcturus Engineering, Inc., Mountain View, CA). Only one neuron was captured onto each cap, and this ratio was confirmed by observing the caps under a microscope. DNA was extracted from individual neurons by overlaying the cells with 14 μl of DNA extraction buffer (0.04% proteinase K, 1% Tween 20, 10 mM Tris-HCl, and 1 mM EDTA, pH 8.0) at 37°C overnight, heating to 95°C for 8 min to inactivate the proteinase K, and cooling to room temperature.
PCR amplification of the HSV-1 tk coding sequence from ganglionic DNA.
To determine the sequences of individual HSV-1 tk genes in patient 708 ganglia, viral tk sequences encompassing the entire ORF (Fig. 1) were amplified from total ganglionic DNA with forward primer tkF1b and reverse primer tkR1, or tkF51 and tkR61 (Table 1), by using the Expand High Fidelity PCR system (Roche, Penzberg, Germany). The final concentrations of reagents in a 50-μl PCR mixture were as follows: 1× Expand High Fidelity PCR buffer, 200 μM deoxynucleoside triphosphate, 300 nM (each) primers, 0.375 μl of the Expand High Fidelity enzyme mix, and 20 to 200 ng of ganglionic DNA. The reaction mixtures were subjected to one cycle of 97°C, 55°C, and 72°C (3 min each) in a DNA thermal cycler (PerkinElmer, Norwalk, CT). This step was then followed by 35 cycles of 97°C for 30 s, 55°C for 1 min, and 72°C for 3 min. Finally, the reaction mixtures were incubated at 72°C for 7 min. Part of each PCR product was resolved on agarose gels to determine the quality of the PCR products. To determine the HSV-1 tk sequence in the seven-G homopolymer region (Fig. 1) in single neurons, 12 μl of DNA extract from each individual neuron was added to PCR mixtures in a final volume of 25 μl with forward primer tkF82 and reverse primer tkR21 (300 nM [each]); other components of the PCR mixture were identical to those listed above.
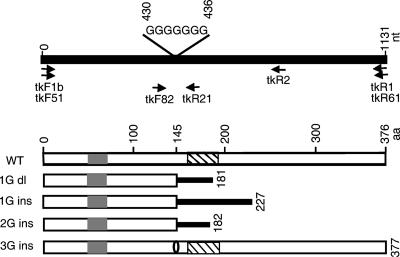
FIG. 1.
Schematic representation of HSV-1 tk gene and mutations in the seven-G homopolymer region. The top panel represents the tk gene, with the locations of the seven-G homopolymer region and the PCR primers shown. The bottom panel represents the wild-type TK protein and the mutated polypeptides predicted to correspond to mutations in the seven-G region. Gray boxes, ATP binding domains; striped boxes, nucleoside binding domains; bold lines, amino acid sequences resulting from frameshifts; aa, amino acids; dl, deletion; ins, insertion. The oval on the bottom polypeptide diagram indicates an insertion of one amino acid.
TABLE 1.
Oligonucleotides used for PCR and sequencing
| Name | Sequence (5′ to 3′) |
|---|---|
| tkF1b | CGCCAAGCTTAAGCCACCATGGCTTCGTACCCCG |
| tkR1 | CCGTTCTAGAGTTAGCCTCCCCCATCT |
| tkF51 | GAAACTCCCGCACCTCTTCGG |
| tkR61 | GGTTCCTTCCGGTATTGTCTCC |
| tkF82 | AAGCGCCCAGATAACAATGG |
| tkR21 | AACACAGGAGGGCGGCGATG |
| tkF5 | CAAGAAGCCACGGAAGTC |
| tkR2 | GCTGTCCCCAATCCTCCCG |
| M13 Forward (−20) | GTAAAACGACGGCCAG |
| M13 Reverse | CAGGAAACAGCTATGAC |
| pc3.1F | AGAGAACCCACTGCTTACTGGC |
| pc3.1R | CTAGAAGGCACAGTCGAGGCTG |
Subcloning of HSV-1 tk coding sequence amplified by PCR.
PCR products amplified with primers tkF1b and tkR1 were digested with restriction enzymes HindIII and XbaI and inserted into the corresponding sites of plasmid pcDNA3.1-V5-His A (Invitrogen, Carlsbad, CA). Escherichia coli DH5α (Invitrogen) was then transformed with the ligation mixtures. The PCR products amplified with primers tkF51 and tkR61 or tkF82 and tkR21 were inserted into TA cloning vector pCRII by following the instructions of the manufacturer (Invitrogen) and then used to transform E. coli TOP10 cells (Invitrogen). Bacteria were grown on agar plates containing 75 μg of ampicillin/ml at 37°C overnight and then stored at 4°C, and most clones were used in 3 days. To produce plasmid DNA for sequencing, single colonies on the agar plates were carefully picked up and cultured in Luria-Bertani medium containing 75 μg of ampicillin/ml overnight and DNA was isolated using a QIAprep spin miniprep kit (QIAGEN, Valencia, CA).
Sequencing and data analysis.
Plasmid DNA clones with full-length HSV-1 tk ORF inserts were sequenced with tk-specific primers tkF5, tkR1, and tkR2 and vector-specific primers M13 Forward (−20), M13 Reverse, pc3.1F, and pc3.1R. Clones containing the short fragment (about 147 bp) encompassing the seven-G homopolymer region of HSV-1 tk from single neurons were sequenced with M13 primers only. Sequencing data were analyzed with Sequencher 4.5 software (Gene Codes Corporation, Ann Arbor, MI) and compared with the HSV-1 strain 17+ sequence in GenBank (accession number X14112). HSV-1 tk nucleotide polymorphisms were determined by comparing the sequences to the data reported by Kudo et al. to distinguish polymorphisms from changes identified as mutations (25). Statistical analyses of the frequency of nucleotide alterations outside the seven-G region and of insertions and deletions in the seven-G region in PCR subclones were performed using the chi-square test and Fisher's exact method, respectively.
RESULTS
An extremely high level of HSV-1 DNA was detected in ganglia from an immunocompromised patient.
