Abstract
Both viral and host factors are thought to influence the pathogenesis of hepatitis C virus (HCV) infection. We studied strain HC-TN (genotype 1a), which caused fulminant hepatic failure in a patient and, subsequently, severe hepatitis in a chimpanzee (CH1422), to analyze the relationship between disease severity, host immune response, viral evolution, and outcome. A second chimpanzee (CH1581) was infected from CH1422 plasma, and a third chimpanzee (CH1579) was infected from RNA transcripts of a consensus cDNA of HC-TN (pHC-TN). RNA transcripts of pHC-TN did not replicate in Huh7.5 cells, which were recently found to be susceptible to infection with another fulminant HCV strain (JFH1). The courses of viremia were similar in the three animals. However, CH1581 and CH1579 developed a less severe acute hepatitis than CH1422. CH1579 and CH1422 resolved the infection, whereas CH1581 became persistently infected. CH1579 and CH1581, despite their differing outcomes, both developed significant intrahepatic cellular immune responses, but not antibodies to the envelope glycoproteins or neutralizing antibodies, during the acute infection. We analyzed the polyprotein sequences of virus recovered at five and nine time points from CH1579 and CH1581, respectively, during the first year of follow-up. High mutation rates and high proportions of nonsynonymous mutations suggested immune pressure and positive selection in both animals. Changes were not selected until after the initial decrease in virus titers and after the development of immune responses and hepatitis. Subsequently, however, mutations emerged repeatedly in both animals. Overall, our results indicate that disease severity and outcome of acute HCV infection depend primarily on the host response.
Acute hepatitis C is often asymptomatic. However, the disease burden of hepatitis C virus (HCV) is very significant, since about 170 million people worldwide are chronically infected, leading to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma in a significant proportion of infected individuals. HCV is now the leading cause of liver transplantation (35). The chimpanzee is the only animal model in which to study the natural history of HCV (4). Experimental infections permit collection of frequent samples, including liver tissue samples, before infection and during the acute phase and thus permit studies of early virological and immunological events that define the outcome. Furthermore, by inoculating chimpanzees intrahepatically with RNA transcripts of infectious HCV clones it is possible to study monoclonal virus infections (20, 45-47), in which virus interaction with the host is not initially influenced by a viral quasispecies. Also, in the study of the association between HCV and pathogenesis, infection from a molecular clone eliminates the possibility that an observed phenotype is caused by a coinfecting agent.
Fulminant hepatitis caused by hepatotropic viruses is a rare but potentially fatal condition. Initially, HCV was not recognized as an etiological agent of fulminant hepatitis (43). However, a significant number of Japanese patients with fulminant hepatitis had evidence of HCV infection (30, 44). Subsequently, the temporal relationship between transfusion-acquired HCV (genotype 1b) infection and development of fulminant hepatitis was described (12). Certain HCV strains, including strain HC-TN, recovered from a patient with fulminant hepatitis, appear to be associated with the development of severe hepatitis (13, 19). To study the relationship between HC-TN and disease phenotype, we transmitted this strain to chimpanzees and constructed an infectious clone to investigate monoclonal infection in a transfected chimpanzee.
The host and viral factors that determine the outcome of primary HCV infection are poorly understood. The host resolves less than 30% of infections. Viral clearance is associated with vigorous cellular immune responses (10, 24, 39), but persistence may be associated with viral escape from such T-cell responses (11). Others found that the development of antibodies to the envelope 2 (E2) hypervariable region 1 (HVR1), which contains a neutralization epitope, was associated with clearance (1, 50). Finally, genetic heterogeneity, in particular in HVR1, might predict the outcome of acute HCV (14).
Previously, we used chimpanzees to study the virological and immunological correlates of disease and outcome of acute HCV infection (16, 37, 38). Animals with viral clearance or with transient clearance followed by persistence at low titers had significant intrahepatic CD4+ and CD8+ T-cell responses, as well as induction of gamma interferon and gamma interferon-induced genes in the liver. Thus, the initial control of HCV is mediated by intrahepatic cellular immune responses. However, it is still unclear why animals with significant intrahepatic responses can have different outcomes. In the present study, we found that two chimpanzees infected with the HC-TN strain had comparable courses of viremia and vigorous host cellular immune responses. However, the infection resolved in only one animal. A detailed sequence analysis of viruses recovered from these animals was undertaken to determine the potential role of virus evolution in the outcome of acute HCV.
MATERIALS AND METHODS
Source of HCV strain HC-TN.
HCV strain HC-TN was from a patient who developed fulminant hepatic failure twice after liver transplantation for cryptogenic liver cirrhosis (13); she apparently became infected with HCV after receiving red blood cells before transplantation. A chimpanzee (CH1422) was inoculated with 100 μl of serum obtained 5 days before the first liver transplantation (see Fig. 1A) (13). (The data in Fig. 1A were adapted from Farci et al. [13], but the qualitative and quantitative HCV RNA tests were performed in this study.) A pool of HC-TN virus was made from plasmapheresis units collected from CH1422 during weeks 4 to 6 postinoculation.
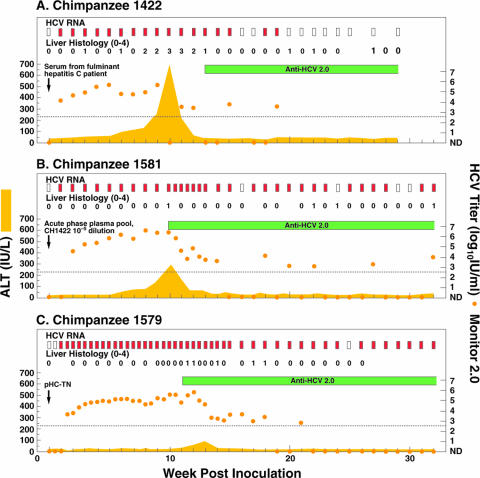
FIG. 1.
Course of infection with HCV strain HC-TN in (A) CH1422 (first-passage polyclonal infection), (B) CH1581 (second-passage quasipolyclonal infection), and (C) CH1579 (pHC-TN monoclonal infection). Serum samples collected once or twice weekly were tested for HCV RNA by an in-house RT-nested PCR with 5′ UTR primers and/or by use of a Roche Monitor 2.0 test. Red rectangle, positive by RT-nested PCR and/or by Monitor; white rectangle, negative by RT-nested PCR in two independent assays. The orange dots represent HCV Monitor titers; samples below the detection limit of 600 IU/ml (indicated by the dotted line) are shown as not detected (ND). Seroconversion in the second-generation ELISA is represented by a green horizontal bar. Yellow-shaded area, serum ALT. For liver histology, necroinflammatory changes of liver biopsy samples are graded 0 (normal), 1 (mild), 2 (mild to moderate), 3 (moderate to severe), or 4 (severe).
Amplification, cloning, and sequence analysis.
RNA extracted from 100-μl aliquots of the HC-TN pool with a TRIzol LS system (Life Technologies, Gaithersburg, MD) was denatured at 65°C for 2 min. HCV cDNA was synthesized at 42°C for 1 h with Superscript II reverse transcriptase (Life Technologies) and specific reverse primers. The cDNA was treated with RNase H and RNase T1 (Life Technologies) (46). To clone the entire open reading frame (ORF), we performed a one-round long PCR using an Advantage PCR polymerase mix (Clontech, Palo Alto, CA) (46). Gel-purified amplicons were A tailed with Taq DNA polymerase (Life Technologies) at 72°C for 1 h and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA). DH5α-competent cells (Life Technologies) were transformed and selected on LB agar plates containing 100 μg/ml ampicillin (Stratagene, La Jolla, CA) and amplified in LB liquid cultures at 30°C (46). A region spanning from nonstructural 5B (NS5B) to the conserved region of the 3′ untranslated region (UTR) was amplified by nested PCR with an Advantage 2 PCR polymerase mix and cloned as described previously (46). Final DNA preparations were sequenced using standard procedures.
