Abstract
Since an outbreak of severe acute respiratory syndrome (SARS) was averted in 2004, many novel coronaviruses have been recognized from different species, including humans. Bats have provided the most diverse assemblages of coronaviruses, suggesting that they may be the natural reservoir. Continued virological surveillance has proven to be the best way to avert this infectious disease at the source. Here we provide the first description of a previously unidentified coronavirus lineage detected from wild Asian leopard cats (Prionailurus bengalensis) and Chinese ferret badgers (Melogale moschata) during virological surveillance in southern China. Partial genome analysis revealed a typical coronavirus genome but with a unique putative accessory gene organization. Phylogenetic analyses revealed that the envelope, membrane, and nucleoprotein structural proteins and the two conserved replicase domains, putative RNA-dependent RNA polymerase and RNA helicase, of these novel coronaviruses were most closely related to those of group 3 coronaviruses identified from birds, while the spike protein gene was most closely related to that of group 1 coronaviruses from mammals. However, these viruses always fell into an outgroup phylogenetic relationship with respect to other coronaviruses and had low amino acid similarity to all known coronavirus groups, indicating that they diverged early in the evolutionary history of coronaviruses. These results suggest that these viruses may represent a previously unrecognized evolutionary pathway, or possibly an unidentified coronavirus group. This study demonstrates the importance of systematic virological surveillance in market animals for understanding the evolution and emergence of viruses with infectious potential.
Since the severe acute respiratory syndrome (SARS) outbreak in early 2003, many new viruses of the family Coronaviridae have been identified in diverse host species, including birds, humans, and other mammals (7, 12-14, 18, 25, 27, 28, 29). Of these hosts, bats have provided the highest genetic diversity of novel coronaviruses (CoVs) (23, 29). Analyses of the genomic organization and phylogenetic relationships of CoVs from bats and other host species indicate that bats are the likely natural reservoirs for all known CoVs from other hosts (23, 26). This research also suggests that CoVs resident in bats may be introduced into other animals and subsequently become established in these hosts.
Interspecies transmission of CoV from its natural reservoir to another species might result in disease outbreaks in naive hosts. The emerging pathway of SARS CoV in humans and market civets is an example of this (4, 5). Accumulated information has demonstrated that many SARS-like CoVs are harbored in bat species (12, 13, 23). However, the cross homologies between civet and human SARS CoV and bat SARS-like CoVs are still relatively low (about 92%), even in the most conserved regions of the replicase gene (13). Phylogenetic analyses and divergence dates of these CoVs indicate that the likely intermediate host of SARS CoV has still not been determined (26). Thus, the evolutionary pathway of SARS CoV has still not been fully determined.
The SARS outbreak in humans abruptly showed that previously unknown CoVs resident in wild animals are capable of interspecies transmission and can cause severe disease in humans within a short period of time (4, 5, 17, 26). Reviewing the emergence of SARS in China shows that this outbreak was caused by the evolution of the CoV ecosystem, i.e., many exotic wild species being farmed and brought into live-animal markets located in urban areas (5). Of concern is that this ecosystem has not been affected despite the implementation of a ban on the sale of wild animals in Guangdong province soon after a second SARS outbreak was averted in early 2004 (5). Different wild-animal species, including farmed or untamed wild animals, are still being smuggled and sold in many regions within China, especially in the south. This raises the risk of the reemergence of SARS and the emergence of other infectious diseases.
To further understand the evolutionary pathway of SARS CoV, and to detect possible emerging diseases, systematic virological surveillance of wild animals has been conducted at illegal-trade live-animal markets from 2005 onward. A variety of CoVs have been detected in the wild animals sampled. Here we report the detection of a novel and relatively ancient CoV. Phylogenetic analyses revealed that the major replication and structural genes of this novel CoV were most closely related to group 3 CoVs, represented by avian infectious bronchitis virus, while the spike protein gene clustered with group 1 CoVs from mammals. However, the viruses were genetically highly divergent and fell into an outgroup position with respect to all of the known CoV groups in all of the genes tested, indicating that they diverged early in the evolutionary history of CoVs. As this novel CoV was detected in two different species of wild animal at the same market on the same sampling occasion, it is possible that these CoVs were recently transmitted from other unknown species, raising concern about further interspecies transmission to other species, including humans.
MATERIALS AND METHODS
Sampling.