An 18-year-old woman (patient 708) presented at the Clinical Center, National Institutes of Health, Bethesda, MD, with an undefined disorder of cellular immunity under a protocol approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases. She had had multiple episodes of HSV infection, beginning at 17 months of age, which included eczema herpeticum and herpetic keratitis, and subsequently had recurrent HSV-1 skin infections over diffuse areas of her body (Fig. 2). She also had a history of disseminated molluscum contagiosum and recurrent Staphylococcus and Pseudomonas pneumonias. Upon presentation at the Clinical Center, she had ulcerative lesions on her buttocks, perianal region, and distal calves and a blister on her upper lip. Molluscum contagiosum lesions and dry, scaly erythema were present over large areas of her body. She was treated with multiple courses of antiviral therapy, including ACV and foscarnet, but her skin lesions never resolved completely. While several HSV-1 isolates from skin and mucocutaneous lesions were sensitive to ACV, others were resistant (Table 2). She died of cutaneous T-cell lymphoma at age 18.
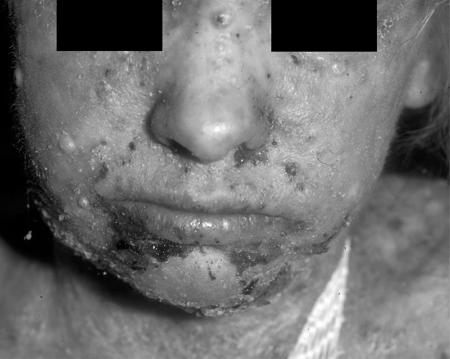
FIG. 2.
Diffuse skin lesions on patient 708. This photograph of patient 708 was taken when the patient was 13 years old and shows diffuse skin lesions on the face and neck. A swab sample collected from the face on the day of the photograph grew HSV-1 that was sensitive to ACV (IC50 = 0.22 μg/ml).
TABLE 2.
Results of drug sensitivity testing of HSV-1 clinical isolates from patient 708
| Patient agea (yrs) | Site of swab | IC50 (μg/ml)b of:
|
Last recorded therapy | |
|---|---|---|---|---|
| ACV | Foscarnet | |||
| 18.84 | Skin | 13 | 22 | ACV and foscarnet |
| 18.79 | Left inguinal area | 0.5 | 14 | Foscarnet |
| 14.39 | Labium | 3.2 | 19 | ACV |
| 14.39 | Abdomen | 1.9 | 16 | ACV |
| 13.67 | Chin | 0.1 | 27 | Foscarnet |
| 13.62 | Face | 0.2 | 48 | Foscarnet |
| 13.10 | Vagina | 3.9 | NR | ACV |
| 13.10 | Vagina | 1.8 | NR | ACV |
Age at which the corresponding swab sample was taken.
Resistance is defined as an IC50 of ACV of ≥2 μg/ml and an IC50 of foscarnet of ≥150 μg/ml. NR, not recorded.
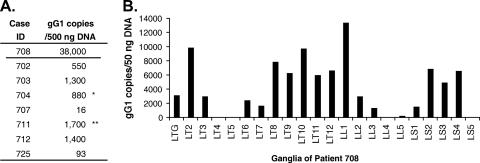
To estimate the levels of HSV-1 DNA in the TG and DRG of patient 708, HSV-1 DNA among the total DNA isolated from ganglia was quantified by real-time quantitative PCR with primers and a probe specific for the HSV-1 gG gene (41). DNA from TG of patient 708 was examined in parallel with TG DNA from seven other HSV-1-seropositive patients without chronic Acvr infection. Copies of HSV-1 DNA at 38,000 per 500 ng of total TG DNA were detected in the LTG from patient 708. The viral DNA loads in the other seven patients ranged from 16 to 1,700 copies per 500 ng of total TG DNA, with a median of 880 copies. The viral DNA load in TG from patient 708 was 21-fold higher than the maximum, and 42-fold higher than the median, viral DNA load in the TG of the seven other patients (Fig. 3A). Since Patient 708 had herpetic lesions over the surface of her whole body, including the genital area, it was expected that all of her DRG would be HSV DNA positive. In fact, the HSV-1 DNA copy numbers in several of her DRG were much higher than that in her TG (Fig. 3B). Of the 21 DRG (the LT2 to LS5 ganglia) tested, only 1 ganglion (the LT4 ganglion) had undetectable HSV-1 DNA (<5 copies per 50 ng of DNA). Four ganglia (the LT5, LL4, LL5, and LS5 ganglia) had low viral DNA loads, with 6 to 260 copies per 50 ng of DNA. Six ganglia (the LT3, LT6, LT7, LL2, LL3, and LS1 ganglia) had viral DNA loads comparable to or slightly lower than that in the TG, ranging from 1,400 to 3,000 HSV-1 genome copies per 50 ng of DNA. In the remaining 10 DRG, viral DNA loads were even higher than that in the TG, ranging from 5,000 to 13,000 HSV-1 genome copies per 50 ng of DNA. These findings indicate that in this immunocompromised patient, HSV-1 established latent infection in the DRG, including the sacral ganglia, at rates similar to or greater than that in the TG. Assays to detect reactivation in these ganglia, including reverse transcription-PCR for the identification of transcripts of HSV-1 late genes, immunohistochemistry staining for HSV-1 antigens, and the culture of ganglion homogenates for infectious virus, were all negative (data not shown). Thus, there was likely little or no virus replication in the ganglia when the patient died. Patient 708 was seronegative for HSV-2, and all ganglia tested, including sacral ganglia, were negative for HSV-2 DNA by PCR.
FIG. 3.
Extremely high HSV-1 DNA loads in TG and DRG of patient 708. Total DNA isolated from ganglia was quantified by real-time PCR with primers and a probe specific for the HSV-1 gG gene. (A) DNA samples isolated from TG of patient 708 and seven other patients were each used as templates in PCRs (500 ng/reaction). *, median; **, maximum. (B) DNA samples isolated from the left TG and left DRG of patient 708 were each used as templates in PCRs (50 ng/reaction).
Diverse HSV-1 tk mutations in the seven-G homopolymer region were detected in individual ganglia.
To determine if HSV-1 tk mutants can infect and establish latency in human ganglia, we tried to recover infectious virus from ganglia by explant cocultivation but failed to recover virus (data not shown). Therefore, we performed PCR amplification of HSV-1 tk sequences from ganglionic DNA. We first tried to directly sequence the PCR products from the LT6 ganglion. No definite mutation was found in the tk sequence, and the quality of the sequencing data for the seven-G region was suboptimal.