The 5′ terminus was amplified from serum by 5′ rapid amplification of cDNA ends (RACE) with dC or dA tailing (Life Technologies) and three antisense C primers (615R [5′-CGCAACCCTCATTGCCATAG-3′] for reverse transcription [RT], 519R [5′-CTCGAGGTTGCGACCGCTCGGAAG-3′] for the first PCR, and 433R+Kpn-I [5′-CGGGGTACCACGATCTGACCGCCACCCGGGAAC-3′] for the second PCR). To determine the 3′-terminal sequence, the 5′ end of the negative-strand HCV RNA extracted from liver homogenate obtained from CH1581 was amplified by 5′ RACE with dC tailing and specific primers (−351R [5′-TGGTTCACGGCTGGCTACAG-3′] for RT, −334R [5′-CAGCGGGGGAGACATTTATCACAG-3′] for the first PCR, and −315R [5′-CACAGCGTGTCTCATGCCCGGCCC-3′] for the second PCR). The PCR products were cloned into pCR2.1-TOPO (Invitrogen).
To determine the consensus sequence of the entire ORF of HC-TN recovered from chimpanzees, we used two procedures. In serum samples with titers of >105 IU/ml, we performed long RT-PCR followed by nested PCR with genotype 1a-specific primers of 10 fragments (46). In samples with titers of <105 IU/ml, we performed RT-nested PCR, with Taq Gold DNA polymerase (Perkin Elmer, Wellesley, MA) (5), of 19 fragments by use of 1a-specific primers. The numbers of observed synonymous substitutions (ds) and nonsynonymous substitutions (dn) and the ratios of synonymous to nonsynonymous substitutions (ds/dn) were calculated using the Syn-SCAN program (http://hivdb6.stanford.edu/synscan/synscancgi).
Full-length consensus cDNA clone of HC-TN.
pHC-TN was constructed by standard molecular techniques using three clones that contained the ORF, one clone that contained the variable and poly(U-UC) regions of the 3′ UTR, and pCV-H77C (46). Large-scale preparation of a single clone was performed with a QIAGEN (Valencia, CA) Endofree maxi kit (46). The final DNA had the expected sequence.
Experimental infection of chimpanzees.
The housing and care of chimpanzees were in compliance with relevant guidelines and requirements (32). CH1581 was inoculated intravenously with dilutions of the CH1422 pool. CH1579 was inoculated intrahepatically by a percutaneous procedure (47) with RNA transcribed by T7 RNA polymerase (Promega, Madison, WI) from 20 μg of XbaI-digested pHC-TN (46). Serum samples were collected once or twice weekly and tested for HCV RNA (Monitor 2.0; Roche Diagnostics, Indianapolis, IN), HCV antibodies (ELISA 2.0; Abbott, Chicago, IL), and alanine aminotransferase (ALT) (Anilytics, Gaithersburg, MD). Monitor-negative samples were tested by a more sensitive RT-nested PCR (5). Samples obtained by weekly liver biopsies were examined for necroinflammatory changes (7).
We tested for anti-E1 by use of an enzyme-linked immunosorbent assay (ELISA) with recombinant E1 protein (amino acids [aa] 192 to 329) expressed from strain H77 (2, 29) and for anti-E2 by use of an ELISA with recombinant E2 protein (aa 388 to 664) of strain H, provided by I. K. Mushahwar (Abbott) (25, 29). Antibodies against E2 HVR1 were assayed with an ELISA using a biotinylated HC-TN-specific peptide (aa 384 to 410) (29). The percent neutralization in postinfection sera, compared with that in the preinoculation sample, was determined with a retroviral HCV pseudovirus assay using ppH77(1a) (provided by Francois-Loic Cosset, Ecole Normale Superieure de Lyon, Lyon, France), as described in detail previously (29).
The details of protocols used to detect cellular immune responses were published previously (38, 39). Peripheral blood mononuclear cells (PBMC) were isolated from 40 ml of blood. Liver-infiltrating lymphocytes were isolated from liver tissue obtained by needle biopsy. Cell suspensions were incubated with magnetic beads coupled to anti-CD4 or anti-CD8, and bound CD4+ or CD8+ T cells were isolated using a magnetic particle concentrator and next expanded for 2 weeks. PBMC or polyclonally expanded CD4+ T cells were tested for HCV-specific proliferative capacity after 6 days of culture with HCV-1 proteins (C22, C33-c, c100, and NS5), provided by M. Houghton (Chiron, Emeryville, CA). 3H[thymidine] was added for 16 h, and the mean levels of thymidine incorporation in the HCV protein-stimulated and control cultures were used to calculate the stimulation indexes (SI); values of >2.0 were considered positive. Polyclonally expanded CD8+ T cells were tested by intracellular gamma interferon staining after 5 h of stimulation with autologous Epstein-Barr virus-immortalized B-cell lines that were infected with recombinant HCV H77-encoding vaccinia viruses vHCV(1-1488) or vHCV(827-3011) together with VTF7, provided by C. M. Rice (Rockefeller University, New York, NY), or with VTF7 alone. The frequency of HCV-specific CD8+ T cells was defined as the percentage of CD8+ T cells that produced gamma interferon in response to stimulation by B-cell lines coinfected by vHCV and VTF7 after subtraction of the gamma interferon-positive, CD8+ T cells detected after stimulation in the absence of vHCV.
Transfection of Huh7.5 cells with RNA transcripts from pHC-TN.
Huh7.5 cells, provided by C. M. Rice (Rockefeller University, New York, NY), were maintained in growth medium consisting of complete Dulbecco's modified Eagle's medium (Gibco BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum, 50 IU/ml penicillin G, and 50 μg/ml streptomycin. Cells were incubated at 37°C in a humidified 5% CO2 incubator. RNA was transcribed, as described above, from pJFH1 and pHC-TN digested with XbaI; the pJFH1 plasmid was provided by Takaji Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan).
Transfection was performed using a DMRIE-C reagent (Invitrogen) in six-well plates (4 × 105 Huh7.5 cells/well). Briefly, cells were washed with 2 ml of Opti-MEM I medium (Gibco). Eight microliters of DMRIE-C reagent was first diluted in 1 ml of Opti-MEM I medium before the addition of a transcription mixture containing approximately 3 μg of RNA transcripts (based on gel analysis). Finally, the complexed RNA was incubated with the washed Huh7.5 cells at 37°C for 4 h and the medium was replaced with complete growth medium. For immunofluorescence staining with mouse anti-HCV core protein monoclonal antibody (B2) (Anogen, Mississauga, Ontario, Canada), the Huh7.5 cells were trypsinized, transferred to eight-well chamber slides, and incubated at 37°C overnight. The cells were washed twice with phosphate-buffered saline (PBS) and fixed and permeabilized with acetone for 3 min. Twenty-five microliters of a 1/200 dilution (in 5% bovine serum albumin in PBS) of the HCV anticore antibody was added to each grid and incubated at room temperature for 20 min. After a wash with PBS, a 1/100 dilution of the secondary antibody, anti-mouse immunoglobulin G (heavy plus light chains) fluorescein isothiocyanate-conjugated antibody (Pierce), was added to each grid and incubated at room temperature for 3 min. A drop of VectaShield containing DAPI (4′,6′-diamidino-2-phenylindole) was added to each grid of cells to stain nuclei. Slides were read with an Axioscope 2 Plus fluorescence microscope (Zeiss).