Since February 2005, illegally traded wild animals were sampled systematically at live-animal markets in Guangdong and Guangxi Provinces, China. Rectal swabs were collected from 24 species of wild mammals. Specimens were placed in transport medium and kept in liquid nitrogen for transportation to the laboratory and then stored at −80°C.
Virus detection and isolation.
Viral RNA was extracted from the rectal swab material with the QIAamp viral RNA minikit (QIAGEN, Westburg, The Netherlands) and used as the template for reverse transcription (RT)-PCR detection of the CoV putative RNA-dependent RNA polymerase (RdRp) domain of the replicase gene as previously described (23). Primers conserved for all known CoVs were designed for RT-PCR detection (23). The sequences of the primers used are available upon request. The RdRp PCR products were gel purified with the QIAquick PCR purification kit (QIAGEN) and sequenced to confirm virus detection. The sequences of the primers used are available upon request. Virus isolation was attempted for representatives of the PCR-positive specimens; however, this was unsuccessful with multiple cell lines (FRHK4, Vero E6, and CV1), as previously described (23). Consequently, our investigations focused on decoding the genomes of the CoVs from the different positive samples.
Genome analysis.
The nucleotide data from sequencing of the RdRp diagnostic PCR fragments were analyzed by using the CoV sequences available in GenBank to determine the diversity of the CoVs detected. This analysis identified three CoVs from a previously unidentified lineage that were selected for genome sequencing. RNA extraction, cDNA amplification, and sequencing were conducted as previously described (23).
Open reading frames (ORFs) of the partial genome of the novel CoV were identified with the program fgenesV (Softberry Inc., Mount Kisco, NY) and mapped with SeqBuilder (Lasergene version 7; DNAStar, Madison, WI). Homology searches of identified ORFs against other known CoVs were conducted in GenBank.
Sequence similarity.
To further assess the genomic similarity of the novel CoVs with other major lineages of CoVs, the full-length amino acid alignments of each of the major gene products were used to calculate their similarity (p distances) with MEGA3 (10). The virus sequences used in this analysis are the same as those in the phylogenetic trees.
Phylogenetic analysis.
To evaluate the phylogenetic relationship of this novel CoV, the helicase (HEL) domain of the replicase gene and the spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) genes were chosen for analysis. Nucleotide sequence alignments were carried out with TransAlign (1) by using ClustalX (24) to conserve codon positions and manually optimized wherever necessary with Se-Al version 2.0 (19). All ambiguously aligned regions were removed prior to phylogenetic analyses. Phylogenetic trees were built by neighbor joining in PAUP* version 4.0b (22) with the appropriate model of sequence evolution selected by MrModeltest version 2 (16). The same model of evolution was used to calculate Bayesian posterior probability in MrBayes version 3.1 (6). Gaps were treated as missing data in all analyses. Multiple sequence alignments of all data sets have been deposited in TreeBASE (www.treebase.org).
Currently, the International Committee on Taxonomy of Viruses classifies the members of the family Coronaviridae into three groups (2, 3). However, due to the increasing diversity of the family Coronaviridae, in this study we used a system that includes putative groups 4 and 5, as previously described (23, 26).
Detection of recombination.
The Genetic Algorithm for Recombination Detection in the program package HYPHY was used to determine if this novel CoV contains recombinant genes (8, 9). It must be noted that recombination analysis was conducted only within gene segments. Breakpoints identified within the gene sequence were assessed by testing the congruence of neighbor-joining trees generated by the Genetic Algorithm for Recombination Detection for each fragment with the Shimodaira-Hasegawa test in PAUP* (20, 22).
Nucleotide sequence accession numbers.
The sequences generated in this study have been deposited in GenBank under accession numbers EF584902 to EF584914.
RESULTS
Surveillance and prevalence.
Of the 24 species tested, 8 were CoV positive. However, only Asian leopard cats (Prionailurus bengalensis; positivity rate, 2.4%) and Chinese ferret badgers (Melogale moschata; positivity rate, 1.1%) regularly tested positive for CoV on multiple sampling occasions (Table 1). Furthermore, these species were present almost year round in the live-animal markets and could be sampled each month (Table 1). The remaining six species that tested positive for CoV on just a single occasion each were Mustela kathiah (yellow-bellied weasel), M. sibirica (Siberian weasel), Paguma larvata (masked palm civet), Rhizomys sinensis (Chinese bamboo rat), Viverricula indica (lesser Indian civet), and Petaurista sp. (flying squirrel). It is noteworthy that most of the positive samples were obtained in the winter. This corresponds to a marked increase in the number of animals and species available in these markets due to increased consumer demand in the winter (Table 1).