Since the frequency of tk nucleotide substitution mutations in the ganglia might be very low and because insertion or deletion mutations in the seven-G region might be present, the direct sequencing of PCR products was considered an insensitive method for the detection of these tk mutants. Thus, we subcloned PCR products and sequenced individual clones. Total DNA isolated from 12 ganglia was amplified by PCR with primers specific for HSV-1 tk so that the entire 1,131-bp tk ORF was obtained (Fig. 4A). PCR products were subcloned into plasmid vectors, and DNA samples prepared from single clones were sequenced (Fig. 4B). HSV-1 tk mutations in the seven-G homopolymer region (nt 430 to 436) were found in 11 of the 12 ganglia tested, with frequencies ranging from 4.2 to 76% (Table 3). In total, 89 (25%) of the 354 clones sequenced were identified as mutants.
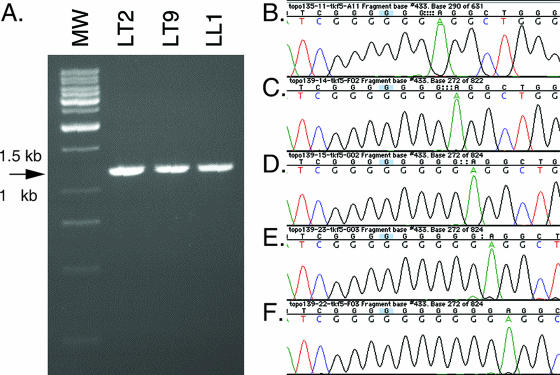
FIG. 4.
PCR subcloning and sequencing of HSV-1 tk DNA samples from ganglia of patient 708. Total DNA samples isolated from ganglia were amplified by PCR with primers specific for HSV-1 tk so that the product encompassed the entire ORF of tk. The PCR products were then subcloned into plasmid vectors, and DNA samples prepared from single bacterial colonies were sequenced. (A) Representative gel showing the 1.2-kb PCR products from the LT2, LT9, and LL1 DRG, indicated by an arrow. MW, molecular weight standard. (B to F) Chromatograms showing the seven-G homopolymer regions of tk clones from patient 708 with a variety of mutations: one-G deletion (B), seven-G WT sequence (C), one-G insertion (D), two-G insertion (E), and three-G insertion (F).
TABLE 3.
Diverse mutations in the HSV-1 tk seven-G homopolymer region detected in individual ganglia from patient 708
| Source of DNA | No. of clones sampled | No. of clonesa with:
|
% of mutant clones | ||||
|---|---|---|---|---|---|---|---|
| 7-G (WT) sequence | 1-G del | 1-G ins | 2-G ins | 3-G ins | |||
| LTG | 48 | 46 | 2 | 0 | 0 | 0 | 4.2 |
| LT2 ganglion | 23 | 16 | 0 | 0 | 6 | 1 | 30 |
| LT6 ganglion | 75 | 59 | 0 | 2 | 12 | 2 | 21 |
| LT7 ganglion | 23 | 20 | 1 | 2 | 0 | 0 | 13 |
| LT8 ganglion | 23 | 12 | 0 | 2 | 6 | 3 | 48 |
| LT9 ganglion | 23 | 10 | 0 | 1 | 10 | 2 | 57 |
| LL1 ganglion | 23 | 15 | 0 | 7 | 0 | 1 | 35 |
| LL3 ganglion | 21 | 5 | 0 | 1 | 12 | 3 | 76 |
| LS1 ganglion | 24 | 19 | 0 | 0 | 5 | 0 | 21 |
| LS2 ganglion | 24 | 18 | 1 | 2 | 2 | 1 | 25 |
| RT6 ganglion | 23 | 21 | 2 | 0 | 0 | 0 | 8.7 |
| RS1 ganglion | 24 | 24 | 0 | 0 | 0 | 0 | 0.0 |
| Total | 354 | 265 | 6 | 17 | 53 | 13 | 25 |
del, deletion; ins, insertion.
To determine if the mutations in the seven-G region of HSV-1 tk detected in the ganglia from patient 708 might be due to PCR errors or mutations occurring during plasmid propagation in bacteria, DNA from the LT6 ganglion of patient 708 and from a plasmid clone with HSV-1 WT tk were amplified by PCR under identical conditions and the products were cloned and sequenced. While 8 (17%) of 46 clones from the LT6 ganglion of patient 708 had mutations in the seven-G homopolymer region, none of 43 clones from the WT plasmid had mutations at this site (P = 0.006) (Table 4). Furthermore, DNA from the TG of another subject, patient 725, who had no history of ACV-resistant HSV-1 infection, was amplified by PCR and subcloned, and no mutations were found in the seven-G homopolymer regions of 24 clones (P = 0.04) (Table 4). These data indicate that the tk mutations in the seven-G homopolymer region detected in the ganglia from patient 708 are unlikely to be attributable to PCR errors or mutations occurring during bacterial culture.
TABLE 4.
Determining the PCR subcloning fidelity of the seven-G homopolymer region in WT HSV-1 tk
| PCR template | No. of clones sampled | No. of clonesa with:
|
% of mutant clones | ||||
|---|---|---|---|---|---|---|---|
| 7-G (WT) sequence | 1-G del | 1-G ins | 2-G ins | 3-G ins | |||
| LT6 DRG DNA from patient 708 | 46 | 38 | 0 | 2 | 5 | 1 | 17 |
| Plasmid with WT tk | 43 | 43 | 0 | 0 | 0 | 0 | 0.0b |
| TG DNA from patient 725 | 24 | 24 | 0 | 0 | 0 | 0 | 0.0c |
del, deletion; ins, insertion.
The difference between the results for the WT tk control template and the template from patient 708 were statistically significant (P = 0.006).
The difference between the results for the template from patient 725 and that from patient 708 were statistically significant (P = 0.04).