Nucleotide sequence accession number.
The nucleotide sequence of pHC-TN (an infectious clone of strain HC-TN) has been deposited in the GenBank database under the accession number EF621489.
RESULTS
We studied a genotype 1a strain (HC-TN) of HCV recovered from a patient who developed fulminant hepatic failure (13). Following transmission to CH1422, it caused severe hepatitis, with an ALT peak of 744 IU/liter and with an unusually pronounced necroinflammatory activity in liver biopsy samples (Fig. 1A). Serum HCV titers reached 105 to 105.5 IU/ml during weeks 4 to 9, followed by a dramatic decrease and clearance during week 20. We did not have any remaining sera from the patient used to inoculate CH1422. Thus, to further investigate the phenotype of the HC-TN strain, we performed a second passage, this time to CH1581 (Fig. 1B). Furthermore, we constructed a full-length consensus cDNA clone, the infectivity of which was confirmed in CH1579 (Fig. 1C), to study the phenotype of monoclonal infection with this particular HCV strain.
Genetic analysis of strain HC-TN.
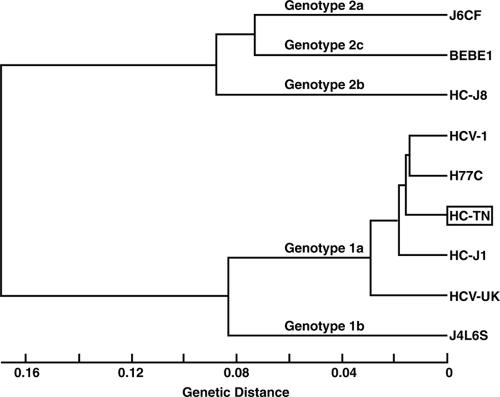
We analyzed the HC-TN sequence from the CH1422 plasma pool. To determine the consensus ORF, we directly sequenced amplicons obtained by long RT-PCR followed by PCR of 10 overlapping fragments. In addition, we analyzed three clones obtained from the long-RT-PCR amplicons. The ORF consisted of 9,033 nt encoding 3,011 aa. The genome population in CH1422 was virtually homogeneous, since heterogeneity was found among the three clones at only 26 (0.29%) nucleotide and 20 (0.66%) amino acid positions. Also, the clones had identical sequences within HVR1. The consensus sequence deduced from the ORF clones was identical to that obtained by direct sequencing. It differed from those of other 1a strains (9, 23, 33, 46) by 4.3% to 8.0% and by 2.9% to 5.4% at the nucleotide and amino acid levels, respectively (Table 1). A tree analysis of the polyprotein sequence of representative HCV isolates (9, 23, 31, 33, 34, 45-47) showed that HC-TN was most closely related to the prototype strains HCV-1 and H77 (Fig. 2). Since the polyprotein cleavage sites were highly conserved among the 1a strains, the HC-TN gene products are predicted to be the same as those of strain H77 (17, 26).
TABLE 1.
Differences in nucleotide and predicted amino acid sequences between HC-TN and other genotype 1a strains
| Genomic region | nt positiona | No. (%) of nt differences
|
aa positiona | No. (%) of aa differences
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H77C | HCV-1 | HC-J1 | HCV-UK | H77C | HCV-1 | HC-J1 | HCV-UK | |||
| 5′ UTR | 1-341 | 1 (0.3) | 0 (0.0) | 2 (0.6) | 2 (0.6) | |||||
| Core | 342-914 | 7 (1.2) | 13 (2.3) | 11 (1.9) | 15 (2.6) | 1-191 | 1 (0.5) | 3 (1.6) | 3 (1.6) | 3 (1.6) |
| E1 | 915-1490 | 24 (4.2) | 34 (5.9) | 43 (7.5) | 39 (6.8) | 192-383 | 6 (3.1) | 9 (4.7) | 11 (5.7) | 9 (4.7) |
| E2 | 1491-2579 | 70 (6.4) | 76 (7.0) | 81 (7.4) | 150 (13.8) | 384-746 | 25 (6.9) | 28 (7.7) | 28 (7.7) | 48 (13.2) |
| HVR1 | 1491-1571 | 12 (14.8) | 13 (16.0) | 15 (18.5) | 32 (39.5) | 384-410 | 7 (25.9) | 9 (33.3) | 8 (29.6) | 15 (55.6) |
| p7 | 2580-2768 | 9 (4.8) | 10 (5.3) | 11 (5.8) | 22 (11.6) | 747-809 | 2 (3.2) | 3 (4.8) | 4 (6.3) | 5 (7.9) |
| NS2 | 2769-3419 | 36 (5.5) | 42 (6.5) | 47 (7.2) | 65 (10.0) | 810-1026 | 7 (3.2) | 8 (3.7) | 9 (4.1) | 18 (8.3) |
| NS3 | 3420-5312 | 86 (4.5) | 90 (4.8) | 109 (5.8) | 143 (7.6) | 1027-1657 | 12 (1.9) | 14 (2.2) | 16 (2.5) | 18 (2.9) |
| NS4A | 5313-5476 | 8 (4.8) | 9 (5.5) | 9 (5.5) | 17 (10.4) | 1658-1711 | 1 (1.9) | 1 (1.9) | 1 (1.9) | 3 (5.6) |
| NS4B | 5477-6257 | 32 (4.1) | 27 (3.5) | 44 (5.6) | 67 (8.6) | 1712-1972 | 6 (2.3) | 4 (1.5) | 9 (3.4) | 11 (4.2) |
| NS5A | 6258-7600 | 67 (5.0) | 74 (5.5) | 86 (6.4) | 108 (8.0) | 1973-2420 | 23 (5.1) | 21 (4.7) | 25 (5.6) | 30 (6.7) |
| NS5B | 7601-9374 | 53 (3.0) | 62 (3.5) | 73 (4.1) | 101 (5.7) | 2421-3011 | 5 (0.8) | 9 (1.5) | 8 (1.4) | 19 (3.2) |
| ORF | 342-9374 | 392 (4.3) | 437 (4.8) | 514 (5.7) | 727 (8.0) | 1-3011 | 88 (2.9) | 100 (3.3) | 114 (3.8) | 164 (5.4) |
| 3′ UTR | 9375-9599 | |||||||||
| Variable region | 9375-9417 | 1 (2.3) | 0 (0.0) | 3 (7.0) | NAb | |||||
| Conserved region | 9499-9599 | 0 (0.0) | NA | NA | NA | |||||
The nucleotide and predicted amino acid positions correspond to those of pHC-TN.
NA, not available.
FIG. 2.
Tree analysis of predicted polyprotein sequences of HC-TN (boxed) and other HCV strains. The multiple sequence alignment and tree analysis were performed with GeneWorks (6).
To analyze the 5′ UTR of HC-TN, we performed 5′ RACE and sequenced 10 and 3 clones (nucleotides [nt] 1 to 409) obtained following dC and dA tailing, respectively. All clones had identical 5′-terminal sequences, and the remainder of the 5′ UTR was highly conserved. The HC-TN 5′ UTR sequence was identical to that of HCV-1 (18). To analyze the 3′ UTR, we first sequenced 12 clones of PCR products, which included the variable and the poly(U-UC) regions. The variable region consisted of 43 nt (nt 9375 to 9417), including two in-frame termination codons. All clones had identical sequences except at position 9391 (11 C, 1 T). The poly(U/UC) regions varied in length (76 to 148 nt), entirely due to variation of the poly(U) regions (41 to 113 nt). The poly(UC) regions had the same length (35 nt), and the sequences (5′-CUUUUUCCCUCUUUUUCUUCUCUUUUUCCUUCUUU-3′) were identical in all 12 clones except at position 3 (11 U, 1 C). Furthermore, we found that this region had the same sequence in the five clones from negative-strand RNA extracted from CH1581 liver. The sequence of the conserved region was determined by 5′ RACE (dC tailing) on the negative-strand RNA extracted from CH1581 liver homogenate collected at week 8. Five clones (nt 9410 to the 3′ terminus) analyzed had the same 3′-terminal sequences, and the consensus sequence (101 nt) was identical to that of strain H77 (21).