TABLE 1.
Detection of CoV in specimens from Asian leopard cats and Chinese ferret badgers in illegal live-animal markets
| Yr and mo | No. of samples positive/no. tested (% positive)
|
|
|---|---|---|
| Asian leopard cat (P. bengalensis) | Chinese ferret badger (M. moschata) | |
| 2005 | ||
| February | 0/34 | 0/7 |
| March | 0/59 | 0/41 |
| April | 0/84 | 2/87 |
| May | 0/31 | 0/19 |
| June | 0/24 | 0/6 |
| July | 0/37 | 0/0 |
| August | 0/39 | 0/4 |
| September | 0/41 | 0/44 |
| October | 0/38 | 0/136 |
| November | 2/149 | 1/206 |
| December | 14/245 | 6/128 |
| 2006 | ||
| January | 16/308 | 0/78 |
| February | 2/219 | 0/71 |
| March | 1/145 | 2/107 |
| Total | 35/1,453 (2.4) | 11/934 (1.1) |
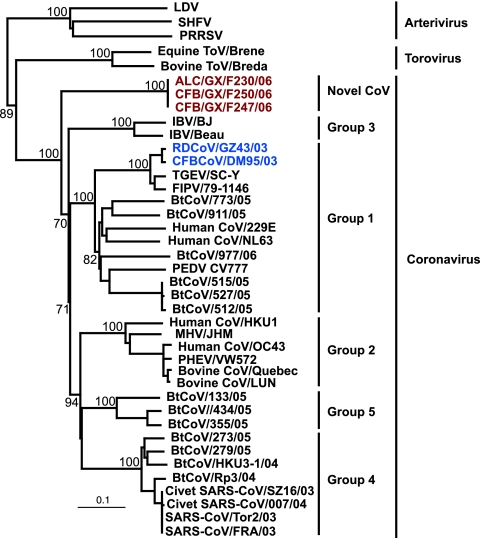
Among the positive samples, preliminary sequence analysis of the RdRp fragment obtained by RT-PCR detection revealed that all but three of the CoVs detected from wild animals shared high genetic homology to previously identified CoVs from wild animals, represented by the Chinese ferret badger/Guangdong/DM95/2003 (CFB/GD/DM95/03), which belonged to group 1 (Fig. 1). However, three viruses, Asian leopard cat/Guangxi/F230/2006 (ALC/GX/F230/06), Chinese ferret badger/Guangxi/F247/2006 (CFB/GX/F247/06), and Chinese ferret badger/Guangxi/F250/2006 (CFB/GX/F250/06), formed a unique phylogenetic group most closely related to group 3 CoVs (Fig. 1). It is noteworthy that these three viruses were the outgroup with respect to all other known CoVs. However, comparison with Arterivirus and Torovirus clearly indicated that these three viruses belong to the family Coronaviridae, suggesting a previously unidentified lineage of this virus family (Fig. 1). In the present study, we therefore focused on decoding the evolutionary history of this unknown CoV lineage, represented by ALC/GX/F230/06, by using the available CoV sequences.
FIG. 1.
Phylogenetic relationships of the RdRp genes of novel CoVs isolated from wild animals in China. The tree was generated by the neighbor-joining method in the PAUP* program. Analyses were based on 440 nucleotides of the RdRp. Values above branches are bootstrap values from 1,000 replicates. The tree was rooted to Okavirus (accession no. AF227196). Scale bar, 0.1 substitution per site. Red text indicates viruses characterized in this study. Abbreviations: LDV, lactate dehydrogenase-elevating virus; SHFV, simian hemorrhagic fever virus; PRRSV, porcine respiratory and reproductive syndrome virus; ToV, torovirus; IBV, infectious bronchitis virus; TGEV, transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; PEDV, porcine epidemic diarrhea virus; MHV, murine hepatitis virus; PHEV, porcine hemagglutinating encephalomyelitis virus; BtCoV, bat coronavirus.
Genome organization.