Various mutations in the seven-G homopolymer region were found, including the deletion of one G and insertions of one, two, and three G residues. The most common mutation was the two-G insertion, which accounted for 59.6% of the total mutant clones. The percentages of clones with the one-G deletion, the one-G insertion, and the three-G insertion were 6.7, 19.1, and 14.6%, respectively. These diverse mutations were found not only in different ganglia, but also within individual ganglia as well. While the majority of mutants in LT2, LT6, LT8, LT9, LL3, and LS1 ganglia had the two-G insertion, the one-G insertion was the most common mutation in the LT7 and LL1 ganglia, and the one-G deletion was the only mutation detected in LTG and RT6 ganglia. These findings indicate that HSV tk mutants with mutations in the seven-G homopolymer region can establish latency and reach high frequencies in human ganglia.
The two-G insertion in the seven-G homopolymer region of HSV-1 tk is unstable during PCR amplification and culture in bacteria.
In contrast to the PCR and subcloning fidelity of the WT HSV-1 tk seven-G homopolymer region, a plasmid containing an HSV tk gene with a two-G insertion in the seven-G homopolymer region from a ganglion of patient 708 was unstable during PCR and propagation in bacteria. Two plasmid clones, Topo143-2.7 and Topo143-4.11, which were originally isolated from the LT9 ganglion and had a nine-G sequence in the homopolymer region, had been colony purified twice and then amplified by PCR, subcloned, and sequenced. Four of 32 clones from Topo143-2.7 and 3 of 12 clones from Topo143-4.11 lost at least one G in the homopolymer region (Table 5). The rate of mutation in the nine-G string after PCR amplification and propagation in bacteria was 16% (7 of 44 clones). Of the seven clones no longer carrying a nine-G string, four showed a reversion to the seven-G sequence, two had a string of eight G residues, and one clone contained a mixture of nine- and eight-G sequences. The mixed nine- and eight-G sequences were likely generated when the plasmid was amplified in bacteria during overnight culture. These data suggest that the addition of the two-G insertion to the seven-G homopolymer region reduces the fidelity of DNA replication in this region. If this effect occurs in human tissues, a significant percentage of Acvr HSV-1 mutants with the two-G insertion in the seven-G homopolymer region may revert to ACV sensitivity over time in the absence of ACV selection.
TABLE 5.
Determining the mutation rate during PCR subcloning of HSV-1 tk plasmid clones from patient 708 with nine-G sequences in the seven-G homopolymer region
| PCR template | No. of clones sampled | No. of clones with:
|
% of mutant clones | |||
|---|---|---|---|---|---|---|
| 9-G sequence | 9- and 8-G sequences | 8-G sequence | 7-G sequence | |||
| Topo143-2.7 | 32 | 28 | 1 | 1 | 2 | 13 |
| Topo143-4.11 | 12 | 9 | 0 | 1 | 2 | 25 |
| Total | 44 | 37 | 1 | 2 | 4 | 16 |
Mutations outside the seven-G homopolymer region occur at frequencies at or below the limit of detection.
To determine if nucleotide mutations occurred outside the seven-G homopolymer region of the HSV-1 tk gene in the ganglia of patient 708, sequences of the entire ORF of each of 354 tk clones from the ganglia were compared with the published tk sequence of HSV-1 strain 17+. Thirteen nucleotide changes were found identically in 332 of the 354 clones: A68G, A106G, C171T, C261T, G266A, T271C, C513T, T717C, G719A, G793A, T933C, T969C, and A1065C (Table 6, column 3). The remaining 22 clones were identical to one another, but all had 5-nt changes compared to the consensus sequence of the other 332 clones (Table 6, column 4). All of the nucleotide changes either were in locations at which HSV-1 tk is known to be polymorphic, based on data from Kudo et al. (25), or do not result in amino acid changes. Thus, these nucleotide changes are not considered to be mutations but polymorphisms, and they suggest that the patient may have been infected with two different strains of HSV-1.
TABLE 6.
Polymorphisms in HSV-1 tk clones from patient 708
| Nucleotide position | Nucleotide at indicated position in:
|
||
|---|---|---|---|
| HSV-1 17+ | 332 HSV-1 tk clones from ganglia | 21 tk clones from RT6 and 1 clone from RS1 | |
| 68 | A | G | G |
| 102 | A | A | G |
| 106 | A | G | G |
| 171 | C | T | T |
| 261 | C | T | C |
| 266 | G | A | A |
| 271 | T | C | C |
| 513 | C | T | C |
| 717 | T | C | C |
| 719 | G | A | G |
| 793 | G | A | A |
| 915 | T | T | C |
| 933 | T | C | C |
| 969 | T | C | C |
| 1065 | A | C | C |
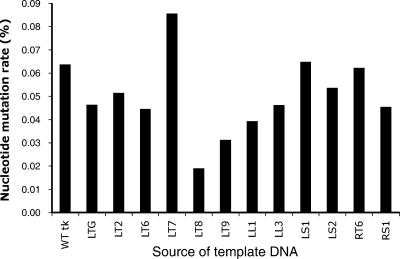
In addition to the nucleotide changes described above, other single-nucleotide substitutions, deletions, and insertions outside the seven-G homopolymer region of the HSV-1 tk clones from ganglia of patient 708 were detected at very low rates. The nucleotide mutation rate in HSV-1 tk clones from each ganglion (number of nucleotides mutated/total number of nucleotides sequenced from clones from each ganglion) ranged from 0.019 to 0.086% per ganglion (Fig. 5). To determine if these low mutation rates reflected changes in the tk gene of the viral genome or were within the range of error due to PCR amplification by the DNA polymerase and cloning into bacteria, plasmid DNA with a WT HSV-1 tk sequence was amplified by PCR, cloned into bacteria, and sequenced using the same conditions as those used for the HSV-1 tk clones from ganglia of patient 708. The mutation rate for tk sequences in the clones amplified from the WT tk plasmid was 0.064% based on the sequencing of 43 clones. Thus, the rates of nucleotide mutations outside the seven-G homopolymer region in 11 of 12 ganglia from patient 708 were lower than the rate for the WT tk control plasmid. The tk clones from the LT7 DRG had the highest nucleotide mutation rate, but the difference from that for the control WT tk plasmid was not statistically significant (P = 0.44). In addition to the nucleotide mutation rates in clones from each ganglion, the frequency of mutations at each nucleotide of the tk gene in all the clones sequenced was analyzed, under the assumption that a mutation seen in multiple clones would have a higher chance of being derived from a mutation in the viral tk rather than an event during PCR and cloning into bacteria. Most of the mutations outside the seven-G homopolymer region in the tk clones from ganglia occurred in only 1 of the 354 clones that were sequenced; 25 of the mutations were each found in two clones (e.g., G459A was found in one clone from the LTG and one clone from the LT7 ganglion), and 3 nucleotide mutations were found in three or four clones. Thus, the low frequency of nucleotide mutations found outside the seven-G homopolymer region of the tk clones from the patient's ganglia was within the range of error due to PCR amplification and cloning into bacteria. If any of these mutations were present in HSV-1 genomes in the patient's ganglia, they were at a frequency too low to be differentiated from PCR errors.