Infectivity titration of HC-TN plasma pool.
The pool collected from CH1422 during weeks 4 to 6 had an HCV genome titer of ∼105 IU/ml (Monitor 2.0, 105.3 IU/ml; Versant HCV RNA b-DNA 3.0 [Bayer, Tarrytown, NY], 105.0 IU/ml). Its infectivity titer was determined by reverse titration in CH1581. The 10−6 dilution was noninfectious. However, HCV was transmitted to CH1581 by inoculation of 1 ml of a 10−5 dilution (Fig. 1B), indicating an infectivity titer of ∼105 chimpanzee infectious doses/ml. We analyzed the ORF of HCV recovered from the serum of CH1581 at week 8. Differences between the CH1581 sequence and the consensus sequence of the CH1422 virus used as the inoculum were found at only two nucleotide positions (A1535G and G6531A) and resulted in one NS5A amino acid change (A2064T). Both substitutions were also found at week 1.
In CH1581, HCV RNA titers peaked at ∼106 IU/ml during weeks 5 to 10, followed by a 3-log10 decrease in viremia titers during weeks 10 to 15 (Fig. 3A). The titers were frequently below the detection limit of the Monitor test (102.8 IU/ml) during weeks 15 to 37. Furthermore, a sensitive RT-nested PCR test was negative at weeks 16, 24, 29, and 30, and during weeks 38 to 52, the HCV titers remained at 103 to 104 IU/ml. The second-generation ELISA became positive at week 10. However, the animal did not develop anti-E1, anti-E2, or anti-HVR1 until after 45 weeks of follow-up (29). Furthermore, the chimpanzee did not develop significant (≥50% neutralization) neutralizing antibodies during the first year of follow-up (29) (Fig. 3A). The chimpanzee developed acute hepatitis, with a peak ALT level of 296 IU/liter (week 10). Mild necroinflammatory changes were detected in liver biopsy samples at weeks 10, 22, and 24, as well as during the persistent phase of infection.
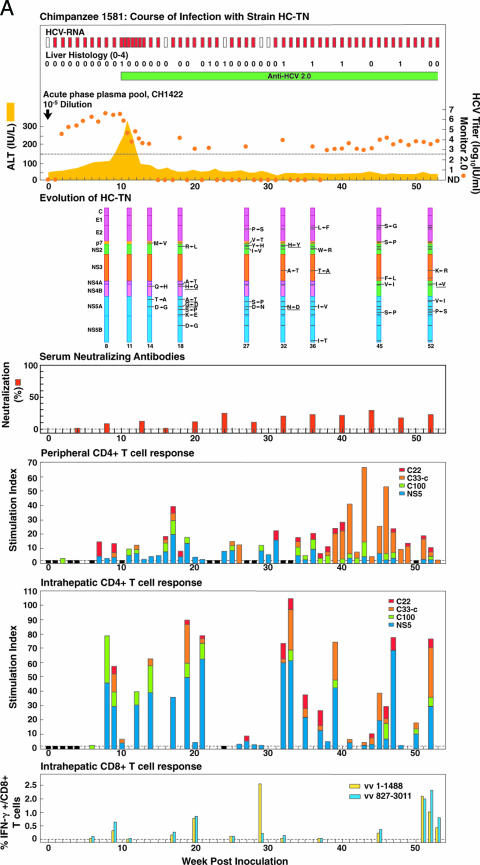
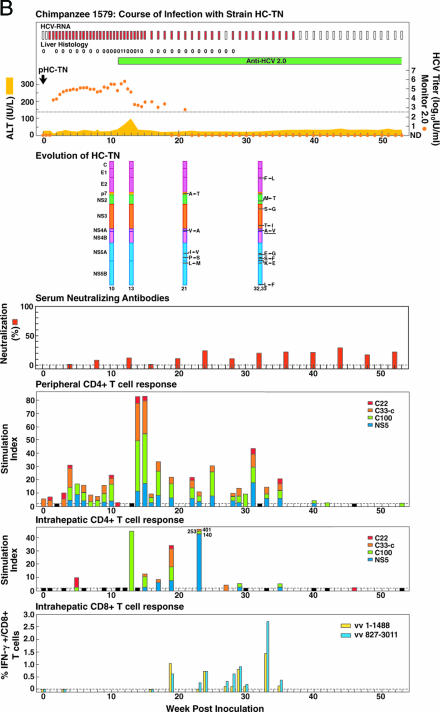
FIG. 3.
Course of infection, viral evolution, and immune responses in (A) CH1581 and (B) CH1579, which were infected with HCV strain HC-TN (quasipolyclonal and monoclonal infections, respectively). See the legend for Fig. 1 for the details of the course of infection. Representing the evolution of HC-TN, the HC-TN genome is shown as a vertical bar with the core (C) at the top and NS5B at the bottom. Solid black lines with capital letters indicate new amino acid changes that were identified when a sequence was compared with the sequence obtained at the previous time point. Underlined capital letters indicate mutations that had occurred by one time point but had changed back to the original sequence by the next time point analyzed. Solid black lines without capital letters represent amino acid changes that persisted. The week the sequence was analyzed is indicated at the bottom of each genome. For neutralizing antibodies, the percent neutralization of retroviral pseudovirus particles bearing the HCV envelope proteins (>50% was considered significant) is shown. The peripheral and intrahepatic CD4+ T-cell responses to core (red), NS3 (orange), NS3-NS4 (green), and NS5 (blue) are shown as specific SI. A specific SI of >2 was considered significant. At weeks tested in which the SI was ≤2 against all four antigens, the negative result is indicated by a black bar (with a value of 2). The intrahepatic CD8+ T-cell response is represented as the percentage of intrahepatic CD8+ T cells that produced gamma interferon (IFN-γ) after stimulation with transiently expressed HCV proteins, as described in Materials and Methods. vv 1-1488, vaccinia virus vHCV(1-1488); vv 827-3011, vaccinia virus vHCV(827-3011).
RNA transcripts from a consensus clone (pHC-TN) are infectious in vivo but not in vitro.
In pHC-TN, the core sequence of the T7 promoter and the consensus sequence of HC-TN (9,599 nt) were contained in the NotI/XbaI-digested pGEM-9Zf vector. It contained a 5′ UTR of 341 nt, an ORF of 9,033 nt, and a 3′ UTR with a variable region of 43 nt followed by an 81-nt poly(U/UC) tract (46 U, 35 UC) and a 3′-terminal conserved sequence of 101 nt. RNA transcripts of pHC-TN were percutaneously injected into the liver of CH1579. For unknown reasons, the first in vivo transfection failed. However, following a subsequent transfection 4 weeks later with new RNA transcripts, the animal became infected (Fig. 3B). The HCV ORF sequence recovered from week 10 sera was identical with that of pHC-TN. The HCV titers reached peak levels of 105 to 105.5 IU/ml during weeks 4 to 13, followed by a significant decrease. The quantitative Monitor test was negative during weeks 22 to 53. However, the qualitative RT-nested PCR test remained positive until week 37. Thus, this chimpanzee had prolonged acute resolving HCV infection. The animal became positive in the second-generation ELISA at week 11 but did not develop anti-E1, anti-E2, or anti-HVR1 antibodies. Furthermore, the animal did not develop neutralizing antibodies (29) (Fig. 3B). It had elevated ALT only during weeks 12 to 13 (peak, 90 IU/liter) and mild histological changes in liver biopsy samples during weeks 11 to 18.