As the novel lineage is phylogenetically most closely related to either group 1 or group 3 CoVs (see below), a comparison of the partial genome organization of ALC/GX/F230/06 and representative strains of these groups is presented in Fig. 2. The virus ALC/GX/F230/06 had a typical CoV genome order, with the exception of ORF1a (data not available). The remaining genome was arranged in the order 5′-ORF1b, S, E, M, and N. However, the organization of putative accessory genes in ALC/GX/F230/06 is unique among CoVs. In particular, ORF3, which is present in all known CoVs, was not identified in ALC/GX/F230/06 (Fig. 2). However, ORF6 (94 amino acids), between the M and N genes, and ORF7 (200 amino acids), overlapping the N gene farther downstream, were found in the ALC/GX/F230/06 genome (Fig. 2).
FIG. 2.
Schematic representation of the ORFs of Asian leopard cat/Guangxi/F230/2006 and representative group 1 and group 3 CoVs. Putative accessory proteins are indicated by gray boxes. For definitions of abbreviations, see the legend to Fig. 1.
Sequence similarity.
To understand the similarity of the novel CoV to other known CoVs, we compared the major genes for amino acid sequence similarity (Table 2). The virus ALC/GX/F230/06 had the highest homology with group 3 in the HEL domain and the E and N genes and the highest homology with group 1 in the S gene (Table 2). However, the M gene of ALC/GX/F230/06 was similar to those of both group 2 and group 3 viruses (Table 2). As the similarities between ALC/GX/F230/06 and other CoV groups were much lower than those between groups, this virus may represent a previously unidentified CoV group (23). It is noteworthy that the RdRp domain and S gene of the three novel CoVs shared 100% amino acid homology (data not shown), suggesting that they were recently introduced from a common source.
TABLE 2.
Amino acid similarities between Asian leopard cat/Guangxi/F230/2006 and other known CoVs
| Gene product | Similarity (p distance)a
|
||||
|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | |
| Helicase | 50.5 | 45.8 | 57.6 | 47.7 | 46.1 |
| Spike | 44.4 | 25.9 | 29.3 | 24.9 | 26 |
| Envelope | 14.4 | 14.1 | 24.4 | 18.7 | 15.8 |
| Membrane | 22.9 | 25.7 | 25.3 | 22.5 | 24.9 |
| Nucleocapsid | 19.4 | 24.9 | 33.7 | 26.5 | 22.2 |
Full-length amino acid sequence alignments of each of the major gene products were used to calculate similarities with the program MEGA3. The alignments are the same as those used to generate the phylogenetic trees in Fig. 3 and 4 and have been deposited in TreeBase (http:/treebase.org).
Phylogenetic analysis.
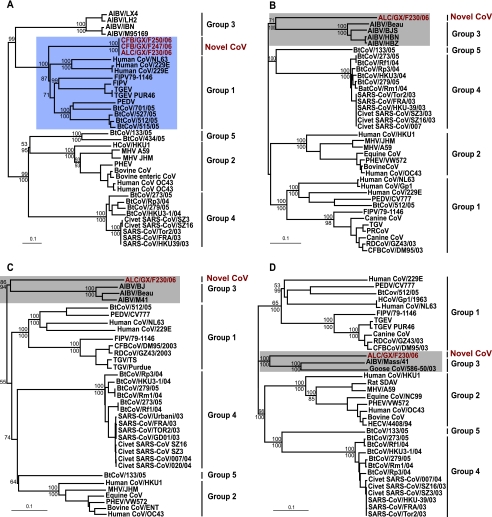
All of the major genes, including the S, E, M, and N genes, along with the HEL domain, were phylogenetically analyzed (Fig. 3 and 4). It is interesting that the S gene of ALC/GX/F230/06 clustered with group 1 (Fig. 3A), while the HEL domain and the E, M, and N genes clustered with group 3 CoVs (Fig. 3B to D and 4). Moreover, for all of the genes analyzed, ALC/GX/F230/06 is highly divergent and consistently falls into an outgroup position with respect to all known CoV groups. Due to the consistent grouping of the ALC/GX/F230/06 genes with those of group 3 CoVs, except for the S gene, it is very likely that they have an ancestor in common. This novel CoV is the first recognized in a mammalian host that has genes phylogenetically associated with those of group 3. The difference in the phylogeny of the S gene and the remaining genes is difficult to interpret and may be due to the early divergence of the virus, recombination, or possibly host adaptation.
FIG. 3.