FIG. 5.
Comparison of the frequencies of nucleotide mutations in clones obtained by PCR amplification from ganglia of patient 708 and from a plasmid with WT tk. The nucleotide mutation rates (calculated as the number of nucleotides mutated divided by the total number of nucleotides sequenced from tk clones derived from individual ganglia) were determined for clones from patient 708 ganglion DNA and for clones from a plasmid containing WT tk. The nucleotide mutation rates for clones from 11 of 12 ganglia were lower than that for the WT tk control plasmid.
HSV-1 tk mutants and WT virus can coinfect a single neuron.
To determine if HSV-1 tk mutant and WT virus can coinfect single neurons, individual neurons were isolated from ganglion tissue sections by LCM (41). DNA extracted from single neurons was amplified by PCR to obtain a 147-bp fragment encompassing the seven-G homopolymer region. Fourteen neurons from the LT9 ganglion were HSV-1 tk positive by PCR. PCR products from each neuron were cloned separately, and multiple clones from each neuron were sequenced. Of the HSV-1 DNA-positive neurons, 29% (4 of 14) contained only WT HSV-1 tk DNA, 29% (4 of 14) had only mutant HSV-1 tk DNA, and 43% (6 of 14) had both WT and mutant HSV tk DNA (Table 7). These data indicate that in human ganglia, HSV-1 tk mutants and WT virus can coinfect neurons and HSV-1 tk mutants can infect neurons and establish latency in neurons in the absence of WT virus.
TABLE 7.
HSV-1 WT and tk mutants detected in individual neurons from a ganglion of patient 708
| Composition of virus population in single neurons (% of total no. of neurons) | Single neurona identification | No. of clones sampled | No. of clonesb with:
|
% of mutant clones | ||||
|---|---|---|---|---|---|---|---|---|
| 7-G (WT) sequence | 1-G del | 1-G ins | 2-G ins | 3-G ins | ||||
| Both WT and mutant clones in | sn-1 | 16 | 12 | 4 | 25 | |||
| each neuron (42.9) | sn-2 | 17 | 16 | 1 | 6 | |||
| sn-3 | 16 | 12 | 4 | 25 | ||||
| sn-4 | 21 | 7 | 14 | 67 | ||||
| sn-5 | 24 | 5 | 1 | 16 | 2 | 79 | ||
| sn-6 | 17 | 16 | 1 | 5.9 | ||||
| WT only in each neuron (28.6) | sn-7 | 13 | 13 | 0.0 | ||||
| sn-8 | 6 | 6 | 0.0 | |||||
| sn-9 | 23 | 23 | 0.0 | |||||
| sn-10 | 10 | 10 | 0.0 | |||||
| Mutants only in each neuron (28.6) | sn-11 | 13 | 13 | 100 | ||||
| sn-12 | 21 | 18 | 3 | 100 | ||||
| sn-13 | 22 | 2 | 20 | 100 | ||||
| sn-14 | 10 | 1 | 9 | 100 | ||||
Single neurons were retrieved from LT9 DRG tissue sections by LCM.
del, deletion; ins, insertion.
DISCUSSION
Acvr HSV clinical isolates from human mucocutaneous lesions have been studied extensively; however, Acvr HSV in the human nervous system has not been reported previously. Here we describe HSV tk sequences in TG and DRG from an immunocompromised patient with chronic HSV-1 cutaneous infection; many cultures from her skin swabs were positive for Acvr virus. While we were unable to recover live virus from her ganglia, PCR amplification and sequencing indicated that a variety of mutations were present within a sequence of seven G residues at nt 430 to 436 of the tk ORF. This seven-G region is one of two homopolymer regions that are hot spots for mutations (19, 33), and this region accounts for about 50% of HSV tk mutant isolates from Acvr HSV human skin lesions (16, 26, 33, 36).
We sequenced the entire tk ORFs from 354 HSV-1 tk clones obtained from ganglia of patient 708 and found four different mutations in the seven-G homopolymer region of the HSV-1 tk gene. The HSV tk mutant populations in individual ganglia of patient 708 may have derived their diversity partially from one another due to the increased instability of the insertion mutations. But the results summarized in Table 3 show that different mutations (the two-G insertion, the one-G insertion, and the one-G deletion) predominated in some ganglia, thus suggesting that the HSV-1 tk mutants in this patient may have arisen independently during replication in skin lesions and then established latency in the corresponding ganglia. Stranska and colleagues reported that sequential Acvr isolates from skin lesions of individual immunocompromised patients had mutations in different regions of the HSV tk gene (34, 35), supporting the hypothesis that continuing ACV selection can result in the emergence of new HSV tk mutants. However, if this hypothesis is correct, mutations in the HSV-1 tk gene in the ganglia of patient 708 would be expected to occur in multiple regions, since a vast array of HSV tk mutants from peripheral lesions of immunocompromised patients have been reported (10, 16). This assumes, however, that all HSV tk mutants are able to migrate to, and establish latency in, ganglia with equal levels of efficiency.