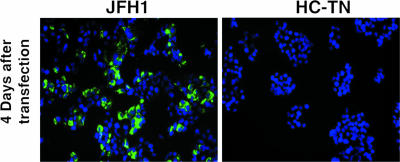
A molecular clone of strain JFH1, also recovered from a patient with fulminant hepatitis C, was recently found to be infectious in Huh7 and Huh7.5 cells (41, 49). We transfected Huh7.5 cells with RNA transcripts from pJFH1 and pHC-TN (Fig. 4). We also included JFH1 chimeras, which contain core through NS2 from HC-TN (data not shown). Clear evidence of replication was observed with JFH1 and TN/JFH1 chimeras, but there was no evidence of HCV replication with HC-TN. The fact that replication of the TN/JFH1 chimeras could be detected by staining for core proved that the anticore used for immunofluorescence staining could readily detect the HC-TN strain; therefore, the lack of staining in cells transfected with pHC-TN indicated that this virus could not replicate in these cells. Following one transfection with RNA transcripts of pHC-TN, the culture was monitored for more than 20 weeks with no evidence of HCV replication.
FIG. 4.
Testing for replication of pHC-TN in vitro. Huh7.5 cells were transfected with RNA transcripts of pJFH1 and pHC-TN. Immunofluorescence staining was performed with an HCV core-specific mouse monoclonal antibody.
T-cell responses in HC-TN-infected chimpanzees irrespective of the final outcome.
Analyses of the cellular immune responses were performed with CH1581 and CH1579 (Fig. 3). Cells had not been collected from CH1422. CD4+ T cells from PBMC or polyclonally expanded from liver specimens were tested for HCV-specific proliferative responses with HCV-1 proteins (C22, C33-c, c100, and NS5). Polyclonally expanded liver CD8+ T cells were tested for intracellular gamma interferon staining after coculture with autologous B cells expressing the entire H77 polyprotein from vaccinia viruses.
In CH1581, HCV-multispecific peripheral and intrahepatic CD4+ T-cell responses were detected in the liver beginning at weeks 7 and 8, respectively (Fig. 3A) (38). These responses decreased during the period with low-titer viremia (weeks 22 to 31). Intrahepatic CD8+ T-cell responses were detected at week 9 and in the available samples tested thereafter, including samples tested during the period with low-titer viremia (Fig. 3A) (38). In CH1579, the peripheral HCV-multispecific CD4+ responses appeared earlier and were more vigorous than the intrahepatic CD4+ responses (Fig. 3B). In fact, multispecific CD4+ responses against core, NS3, NS4, and NS5 proteins were detected during weeks 1 to 35. In contrast, intrahepatic multispecific CD4+ T-cell responses against core, NS3, NS4, and NS5 proteins were detected primarily during the initial decrease of viremia titers during weeks 13 to 23. The strongest intrahepatic CD4+ response observed in the present study occurred at week 23 in CH1579. Weaker intrahepatic CD4+ responses were observed during the following period with low-titer viremia. Vigorous intrahepatic CD8+ T-cell responses were observed during the low-titer-viremia period that preceded viral clearance (Fig. 3B). We tried but failed to detect a reproducible CD8+ T-cell response against selected epitopes in either animal, owing to the apparent low frequency of that response in the periphery (data not shown). Overall, we found that both animals had vigorous HCV-specific T-cell responses during the dramatic decrease in HCV titers and that these responses were sustained while viremia was present.
Repeated emergence of new virus variants during the host immune response.
We sequenced the entire ORF of viruses recovered from the three chimpanzees, each at multiple time points (Tables 2, 3, and 4). For CH1422, we analyzed the HCV sequence from the pool taken at peak viremia titers (weeks 4 to 6) as well as that from a serum sample taken at week 19, after the virus became transiently undetectable at weeks 16 and 17 (Fig. 1A). We detected 78 nucleotide and 17 amino acid substitutions at week 19. The relatively high mutation rate and ds/dn ratio compared with those of CH1581 and CH1579 (Table 2) suggested that the virus that reemerged represented the selection of a preexisting minor variant. The amino acid changes were located in core (G187V), E2 (I438V and S453P), p7 (L765V and L790F), NS2 (V873I, V879I, and K927N), NS3 (L1504P), NS4B (A1832T), NS5A (K2016R, Q2095R, E2228G, L2340P, and K2414E), and NS5B (H2483Y and T2810I).
TABLE 2.
Nucleotide and amino acid substitutions observed in chimpanzees infected with HC-TN
| Chimpanzee | Time point (wk) | Mutation rate (103)/site/yr
|
No. of ds | No. of dn | ds/dn ratio | |
|---|---|---|---|---|---|---|
| nt | aa | |||||
| CH1422a | 19 | 23.63 | 17.27 | 61 | 17 | 11.1 |
| CH1581b | 8 | 0 | 0 | 0 | 0 | NAd |
| 11 | 0 | 0 | 0 | 0 | NA | |
| 14 | 11.51 | 23.03 | 2 | 4 | 1.52 | |
| 18 | 14.39 | 34.53 | 2 | 8 | 0.76 | |
| 27 | 6.40 | 11.51 | 3 | 7 | 1.30 | |
| 32 | 3.45 | 6.91 | 1 | 2 | 1.52 | |
| 36 | 24.47 | 25.91 | 11 | 6 | 5.58 | |
| 45 | 3.38 | 9.59 | 1 | 5 | 0.61 | |
| 52 | 4.11 | 9.87 | 1 | 4 | 0.76 | |
| 52c | 3.65 | 6.64 | 13 | 20 | 1.98 | |
| CH1579b | 10 | 0 | 0 | 0 | 0 | NA |
| 13 | 0 | 0 | 0 | 0 | NA | |
| 21 | 4.32 | 10.79 | 1 | 5 | 0.61 | |
| 32 | 5.76 | 14.13 | 2 | 9 | 0.68 | |
| 32c | 2.34 | 6.48 | 1 | 12 | 0.25 | |
The week 19 sequence was compared to the HCV sequence obtained from an acute-phase plasma pool of CH1422.
At each time point, the HC-TN sequence was compared with that obtained at the previous time point.
The HC-TN sequence obtained at week 52 from CH1581 and that obtained at week 32 from CH1579 were also compared with that of the inoculum.
NA, not applicable.
TABLE 3.
