Abstract
The Tyr-X-X-Leu (YxxL) motif of the vaccinia virus F13L protein was examined for late (L) domain activity. The ability of an F13L deletion virus to form plaques was restored by PCR products containing single alanine substitutions within the motif and a YAAL construct but not by constructs lacking both the Y and L residues. Recombinant viruses possessing alanine substitutions in place of the tyrosine or the leucine residue in the YxxL motif demonstrated small, asymmetrical plaques. RNA interference-dependent depletion of Alix and TSG101 (host proteins involved in L domain-dependent protein trafficking) diminished extracellular enveloped virion production to various degrees, suggesting that the YxxL motif is a genuine L domain.
Viral late assembly domains (L domains) are tetrapeptide motifs thought to mediate the assembly and egress of viral particles. The Pro-Thr-Ala-Pro (PTAP) motif was first identified in the p6 Gag protein of human immunodeficiency virus type 1 (HIV-1) as a requirement for viral budding (14, 22). Since then, two additional retroviral L domain motifs, Pro-Pro-X-Tyr (PPxY) and Tyr-X-X-Leu (YxxL), have been discovered (reviewed in reference 11). Mutation of these motifs reduces secretion of HIV particles in certain cell types by arresting virus budding through the plasma membrane. Electron microscopy shows virus particles tethered to the plasma membrane on “stalk-like” structures, consistent with a defect in the assembly/budding process. Additional studies have revealed several cellular components involved in protein trafficking that interact with L domains, suggesting that the assembly of virus particles utilizes established host pathways for egress (reviewed in references 10 and 30). Furthermore, it is now accepted that L domains are likely important for viruses outside the Lentiviridae family as well (8, 9, 32, 34, 39).
Vaccinia virus (VV), a member of the Orthopoxvirus genus, is among the largest of the DNA viruses. During replication, VV undergoes three distinct stages of gene expression, the products of which are referred to as early, intermediate, and late proteins. It is mainly the late proteins that provide the virion with structural elements, the trafficking and assembly of which are regulated by modifications such as acylation, myristoylation, and palmitoylation as well as processing by host cell and virally encoded proteases (1, 5, 7, 16, 17, 18, 21, 26, 42).
Mature VV assumes four infectious forms: the intracellular mature virus (IMV), the intracellular enveloped virus (IEV), the cell-associated enveloped virus (CEV), and the extracellular enveloped virus (EEV). Both CEV and EEV originate from IEV, which are IMV particles swathed in membranes derived from trans-Golgi or endosomal cisternae (reviewed in references 36 and 37). The major antigen located on the surface of IEV is F13L, the product of the F13L open reading frame (ORF) (3, 19, 20). F13L is a 372-amino-acid palmitoylprotein (15) that contains a centrally located YxxL L domain-like motif (residues 153 to 156). F13L has been shown to be involved in viral envelopment and egress (15). The YPPL sequence is 100% conserved in the F13L protein of all strains of VV, as well as in the F13L homologs within the Orthopoxvirus genus. It is also highly conserved in F13L homologs throughout the entire Poxviridae family (Table 1), suggesting that this domain may convey an essential biological advantage to the virus.
TABLE 1.
Viruses in which the YxxL motif is conserveda
| Virus | Motif residue position | Conserved motif |
|---|---|---|
| Orthopoxviruses | ||
| Vaccinia virus | 153-156 | VYSDYPPLATDL |
| Variola virus | 153-156 | VYSDYPPLATDL |
| Camelpox virus | 153-156 | VYSDYPPLATDL |
| Monkeypox virus | 153-156 | VYSDYPPLATDL |
| Cowpox virus | 153-156 | VYSDYPPLATDL |
| Taterapox virus | 153-156 | VYSDYPPLATDL |
| Other poxviruses | ||
| Squirrelpox virus | 153-156 | LYSEYAPLARDL |
| Myxoma virus | 153-156 | VYSTYAPLAADL |
| Lumpy skin disease virus | 154-157 | IYSTYAPLALDL |
| Sheeppox virus | 154-157 | IYSTYAPLALDL |
| Swinepox virus | 154-157 | IYSTYKPLATDL |
| Yaba-like disease virus | 153-156 | IYSDYPPLASDL |
| Fowlpox virus | 162-166 | MDLYFRSLDYKI |
| Canarypox virus | 186-189 | LATQYHLLKSHN |
| Bovine papular stomatitis virus | 215-218 | FLGFYRTLDEDL |
| Orf virus | 215-218 | FLGFYRTLDEDL |
| Crocodilepox virus | 162-165 | RFGDYLALARRG |
| Deerpox virus | 157-160 | IYSTFPPLAIDL |
| 218-221 | ILGFYRTLDADV |
Conserved motifs are underlined.