The predilection for mutations in the seven-G homopolymer region of HSV tk in latent neurons from patient 708 may be related to the observation that these HSV tk mutants in mice can acquire additional changes, resulting in a net insertion of three G residues or a reversion to the WT seven-G sequence, or express low TK activity by ribosomal frameshifting (11-14, 20) and thus increase the ability to establish and reactivate from latency. Therefore, it is possible that skin lesions from patient 708 had HSV-1 tk mutations in regions both within and outside the homopolymeric region but that mutations in nonhomopolymeric regions might be less likely to undergo reversion or translational frameshifting and, thus, these HSV-1 mutants might not be able to establish latency in human neurons as efficiently as the mutants we detected in the patient's ganglia. However, there are reports describing the repeated isolation of identical HSV tk mutants with mutations outside the homopolymer region from patient skin lesions (18, 24, 28, 32); these findings favor the hypothesis that these mutants can establish latency in and reactivate from human ganglia. Alternatively, the strain of the virus or the genetic background of our patient may have favored mutations in the seven-G region of the HSV-1 tk gene by some unknown mechanism. Grey et al. compared an HSV-1 clinical isolate with a two-G insertion in the seven-G homopolymer region of the tk gene (strain C4b) with a recombinant virus in which the same mutation was introduced into HSV-1 strain SC16 (11). The authors found that the nine-G string in strain C4b was very unstable and had a much higher rate of mutation to a 10-G sequence than the nine-G string in strain SC16, both in vitro and in mouse ganglia. This indicates that the stability of an insertion mutation in the seven-G homopolymer region may be determined not only by the length of the homopolymer, but also by nucleotide sequences in the virus outside of the homopolymer region.
We found that the average frequency of HSV-1 tk mutant DNA in ganglia from patient 708 was at least 25% of the total latent HSV-1 DNA (Table 3). Considering the extremely high burden of total HSV DNA in ganglia from patient 708, the amount of the HSV-1 tk mutant DNA in her ganglia may be even larger than the amounts of WT HSV-1 DNA in ganglia from immunocompetent patients. For example, in patient 708's TG (which had a modest viral DNA load and a low HSV-1 tk mutation rate compared to her other ganglia), the HSV-1 tk mutant DNA load was approximately 1,500 copies of HSV-1 tk mutant genomes per 500 ng of DNA, derived by multiplying the total load of 38,000 HSV-1 genome copies per 500 ng of DNA (Fig. 2A) by an HSV-1 tk mutant frequency of 4.2% (Table 3). This number is nearly twice as high as the median HSV-1 load (880 HSV-1 genome copies per 500 ng of DNA) in ganglia from the seven other patients without a history of Acvr HSV infection (Fig. 2A).
The high levels of HSV-1 tk mutant DNA in the ganglia of patient 708 seem contradictory to the observations that tk mutants show impaired capacities for replication and the establishment of latency in mouse ganglia; the high frequency of tk mutations in patient 708 may result from a combination of factors. First, the elevated amount of HSV-1 tk mutant DNA may be due to the type of mutations in the tk gene. Among the tk mutant clones we sequenced, mutations were found only in the seven-G homopolymer region. Most of the mutants (60%) had two-G insertions; 19, 15, and 6.7% of the mutants had the one-G insertion, the three-G insertion, and the one-G deletion, respectively. Besides the three-G insertion that adds one amino acid, producing a TK protein that has been reported to have WT TK activity in previous studies of clinical isolates (11, 13), all of the other mutations, the two-G insertion, the one-G insertion, and the one-G deletion, in the seven-G homopolymer region conferred greater ability than that of other tk mutants to reactivate from latency in the mouse model. These mutants may express a low level of TK by ribsomal frameshifting, reversion to the WT tk sequence, or further mutation to a three-G insertion that restores the tk ORF downstream of the insertion (11, 13, 16, 20). In one study, more HSV-1 latency-associated transcript-positive neurons were detected in mouse ganglia latently infected with a tk mutant that had the two-G insertion in the seven-G region than in ganglia latently infected with a tk deletion mutant (11). Together, these findings demonstrate that viruses with tk mutations identical to those present in patient 708 have a greater ability to establish and reactivate from latency in mouse TG than viruses with tk mutations outside of the seven-G string that alter TK activity. Assuming that latent infection with HSV tk mutants in the mouse model mimics latent infection in humans, the HSV-1 tk mutants found in ganglia from patient 708 are more likely to be able to express low TK activity and therefore to be better able to establish, and reactivate from, latency in human ganglia than viruses in which tk mutations are located outside of homopolymeric regions.
A second reason for the high levels of HSV-1 tk mutant DNA in the ganglia of patient 708 was that she was infected with both WT HSV-1 and tk mutants. We postulate that the HSV-1 tk mutants in the patient's skin lesions that had been selected by ACV therapy traveled together with WT virus on a retrograde course from the skin to the ganglia, where the WT virus replicated and produced viral TK. This viral TK could then complement the replication deficiency of the HSV-1 tk mutant(s) in trans and help the mutants to establish latency. Our observation that mutant and WT tk HSV-1 DNA coresided in 42.9% of the virus-infected individual neurons (Table 7) supports this hypothesis. This hypothesis has been tested in mice. Tenser et al. reported that TK− mutants were easily detected in mouse TG 3 days postinfection when mice were infected with a mixture of TK− mutants and the WT KOS strain but that virus was rarely detectable when mice were infected with TK− mutants only (38). Chen et al., however, recently reported that when they inoculated mice with both WT HSV and tk mutant viruses together, the WT virus showed reduced replication in the TG rather than complementing tk mutant replication (4). The authors postulated that the TK-LacZ fusion protein expressed from their HSV tk mutant might have resulted in the dominant negative inhibition of TK activity produced by the WT virus.
A third explanation for the high frequency of HSV-1 tk mutant DNA in the ganglia of patient 708 is that over several years she suffered repeated, prolonged cutaneous HSV-1 infections often due to Acvr HSV-1. The patient's severe cellular immunodeficiency likely allowed a high level of virus replication. Her ganglia were repeatedly infected with Acvr HSV-1, which spread in a retrograde pattern from the skin and resulted in ever-increasing HSV-1 DNA loads in the ganglia. This hypothesis may explain the clinical observation that while initial recurrences in patients treated with foscarnet after their first episode of Acvr virus infection are frequently due to ACV-sensitive virus, subsequent recurrences are more often due to Acvr virus (31). In contrast, in most studies of HSV tk mutants in mice, the animals receive only a single inoculation and are monitored for weeks to months. Mice, unlike humans, do not undergo the spontaneous reactivation of HSV infection and thus would not be expected to undergo multiple episodes of ganglion infection.