Phylogenetic relationships of the spike (A), envelope (B), membrane (C), and nucleocapsid (D) protein genes of novel CoVs isolated from wild animals in China. Trees were generated by the neighbor-joining method in the PAUP* program. Values above and below branches are neighbor-joining bootstrap values from 1,000 replicates and Bayesian posterior probabilities, respectively. Analyses were based on 2,802, 339, 831, and 1,692 nucleotides of the spike, envelope, membrane, and nucleocapsid protein genes, respectively. The trees were midpoint rooted. Scale bar, 0.1 nucleotide substitution per site. Red text indicates viruses characterized in this study. For definitions of abbreviations, see the legend to Fig. 1.
FIG. 4.
Phylogenetic relationship of the complete HEL domain of the replicase gene of the novel CoVs isolated from wild animals in China. Trees were generated by the neighbor-joining method in the PAUP* program. Values above and below branches are neighbor-joining bootstrap values from 1,000 replicates and Bayesian posterior probabilities, respectively. Analyses were based on 1,797 nucleotides. The tree was midpoint rooted. Scale bar, 0.1 nucleotide substitution per site. Red text indicates viruses characterized in this study. For definitions of abbreviations, see the legend to Fig. 1.
Detection of recombination.
Recombination breakpoints were identified in the HEL domain and S and N gene segments (data not shown). However, the Shimodaira-Hasegawa test between the putatively recombinant segments of the genes showed that these alternate topologies were not statistically supported. This may indicate that these potentially recombinant areas are due to substitution rate variation and not recombination between CoVs (23). Furthermore, the phylogenetic position of ALC/GX/F230/06 did not change in the phylogenies derived from these potentially recombinant sections (data not shown).
DISCUSSION
This report describes the detection of novel CoVs from Asian leopard cats (P. bengalensis) and Chinese ferret badgers (M. moschata) in live-animal markets in southern China. Genetic analyses indicate that these viruses have a typical CoV genome organization (11, 15, 21, 23, 28) but a unique phylogeny among all known CoVs (2, 3, 23, 26). All of the genes of the viruses tested were most closely related to those of group 3 CoVs, except the S gene, which was related to that of viruses in group 1. However, the viruses always fell into an outgroup phylogenetic relationship to other CoVs and had low amino acid similarity to all known CoV groups. Taken together, these findings suggest that the novel CoVs represent a previously unrecognized evolutionary pathway or possibly an unidentified CoV group.
The inconsistent phylogenies of the genes from the novel CoVs may be easily interpreted as a result of a recombination event between group 1 and 3 viruses. However, the phylogenetic outgroup relationships in each of the genes analyzed indicate that these viruses diverged at an early stage in the evolutionary history of the family Coronaviridae, making the detection of recombination impossible in the absence of background genetic data. This situation may therefore provide insight into the early diversification of the family Coronaviridae.
As these novel viruses were detected in two different wild-animal species on the same sampling occasion at the same market, and given the 100% homology of the RdRp domain and the S gene, it is likely that these animals were infected very recently in the market from an unknown source. This hypothesis is also supported by the fact that both species were regularly sampled in the market and frequently tested positive for only group 1 CoVs, but these novel CoVs were detected just on this occasion. However, the available genetic and epidemiological information was insufficient to determine the ecology of this virus and it is impossible to determine the source of infection.
Previous studies suggested that bats are the natural reservoir of SARS CoV (12, 13, 23). In addition, a study of population dynamics indicated that CoVs in bats had constant population growth, while viruses from all other hosts had an epidemic-like population increase (26). It was therefore suggested that the hypothetical ancestor of all CoVs might be derived from viruses resident in bats (25). Previous studies have also shown that diverse CoVs are endemic in different bat species and occasionally introduced into other animals and became established in other hosts (23, 26, 29). It is possible that bats also harbor the counterpart of the novel CoVs detected in the present study. However, we cannot exclude the possibility that there is another source from other wild mammalian populations. Among all CoVs, only group 3 CoVs are found in avian hosts (7, 11, 14). The outgroup relationship of the novel CoV with infectious bronchitis virus provides indirect evidence that group 3 viruses may be derived from those resident in mammals, as previously suggested (26).