Evolution of HC-TN polyprotein in CH1581
| Wka | Amino acid sequenceb
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2
|
p7
|
NS2
|
NS3
|
NS4B
|
NS5A
|
NS5B
|
|||||||||||||||||||||
| 401S | 433L | 461P | 767S | 793M | 834Y | 837R | 841W | 861I | 1405A | 1406K | 1583F | 1746V | 1747A | 1751Q | 2064A | 2116V | 2118S | 2220D | 2223D | 2227I | 2278S | 2341P | 2358S | 2414K | 2664D | 3005I | |
| 1 | T | ||||||||||||||||||||||||||
| 8 | S | L | P | S | M | Y | R | W | I | A | K | F | V | A | Q | T | V | S | D/g | D | I | S | P | S | K | D | I |
| 11 | S | L | P | S | M | Y | R | W | I | A | K | F | V | A | Q | T | V | S | D/G | D | I | S | P | S | K | D | I |
| 13 | Y/h | R/H | W/r | I | V | A | Q | T | V | S | G/d* | D | I | S | |||||||||||||
| 14 | S | L | P | S | V* | Y/h | R/H | W | I | A | K | F | V | A/t | H/q* | A/t* | V | S | G/d* | D | I | S | P | S | K/E | D | I |
| 18 | S | L | P | S | V* | Y | L* | W | I | A | K | F | V | T/a* | Q/h | T/a | V | S | D/g | D | I | P* | P | S | E* | G* | I |
| 26 | Y/h | L* | W | V/i* | V | P* | D | N* | I | P* | |||||||||||||||||
| 27 | S | L | S* | S | T* | H/y* | L* | W | V/i* | A | K | F | V | T* | Q | T | V | P* | D | N* | I | P* | P | S | E* | G* | I |
| 31 | T/a* | K | |||||||||||||||||||||||||
| 32 | S | L/F | S* | S | T* | H/Y* | L* | W/R | V* | T* | K | F | V | T* | Q | T | V | P* | D | D/n | I | P* | P | S | E* | G* | I |
| 36 | S | F* | S* | S/p | T* | Y | L* | R* | V* | A | K | F | V/i | T* | Q | T | V | P* | D | D | V/i* | P* | P | S | E* | G* | T* |
| 45 | G/s* | F* | S* | P/s* | T* | Y | L* | R* | V* | A/v/t/i | K/R | L/f* | I/v* | T* | Q | T | V/i | P/S* | D | D | V* | P* | P | P/s* | E* | G* | T* |
| 51 | P* | S/p* | P* | E* | |||||||||||||||||||||||
| 52 | G* | F* | S* | P* | T* | Y | L* | R* | V* | A/t | R/k* | L* | V/i | T* | Q | T/a | I/v* | P* | D | D | V* | P* | S/p* | P* | E* | G* | T* |
The entire ORF sequence of HC-TN was determined at weeks 8, 11, 14, 18, 27, 32, 36, 45, and 52. Only subdomains were analyzed at weeks 1, 13, 26, 31, and 51.
Amino acid positions correspond to those of pHC-TN. The consensus sequence of viruses recovered from a CH1422 acute-phase plasma pool is shown at the top. Dominant sequences recovered from the chimpanzee are shown in capital letters. Minor sequences recovered from the chimpanzee are shown in lowercase letters. Dominant amino acid changes are marked with an asterisk. Note that two consecutive changes occurred at aa 793.
TABLE 4.
Evolution of HC-TN polyprotein in CH1579
| Wka | Amino acid sequenceb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2
|
p7
|
NS2
|
NS3
|
NS4A
|
NS5A
|
NS5B
|
|||||||
| 403F | 789A | 1018M | 1148S | 1563T | 1700V | 2252I | 2263E | 2341P | 2374S | 2414K | 2456L | 3008L | |
| 10 | F | A | M | S | T | V | I | E | P | S | K | L | L |
| 13 | F | A | M | S | T | V | I | E | P | S | K | L | L |
| 20 | M | A/v* | |||||||||||
| 21 | F/l | T* | M/v | S | T | A/v* | V/i* | E/g | S* | S/f | K/T/n | M/l* | L/f |
| 31 | G* | I/t* | |||||||||||
| 32 | L* | T* | T* | G* | I/t* | V | V* | G* | S* | F* | E* | M* | F* |
| 33 | L* | T* | T/M* | G* | I/t* | V | V* | G* | S* | F* | E* | M* | F* |
The entire ORF sequence of HC-TN was determined at weeks 10, 13, 21, 32, and 33. Only subdomains were analyzed at weeks 20 and 31.
Amino acid positions correspond to those of pHC-TN. The sequence of pHC-TN is shown at the top. Dominant sequences recovered from the chimpanzee are shown in capital letters. Minor sequences recovered from the chimpanzee are shown in lowercase letters. Dominant amino acid changes are marked with an asterisk.
During the first 11 weeks, prior to the initial decrease in virus titers, the sequence for CH1581 remained unchanged, but at each subsequent week tested, new variants emerged (Tables 2 and 3; Fig. 3A). The mutation rates observed to occur between two subsequent time points thereafter ranged from 3.38 × 10−3 to 24.47 × 10−3 and 6.91 × 10−3 to 34.53 × 10−3 substitutions/site/year at the nucleotide and amino acid levels, respectively, and the ds/dn ratios were relatively low during the first year. Twenty amino acid changes were maintained by week 52. Only one of these, K2414E, was detected also in CH1422. The first four changes, including a change in p7, occurred at week 14. At week 18, five additional changes occurred in NS2, NS4B, NS5A, and NS5B. It is noteworthy that three changes observed at week 14 within the nonstructural proteins, in which the original sequence was present as a minor species, had reverted to the initial sequence at week 18. To rule out that these changes observed only at week 14 did not represent PCR or sequencing errors, we reamplified and sequenced the genome regions with these mutations from the week 14 sample, as well as from a week 13 sample (Table 3). All three changes were confirmed at week 14, and one change was present also at week 13. A virus with six additional mutations, in E2 (outside HVR1), p7, NS2, and NS5A, emerged at week 27. This included a new change at the p7 position that had occurred at week 14. During weeks 32 to 52, changes were observed at each time point analyzed. They were located within E2, p7, NS2, NS3, NS5A, and NS5B. They included a single HVR1 change at week 45.
Changes were not found in CH1579 during the first 13 weeks of follow-up (Table 4; Fig. 3B). The mutation rates observed thereafter were high, and the ds/dn ratios were low (Table 2). Twelve amino acid changes, located in E2 (within HVR1), p7, NS2, NS3, NS5A, and NS5B, had emerged by week 32, and the identical sequence was also present at week 33. A single mutation in NS5A (K2414E) was also identified in CH1422 and CH1581. Another NS5A mutation (P2341S) was found to occur in CH1581. Since CH1579 was infected from RNA transcripts of an infectious clone, these mutations could not have originated from the original source virus but evolved de novo in this animal. Four mutations, located in p7, NS5A, and NS5B, existed already at week 21; one mutation detected in NS4A changed to the original sequence at week 32. However, we found that this mutation was also present at week 20, confirming that it was not an artifact.
DISCUSSION
It is not known whether infection with particular HCV strains is associated with severe forms of acute hepatitis C. We previously reported that infection with strain HC-TN (genotype 1a) was associated with fulminant hepatic failure and, following transmission to a chimpanzee, caused unusually severe acute hepatitis (13). In the present study, however, two additional chimpanzees infected with strain HC-TN developed typical acute hepatitis, with peak ALT levels of 296 IU/liter and 90 IU/liter and minimal necroinflammatory changes in liver biopsy samples (Fig. 1). The ALT values were similar to the mean peak ALT of 215 ± 122 IU/liter (mean ± standard deviation) observed in >30 chimpanzees acutely infected with other genotype 1 strains (13). Thus, we could not confirm that strain HC-TN was more virulent than other strains in chimpanzees.
Virulence depends upon a complex interplay between the virus and the host and may be influenced by the dose of infecting virus, route of entry, and virus sequence, as well as by the immune status of the host. Virus dose or transmission route could have influenced the liver disease in the HC-TN-infected chimpanzees. However, Feinstone et al. (15) reported that there was no correlation between the infectious HCV dose of the inoculum and the peak ALT among experimentally infected chimpanzees. Furthermore, the course of infection did not differ in animals infected from RNA transcripts and from intravenous inoculation (8, 27, 28, 46).