While the PCR amplification and bacterial cloning of WT HSV-1 tk from either patient ganglia or a plasmid clone resulted in no detectable mutations in the seven-G homopolymer region (Table 4), instability of a two-G insertion in this region during PCR amplification and cloning in bacteria was observed. This instability of the nine-G string was also seen when these mutants were tested with mice. Instead of the gain of an additional G residue, as described previously (11, 13), the mutation we most frequently observed in PCR subcloning was a two-G deletion resulting in a reversion to the WT seven-G sequence. This instability of the nine-G tk mutation may also occur during the replication of these mutant viruses in human tissue and allow the reemergence of WT virus from the Acvr mutant.
In summary, we found that HSV-1 tk mutants with mutations in the seven-G homopolymer region, similar to mutations in some previously reported Acvr clinical isolates, are able to establish latency with high viral DNA copy numbers in neurons of human TG and DRG (including sacral ganglia). The diverse mutant populations in different ganglia, as well as in individual neurons, indicate that various mutations in the seven-G region may arise directly from WT virus in patients who have been on prolonged or intermittent ACV therapy and that these viruses can establish latency in ganglia. Human ganglia can be infected with HSV-1 repeatedly, and individual neurons can be infected with multiple HSV-1 strains. The observation that some neurons harbored only mutant HSV tk DNA (Table 7) indicates that HSV with mutations in the seven-G homopolymer region of tk is able to establish and maintain latency either in the absence or in the presence of a very small amount of WT virus.
Acknowledgments
This research was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases.
We thank Maria Turner for the photograph of patient 708, Jing Qin for help with statistics, and Philip Krause for critical reading of the manuscript.
Footnotes
Published ahead of print on 25 April 2007.
This paper is dedicated to the memory of Stephen E. Straus, who was our mentor and inspiration for this study.
REFERENCES
- 1.Balasubramaniam, N. K., V. Veerisetty, and G. A. Gentry. 1990. Herpesviral deoxythymidine kinases contain a site analogous to the phosphoryl-binding arginine-rich region of porcine adenylate kinase; comparison of secondary structure predictions and conservation. J. Gen. Virol. 71:2979-2987. [DOI] [PubMed] [Google Scholar]
- 2.Boivin, G., C. K. Edelman, L. Pedneault, C. L. Talarico, K. K. Biron, and H. H. Balfour, Jr. 1994. Phenotypic and genotypic characterization of acyclovir-resistant varicella-zoster viruses isolated from persons with AIDS. J. Infect. Dis. 170:68-75. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D. G., R. Visse, G. Sandhu, A. Davies, P. J. Rizkallah, C. Melitz, W. C. Summers, and M. R. Sanderson. 1995. Crystal structures of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and ganciclovir. Nat. Struct. Biol. 2:876-881. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S. H., Y. W. Lin, A. Griffiths, W. Y. Huang, and S. H. Chen. 2006. Competition and complementation between thymidine kinase-negative and wild-type herpes simplex virus during co-infection of mouse trigeminal ganglia. J. Gen. Virol. 87:3495-3502. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S. H., A. Pearson, D. M. Coen, and S. H. Chen. 2004. Failure of thymidine kinase-negative herpes simplex virus to reactivate from latency following efficient establishment. J. Virol. 78:520-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibo, D., J. Druce, J. Sasadeusz, and C. Birch. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antivir. Res. 61:83-91. [DOI] [PubMed] [Google Scholar]
- 7.Czartoski, T., C. Liu, D. M. Koelle, S. Schmechel, A. Kalus, and A. Wald. 2006. Fulminant, acyclovir-resistant, herpes simplex virus type 2 hepatitis in an immunocompetent woman. J. Clin. Microbiol. 44:1584-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elion, G. B. 1983. The biochemistry and mechanism of action of acyclovir. J. Antimicrob. Chemother. 12(Suppl. B):9-17. [DOI] [PubMed] [Google Scholar]
- 9.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat. 5:88-114. [DOI] [PubMed] [Google Scholar]
- 11.Grey, F., M. Sowa, P. Collins, R. J. Fenton, W. Harris, W. Snowden, S. Efstathiou, and G. Darby. 2003. Characterization of a neurovirulent aciclovir-resistant variant of herpes simplex virus. J. Gen. Virol. 84:1403-1410. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths, A., S. H. Chen, B. C. Horsburgh, and D. M. Coen. 2003. Translational compensation of a frameshift mutation affecting herpes simplex virus thymidine kinase is sufficient to permit reactivation from latency. J. Virol. 77:4703-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, A., and D. M. Coen. 2003. High-frequency phenotypic reversion and pathogenicity of an acyclovir-resistant herpes simplex virus mutant. J. Virol. 77:2282-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths, A., M. A. Link, C. L. Furness, and D. M. Coen. 2006. Low-level expression and reversion both contribute to reactivation of herpes simplex virus drug-resistant mutants with mutations on homopolymeric sequences in thymidine kinase. J. Virol. 80:6568-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, R., E. L. Hill, D. McClernon, G. Davis, S. Selke, L. Corey, and A. Wald. 2005. Acyclovir sensitivity of sequential herpes simplex virus type 2 isolates from the genital mucosa of immunocompetent women. J. Infect. Dis. 192:1102-1107. [DOI] [PubMed] [Google Scholar]
- 16.Harris, W., P. Collins, R. J. Fenton, W. Snowden, M. Sowa, and G. Darby. 2003. Phenotypic and genotypic characterization of clinical isolates of herpes simplex virus resistant to aciclovir. J. Gen. Virol. 84:1393-1401. [DOI] [PubMed] [Google Scholar]