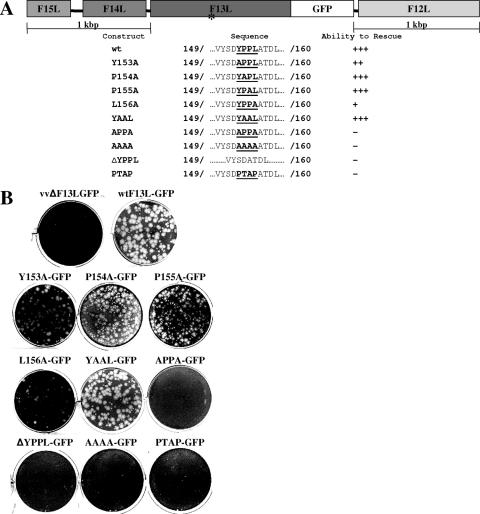
In the present study, we sought to determine whether the tetrapeptide motif located within the VV strain WR F13L protein functions in an L domain-like capacity. VV lacking a functional F13L ORF do not produce EEV and therefore form very small plaques over an extended incubation period (reviewed in reference 31). Thus, the introduction of a PCR product containing a functional F13L ORF into cells infected with an F13L deletion mutant virus (vvΔF13LGFP) should restore the ability of the virus to form plaques. In order to determine the requirement for the YxxL motif, we designed a trans complementation assay in which BSC40 cells were infected with 150 PFU/well of vvΔF13LGFP (the F13L ORF was inactivated by the insertion of green fluorescent protein [GFP] at nucleotide 19) for 1 h at 37°C. Subsequently, the viral inoculum was replaced with fresh medium, and cells underwent liposome-mediated transfection overnight with a PCR product encoding an F13L-GFP fusion protein containing a wild-type or a mutant YxxL motif (Fig. 1A). The medium was then removed and replaced with fresh growth medium containing 1% methylcellulose. At 3 days postinfection, cells were fixed with 5% glutaraldehyde and stained with 0.1% crystal violet. All constructs included a 1-kbp region of flanking DNA on either side of the fusion to allow for transient expression of the fusion protein and to facilitate recombination between the F13L deletion mutant virus and the PCR product. Results from this trans complementation assay demonstrated that the capability of the deletion mutant to form plaques was restored by the wild-type PCR product as well as PCR products containing mutations Y153A, P154A, P155A, and, to a lesser degree, L156A (Fig. 1B). A double-proline mutant also demonstrated wild-type levels of rescue. In contrast, no appreciable rescue was observed for constructs lacking both the Y and L residues as well as for constructs containing the deletion of the YxxL motif. Replacement of the YxxL motif with PTAP also failed to rescue.
FIG. 1.
trans complementation of an F13L deletion mutant virus. (A) Panel of F13L PCR products. *, the approximate location of the YxxL motif within F13L. (B) Six-well plates containing BSC40 cells seeded at a density of 2 × 105 cells/well were infected with an F13L deletion mutant virus for 1 h and then transfected with a PCR product containing one of several F13L YxxL mutations. Twenty-four hours postinfection, the transfection inoculum was replaced with medium containing 1% methylcellulose and incubated for an additional 48 h. The cells were fixed, and plaques were visualized by staining with 0.1% crystal violet.