Single nucleotide or amino acid changes in a virus genome can result in different levels of virulence, as reported, for example, for an amino acid change in the VP4 region of poliovirus (3). We demonstrated that the ORF sequence of virus recovered at peak viremia from CH1579, with mild hepatitis, was identical to the virus sequence recovered from CH1422, with severe hepatitis. It is possible that the sequences of the poly(U/UC) tract of the 3′ UTR, which vary in length and composition among different HCV isolates, differed among the viruses infecting the animals. However, the poly(UC) region of the RNA transcripts used to initiate infection in CH1579 was an exact match with the sequence recovered from CH1422. In fact, the infectious clone of strain HC-TN represents the first true consensus clone of HCV since it did not contain any nucleotide changes, perhaps with the possible exception of the length of the poly(U) stretch of the 3′ UTR.
The virus infecting the chimpanzee with severe hepatitis might have had a higher degree of heterogeneity (quasispecies) than those found in the chimpanzees infected with the lowest possible infectious dose and with the monoclonal virus. However, sequence analysis suggested that the virus recovered from the animal with severe hepatitis was very homogeneous. The courses of viral replication during the early acute phase of infection were very similar in the three animals, suggesting that the viruses infecting the chimpanzees had similar replication capacities. It is noteworthy that exposure to low doses of HCV or RNA transcripts from molecular clones, which did not result in detectable infection, were reported to have primed the host immune response in chimpanzees (22, 36). CH1422, with severe hepatitis, did not have such prior exposure, whereas CH1581 and CH1579, with typical hepatitis, both had such prior exposure. However, the intrahepatic T-cell responses in both of these animals appeared only after about 2 months of active infection. Yet, in CH1579 we did detect a weak peripheral T-cell response in the preinoculation samples.
Host immune responses are thought to determine the outcome of HCV infection. We recently demonstrated that neutralizing antibodies do not appear to play a role in the control of acute HCV in chimpanzees since they do not develop in animals with resolving infection (2, 29). CH1579 did not develop envelope or neutralizing antibodies even though the HC-TN infection resolved (29). A significant peripheral CD4+ T-cell response occurred much earlier in the animal that cleared the infection (CH1579) than in the animal that became chronically infected (CH1581), suggesting that early priming of the T-cell response may be important to the outcome. A significant response was detected at baseline in CH1579 and thus prior to inoculation, maybe reflecting priming through previous inoculations (see above). Both CH1581 and CH1579 developed HCV-specific intrahepatic antiviral CD4+ and CD8+ T-cell responses; the animal (CH1581) with persistence actually appeared to have an earlier appearance of these responses and in general the strength of these responses was greater than those detected in the animal (CH1579) with acute resolving infection (Fig. 3). However, CH1579 had extraordinarily strong CD4 and CD8 responses at weeks 23 and 33, respectively. It is possible that the intrahepatic CD8+ T-cell response, which was more vigorous during the low-titer period in CH1579 than during the corresponding period in CH1581, was efficient enough to eliminate the virus before escape mutants could establish a robust infection in CH1579, which was not the case with CH1581 (see below). The peripheral CD4+ T-cell response waned soon after viral clearance in CH1579, as did the intrahepatic CD4+ and CD8+ T-cell responses, but they all persisted in the chronically infected animal (CH1581), suggesting that persistence of these responses requires continuous antigen stimulation. It was recently reported that HCV-infected chimpanzees with acute resolving infection had an earlier initial decrease in virus titers than animals that developed a persistent infection (27). However, we found the opposite with CH1581 and CH1579 (Fig. 1).
We wondered whether differences in virus evolution in response to the host cellular immune response could explain the different outcomes for CH1579 and CH1581. The emergence of escape mutations in T-cell-targeted epitopes has been documented previously for HCV-infected chimpanzees (11, 42). However, these mutations were not analyzed in the context of coexisting mutations, since only small segments of the genome were sequenced. The cellular immune response against HCV is frequently targeted against multiple epitopes, and escape from a single epitope might not lead to persistence. We analyzed the entire polyprotein sequence of consecutive samples during the acute infection and correlated changes directly with the host humoral and cellular immune responses. In addition, we limited the possibility of selection of preexisting variants in the chimpanzees studied, since CH1581 was inoculated with the lowest possible infectious dose of polyclonal virus and CH1579 was transfected with RNA transcripts from an infectious clone and thus initially had a monoclonal infection. We did not detect any mutations in viruses recovered from CH1581 and CH1579 during the first 11 and 13 weeks of follow-up, respectively. In contrast, during the next 7 and 8 weeks of follow-up we detected six and five amino acid changes, respectively. Thus, despite a high rate of replication and an error-prone RNA-dependent RNA polymerase, mutations were not selected until the initial decrease in HCV titers. The accumulation of minor variants might occur during initial replication, and these variants could be selected by means of host immune pressure or replicative advantages, perhaps as second-site changes compensating for decreased replication fitness caused by other changes. Finally, changes might represent random coselected mutations. Our study does not demonstrate the specific mechanism for the development of mutations but rather a close temporal association with host cellular immune responses. We attempted to analyze the potential escape mechanism, but unfortunately the stored T cells could not be recovered sufficiently to perform the analysis.
Major et al. (28) studied the evolution of monoclonal H77 virus, another genotype 1a strain, in two chimpanzees that became persistently infected. Overall, the mutation rates observed for these animals were lower than those for the HC-TN-infected animals. Both animals developed mutations in p7; one of these mutations (M793V) was observed to occur also in the HC-TN-infected animal that became persistently infected. There was only one other common mutation in the two studies, L2456M, which occurred in the HC-TN-infected animal that cleared HCV. Finally, it should be noted that a similar pattern of development of mutations was observed in chimpanzees infected with monoclonal genotype 1b viruses (7, 40).
Recently, it was found that RNA transcripts from the full-length JFH1 genome (genotype 2a) produced viruses in human liver hepatoma cell lines (41, 49). The JFH1 strain was isolated from a patient with fulminant hepatitis, and it has been a question as to whether that fact was related to the unique ability to grow in cell culture. In contrast, wild-type, full-length HC-TN did not replicate in Huh7.5 cells even though it too was isolated from a patient with fulminant hepatitis. The same was reported previously for strain H77, but it was recently reported that a cell-culture-adapted H77 genome could produce viruses in Huh7.5 cells (48). Given that strains H77 and HC-TN belong to the same HCV subtype and are relatively closely related (Fig. 2), it is possible that these adaptive mutations would also permit replication of the HC-TN strain. Further studies are required to develop a cell culture system for the HC-TN strain.
In conclusion, we have developed an infectious clone of the HC-TN strain. The HC-TN sequence was infectious in vivo, but like other infectious clones of HCV genotype 1, this wild-type sequence was not infectious in Huh7-derived cells. Our in vivo study of the HC-TN strain demonstrates that virulence of HCV depends primarily upon host responses and not the particular virus strain. The cellular immune response against HCV precedes the initial decrease in virus titer and the development of acute hepatitis. The cellular immune responses did not appear to predict the final outcome of the infection, although differences in timing and magnitude of these responses might have played a role. The emergence of new virus variants, in the absence of neutralizing antibodies, is temporally associated with host cellular immune responses. This might play an important role in viral persistence. However, the accumulation of such variants does not assure viral persistence.
Acknowledgments
We thank Marisa St. Claire and Max Shapiro (Bioqual, Inc., Rockville, MD) for animal care and Ronald E. Engle (Hepatitis Viruses Section, National Institutes of Health) and Carola Steiger (Department of Molecular and Experimental Medicine, The Scripps Research Institute) for technical assistance. We thank Isa K. Mushahwar, Charles M. Rice, Takaji Wakita, Michael Houghton, and François-Loïc Cosset for providing reagents.