- 17.Hill, E. L., G. A. Hunter, and M. N. Ellis. 1991. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 35:2322-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horsburgh, B. C., S. H. Chen, A. Hu, G. B. Mulamba, W. H. Burns, and D. M. Coen. 1998. Recurrent acyclovir-resistant herpes simplex in an immunocompromised patient: can strain differences compensate for loss of thymidine kinase in pathogenesis? J. Infect. Dis. 178:618-625. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, C. B., and H. J. Chen. 1995. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene 152:191-193. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, C. B., B. Horsburgh, E. Pelosi, S. Roberts, P. Digard, and D. M. Coen. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA 91:5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson, J. G., K. L. Ruffner, M. Kosz-Vnenchak, C. B. Hwang, K. K. Wobbe, D. M. Knipe, and D. M. Coen. 1993. Herpes simplex virus thymidine kinase and specific stages of latency in murine trigeminal ganglia. J. Virol. 67:6903-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamieson, A. T., G. A. Gentry, and J. H. Subak-Sharpe. 1974. Induction of both thymidine and deoxycytidine kinase activity by herpes viruses. J Gen. Virol. 24:465-480. [DOI] [PubMed] [Google Scholar]
- 23.Katz, J. P., E. T. Bodin, and D. M. Coen. 1990. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J. Virol. 64:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kost, R. G., E. L. Hill, M. Tigges, and S. E. Straus. 1993. Brief report: recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N. Engl. J. Med. 329:1777-1782. [DOI] [PubMed] [Google Scholar]
- 25.Kudo, E., H. Shiota, T. Naito, K. Satake, and M. Itakura. 1998. Polymorphisms of thymidine kinase gene in herpes simplex virus type 1: analysis of clinical isolates from herpetic keratitis patients and laboratory strains. J. Med. Virol. 56:151-158. [DOI] [PubMed] [Google Scholar]
- 26.Morfin, F., G. Souillet, K. Bilger, T. Ooka, M. Aymard, and D. Thouvenot. 2000. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J. Infect. Dis. 182:290-293. [DOI] [PubMed] [Google Scholar]
- 27.Mouly, F., M. Baccard, C. Scieux, L. Schnell, S. Locq-Ebner, F. Morinet, and P. Morel. 1995. Chronic recurrent acyclovir-resistant genital herpes in an immunocompetent patient. Dermatology 190:177. [DOI] [PubMed] [Google Scholar]
- 28.Palu, G., G. Gerna, F. Bevilacqua, and A. Marcello. 1992. A point mutation in the thymidine kinase gene is responsible for acyclovir-resistance in herpes simplex virus type 2 sequential isolates. Virus Res. 25:133-144. [DOI] [PubMed] [Google Scholar]
- 29.Pottage, J. C., Jr., and H. A. Kessler. 1995. Herpes simplex virus resistance to acyclovir: clinical relevance. Infect. Agents Dis. 4:115-124. [PubMed] [Google Scholar]
- 30.Pramod, N. P., S. P. Thyagarajan, K. V. Mohan, and K. Anandakannan. 2000. Acyclovir resistance in herpes simplex virus isolates from keratitis cases: an analysis from a developing country. Microbiol. Immunol. 44:241-247. [DOI] [PubMed] [Google Scholar]
- 31.Safrin, S., C. Crumpacker, P. Chatis, R. Davis, R. Hafner, J. Rush, H. A. Kessler, B. Landry, J. Mills, et al. 1991. A controlled trial comparing foscarnet with vidarabine for acyclovir-resistant mucocutaneous herpes simplex in the acquired immunodeficiency syndrome. N. Engl. J. Med. 325:551-555. [DOI] [PubMed] [Google Scholar]
- 32.Saijo, M., T. Suzutani, K. Itoh, Y. Hirano, K. Murono, M. Nagamine, K. Mizuta, M. Niikura, and S. Morikawa. 1999. Nucleotide sequence of thymidine kinase gene of sequential acyclovir-resistant herpes simplex virus type 1 isolates recovered from a child with Wiskott-Aldrich syndrome: evidence for reactivation of acyclovir-resistant herpes simplex virus. J. Med. Virol. 58:387-393. [PubMed] [Google Scholar]
- 33.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stranska, R., R. Schuurman, E. Nienhuis, I. W. Goedegebuure, M. Polman, J. F. Weel, P. M. Wertheim-Van Dillen, R. J. Berkhout, and A. M. van Loon. 2005. Survey of acyclovir-resistant herpes simplex virus in the Netherlands: prevalence and characterization. J. Clin. Virol. 32:7-18. [DOI] [PubMed] [Google Scholar]
- 35.Stranska, R., A. M. van Loon, R. G. Bredius, M. Polman, E. Nienhuis, M. F. Beersma, A. C. Lankester, and R. Schuurman. 2004. Sequential switching of DNA polymerase and thymidine kinase-mediated HSV-1 drug resistance in an immunocompromised child. Antivir. Ther. 9:97-104. [PubMed] [Google Scholar]
- 36.Stranska, R., A. M. van Loon, M. Polman, M. F. Beersma, R. G. Bredius, A. C. Lankester, E. Meijer, and R. Schuurman. 2004. Genotypic and phenotypic characterization of acyclovir-resistant herpes simplex viruses isolated from haematopoietic stem cell transplant recipients. Antivir. Ther. 9:565-575. [PubMed] [Google Scholar]
- 37.Swetter, S. M., E. L. Hill, E. R. Kern, D. M. Koelle, C. M. Posavad, W. Lawrence, and S. Safrin. 1998. Chronic vulvar ulceration in an immunocompetent woman due to acyclovir-resistant, thymidine kinase-deficient herpes simplex virus. J. Infect. Dis. 177:543-550. [DOI] [PubMed] [Google Scholar]
- 38.Tenser, R. B., S. Ressel, and M. E. Dunstan. 1981. Herpes simplex virus thymidine kinase expression in trigeminal ganglion infection: correlation of enzyme activity with ganglion virus titer and evidence of in vivo complementation. Virology 112:328-341. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, R. L., and N. M. Sawtell. 2000. Replication of herpes simplex virus type 1 within trigeminal ganglia is required for high frequency but not high viral genome copy number latency. J. Virol. 74:965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wade, J. C., C. McLaren, and J. D. Meyers. 1983. Frequency and significance of acyclovir-resistant herpes simplex virus isolated from marrow transplant patients receiving multiple courses of treatment with acyclovir. J Infect. Dis. 148:1077-1082. [DOI] [PubMed] [Google Scholar]
- 41.Wang, K., T. Y. Lau, M. Morales, E. K. Mont, and S. E. Straus. 2005. Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal Ganglia at the single-cell level. J. Virol. 79:14079-14087. [DOI] [PMC free article] [PubMed] [Google Scholar]