The emergence of SARS resulted from the CoV ecology created in live-animal markets in southern China through the housing of many exotic wild-animal species together and by facilitating increased contact with human populations (5). The present study therefore provides a new example of a possible pathway for the emergence of a potential human pathogen. The best way to prevent potential zoonotic infectious diseases is to understand the risk posed by increased human exposure to different animal species within the complex ecosystem of a live-animal market population. While this report was being prepared, many wild animals were still being smuggled and sold in illegal live-animal markets throughout this region, and the possibility remains for continued interspecies transmission to humans. Historically, with the exception of SARS CoV, the independent introductions of the four established human CoVs have not been detected (26). The successful prevention of a second SARS outbreak in early 2004 resulted from early detection of the virus in live-animal markets in southern China (5). This study demonstrates the importance of systematic virological surveillance in market animals for understanding the evolution and emergence of viruses with infectious potential.
Acknowledgments
This work was supported by the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government (no. 03040782), the National Special Task Force Fund for Identification of the Animal Reservoir of SARS CoV, the State Key program (no. 2005CB523004) of the Ministry of Science and Technology, China, and the Li Ka Shing Foundation.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Bininda-Emonds, O. R. P. 2005. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjunes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya, A. E., E. J. Snijder, and W. J. M. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 5.Guan, Y., N. S. Zhong, H. Chen, G. J. D. Smith, and B. J. Zheng. 2006. An averted SARS outbreak, p. 93-100. In J. C. K. Chan and V. C. W. Taam Wong (ed.) Challenges of severe acute respiratory syndrome. Elsevier, New York, NY.
- 6.Huelsenbeck, P. J., and F. R. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 7.Jonassen, C. M., T. Kofstad, I.-L. Larsen, A. Lovland, K. Handeland, A. Follestad, and A. Lillehaug. 2005. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 86:1597-1607. [DOI] [PubMed] [Google Scholar]
- 8.Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk, and S. D. W. Frost. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891-1901. [DOI] [PubMed] [Google Scholar]
- 9.Kosakovsky Pond, S. L., S. D. W. Frost, and S. V. Muse. 2005. Hyphy: hypothesis testing using phylogenetics. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 10.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 11.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviruses, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 12.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. E. Field, P. Daszak, B. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 14.Liu, S., J. Chen, J. Chen, X. Kong, Y. Shao, Z. Han, L. Feng, X. Cai, S. Gu, and M. Liu. 2005. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas). J. Gen. Virol. 86:719-725. [DOI] [PubMed] [Google Scholar]
- 15.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffinth, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petresue, A. G. Tobertson, J. E. Schein, S. Siddiqui, D. E. Smailus, J. M. Stott, and G. S. Yang. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 16.Nylander, J. A. A. 2004. MrModelTest v2. Distributed by the author. Evolutionary Biology Center, University of Uppsala, Uppsala, Sweden.
- 17.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, R. W. Yung, T. K. Ng, K. Y. Yuen, and the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon, L. L. M., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. C. Leung, S. K. Lau, P. C. Y. Woo, K. Y. Suen, K. Y. Yeun, Y. Guan, and J. S. M. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambaut, A. 1996. Se-Al: sequence alignment editor. http://evolve.zoo.ox.ac.uk/.
- 20.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 21.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineages. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, MA.
- 23.Tang, X. C., J. X. Zhang, S. Y. Zhang, P. Wang, X. H. Fan, L. F. Li, G. Li, B. Q. Dong, W. Liu, C. L. Cheung, K. M. Xu, W. J. Song, D. Vijaykrishna, L. L. M. Poon, J. S. M. Peiris, G. J. D. Smith, H. Chen, and Y. Guan. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijaykrishna, D., G. J. D. Smith, J. X. Zhang, J. S. M. Peiris, H. Chen, and Y. Guan. 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 81:4012-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo, P. C. Y., S. K. P. Lau, C.-M. Chu, K.-H. Chan, H.-W. Tsoi, Y. Huang, B. H. L. Wong, R. W. S. Poon, J. J. Cai, W.-K. Luk, L. L. M. Poon, S. S. Y. Wong, Y. Guan, J. S. M. Pieris, and K.-Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo, P. C. Y., S. K. P. Lau, K. S. M. Li, R. W. S. Poon, B. H. L. Wong, H.-W. Tsoi, B. C. K. Yip, Y. Huang, K.-H. Chan, and K. Y. Yuen. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180-187. [DOI] [PMC free article] [PubMed] [Google Scholar]