These studies were supported in part by NIH grant CA76403 (F.V.C.). R.T. was supported by a fellowship from the Cancer Research Institute, New York, and DFG grant TH 719/2-2 (Emmy Noether Program). This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Allander, T., A. Beyene, S. H. Jacobson, L. Grillner, and M. A. Persson. 1997. Patients infected with the same hepatitis C virus strain display different kinetics of the isolate-specific antibody response. J. Infect. Dis. 175:26-31. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J. 2004. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology 39:1469-1475. [DOI] [PubMed] [Google Scholar]
- 5.Bukh, J., C. L. Apgar, R. Engle, S. Govindarajan, P. A. Hegerich, R. Tellier, D. C. Wong, R. Elkins, and M. C. Kew. 1998. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: genetic analysis of the virus and generation of a standardized challenge pool. J. Infect. Dis. 178:1193-1197. [DOI] [PubMed] [Google Scholar]
- 6.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 7.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh, J., R. Thimme, S. Govindarajan, X. Forns, W. Satterfield, E. Eder, K.-M. Chang, M. Yanagi, S. U. Emerson, F. V. Chisari, and R. H. Purcell. 2002. Monoclonal hepatitis C virus infection in chimpanzees, p. 336-340. In H. S. Margolis, M. J. Alter, T. J. Liang, and J. L. Dienstag (ed.), Viral hepatitis and liver disease. International Medical Press, Atlanta, GA.
- 9.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, A. L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A. L. Hughes, and C. M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883-895. [DOI] [PubMed] [Google Scholar]
- 12.Farci, P., H. J. Alter, A. Shimoda, S. Govindarajan, L. C. Cheung, J. C. Melpolder, R. A. Sacher, J. W. Shih, and R. H. Purcell. 1996. Hepatitis C virus-associated fulminant hepatic failure. N. Engl. J. Med. 335:631-634. [DOI] [PubMed] [Google Scholar]
- 13.Farci, P., S. J. Munoz, A. Shimoda, S. Govindarajan, D. C. Wong, A. Coiana, G. Peddis, R. Rubin, and R. H. Purcell. 1999. Experimental transmission of hepatitis C virus-associated fulminant hepatitis to a chimpanzee. J. Infect. Dis. 179:1007-1011. [DOI] [PubMed] [Google Scholar]
- 14.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 15.Feinstone, S. M., H. J. Alter, H. P. Dienes, Y. Shimizu, H. Popper, D. Blackmore, D. Sly, W. T. London, and R. H. Purcell. 1981. Non-A, non-B hepatitis in chimpanzees and marmosets. J. Infect. Dis. 144:588-598. [DOI] [PubMed] [Google Scholar]
- 16.Forns, X., R. Thimme, S. Govindarajan, S. U. Emerson, R. H. Purcell, F. V. Chisari, and J. Bukh. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. USA 97:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334-339. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 21.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, U., T. Tuthill, H. C. Thomas, and J. Monjardino. 2000. Sequence, expression and reconstitution of an HCV genome from a British isolate derived from a single blood donation. J. Viral Hepat. 7:459-465. [DOI] [PubMed] [Google Scholar]
- 24.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesniewski, R. R., S. Watanabe, and S. G. Devare. 1995. Expression of HCV envelope proteins and the serological utility of the anti-E2 immune response. Princess Takamatsu Symp. 25:129-137. [PubMed] [Google Scholar]
- 26.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Major, M. E., H. Dahari, K. Mihalik, M. Puig, C. M. Rice, A. U. Neumann, and S. M. Feinstone. 2004. Hepatitis C virus kinetics and host responses associated with disease and outcome of infection in chimpanzees. Hepatology 39:1709-1720. [DOI] [PubMed] [Google Scholar]
- 28.Major, M. E., K. Mihalik, J. Fernandez, J. Seidman, D. Kleiner, A. A. Kolykhalov, C. M. Rice, and S. M. Feinstone. 1999. Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J. Virol. 73:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muto, Y., J. Sugihara, H. Ohnishi, H. Moriwaki, and K. Nishioka. 1990. Anti-hepatitis C virus antibody prevails in fulminant hepatic failure. Gastroenterol. Jpn. 25:32-35. [DOI] [PubMed] [Google Scholar]
- 31.Nakao, H., H. Okamoto, H. Tokita, T. Inoue, H. Iizuka, G. Pozzato, and S. Mishiro. 1996. Full-length genomic sequence of a hepatitis C virus genotype 2c isolate (BEBE1) and the 2c-specific PCR primers. Arch. Virol. 141:701-704. [DOI] [PubMed] [Google Scholar]
- 32.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 33.Okamoto, H., N. Kanai, and S. Mishiro. 1992. Full-length nucleotide sequence of a Japanese hepatitis C virus isolate (HC-J1) with high homology to USA isolates. Nucleic Acids Res. 20:6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto, H., K. Kurai, S. Okada, K. Yamamoto, H. Lizuka, T. Tanaka, S. Fukuda, F. Tsuda, and S. Mishiro. 1992. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology 188:331-341. [DOI] [PubMed] [Google Scholar]
- 35.Seeff, L. B., F. B. Hollinger, H. J. Alter, E. C. Wright, C. M. Cain, Z. J. Buskell, K. G. Ishak, F. L. Iber, D. Toro, A. Samanta, R. L. Koretz, R. P. Perrillo, Z. D. Goodman, R. G. Knodell, G. Gitnick, T. R. Morgan, E. R. Schiff, S. Lasky, C. Stevens, R. Z. Vlahcevic, E. Weinshel, T. Tanwandee, H. J. Lin, and L. Barbosa. 2001. Long-term mortality and morbidity of transfusion-associated non-A, non-B, and type C hepatitis: a National Heart, Lung, and Blood Institute collaborative study. Hepatology 33:455-463. [DOI] [PubMed] [Google Scholar]
- 36.Shata, M. T., N. Tricoche, M. Perkus, D. Tom, B. Brotman, P. McCormack, W. Pfahler, D. H. Lee, L. H. Tobler, M. Busch, and A. M. Prince. 2003. Exposure to low infective doses of HCV induces cellular immune responses without consistently detectable viremia or seroconversion in chimpanzees. Virology 314:601-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson, M., M. Nascimbeni, S. Gonzales, K. K. Murthy, B. Rehermann, and T. J. Liang. 2001. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology 121:1226-1233. [DOI] [PubMed] [Google Scholar]
- 41.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner, A., A. L. Erickson, J. Kansopon, K. Crawford, E. Muchmore, A. L. Hughes, M. Houghton, and C. M. Walker. 1995. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. USA 92:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, T. L., H. Hsu, E. Donegan, S. Feinstone, H. Greenberg, A. Read, N. L. Ascher, J. P. Roberts, and J. R. Lake. 1991. Hepatitis C virus not found in fulminant non-A, non-B hepatitis. Ann. Intern. Med. 115:111-112. [DOI] [PubMed] [Google Scholar]
- 44.Yanagi, M., S. Kaneko, M. Unoura, S. Murakami, K. Kobayashi, J. Sugihara, H. Ohnishi, and Y. Muto. 1991. Hepatitis C virus in fulminant hepatic failure. N. Engl. J. Med. 324:1895-1896. [DOI] [PubMed] [Google Scholar]
- 45.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 46.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 48.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zibert, A., H. Meisel, W. Kraas, A. Schulz, G. Jung, and M. Roggendorf. 1997. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology 25:1245-1249. [DOI] [PubMed] [Google Scholar]