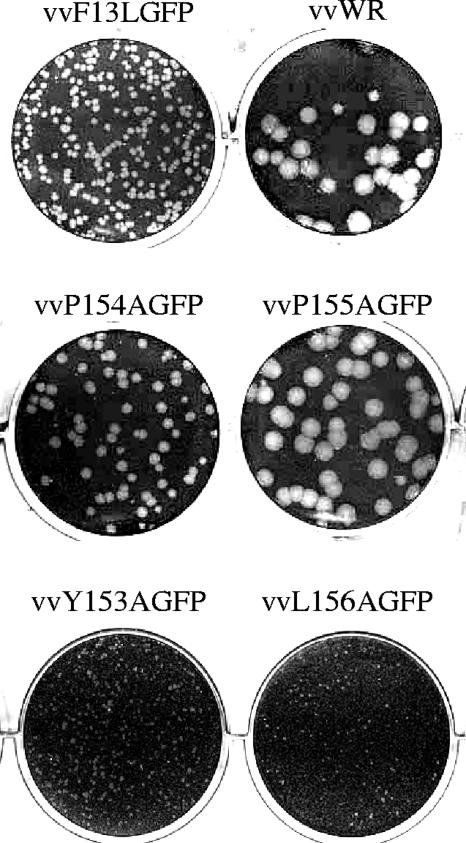
Previous studies of other YxxL L domains have demonstrated that residues Y and L are critical for viral budding (8, 33). To determine if the F13L motif behaved similarly, recombinant viruses were isolated and plaque purified (hereafter referred to as vvY153AGFP, vvP154AGFP, vvP155AGFP, and vvL156AGFP). The plaque phenotype associated with each mutant relative to those of both the wild-type F13L-GFP recombinant virus (vvF13LGFP) and the WR parental strain was then examined. The fusion of GFP to the C terminus of F13L has been shown to be fully functional (13), although virus particles encoding the GFP fusion construct produce slightly smaller plaques. This phenomenon may be due to a less efficient interaction of F13L with other viral factors as a result of the GFP tag, or perhaps the presence of a C-terminal GFP fusion interferes with the viral mechanism of release. In this experiment, BSC40 cells were infected with 50 PFU/well of either a mutant virus or one of the controls for 1 h at 37°C. The viral inoculum was removed and replaced with fresh growth medium containing 1% methylcellulose. At 3 days postinfection, cells were fixed with 5% glutaraldehyde and stained with 0.1% crystal violet. Each mutant virus exhibited a distinct plaque phenotype, with vvP154AGFP and vvP155AGFP producing large plaques reminiscent of those observed for VV WR (Fig. 2). The fact that these mutants produced plaques that are larger than those of the parental vvF13LGFP strain of virus was an unexpected result and is currently under investigation. In contrast, mutants lacking either the Y or the L residue produced extremely small plaques, suggesting a reduction in extracellular virus production. Furthermore, plaque development for each virus was photographed under UV light on a daily basis. Both vvY153AGFP and vvL156AGFP displayed an asymmetric pattern of spread, while the proline-deficient mutants maintained a circular symmetry characteristic of wild-type virus (data not shown). To verify if these observations could be related to decreased stability of the fusion protein as a result of the mutations within the YxxL motif, we conducted an immunoblot analysis of F13L-GFP expression levels for each mutant recombinant virus and compared them to that of vvF13LGFP. Differences in steady-state expression levels were observed for the various mutants (a fivefold reduction for vvY153AGFP, a twofold reduction for vvP154AGFP, a sixfold reduction for vvP155AGFP, and no reduction for vvL156AGFP) (data not shown); however, this does not appear to correlate with the plaque phenotypes. vvL156AGFP, which demonstrated a slight increase in F13L-GFP expression relative to that of the wild-type recombinant virus, rescued the least efficiently and exhibited a small plaque phenotype. Thus, it is unlikely that these amino acid changes lead to the destabilization of protein structure that causes the observed plaque phenotypes.
FIG. 2.
Plaque phenotypes of recombinant F13L YxxL mutant viruses. Six-well plates containing BSC40 cells seeded at a density of 3 × 105 cells/well were infected with virus for 1 h, overlaid with growth medium containing 1% methylcellulose, and incubated for 3 days. Cells were fixed and plaques visualized by staining with 0.1% crystal violet. vvWR, VV WR strain.
The plaque phenotypes coupled with the rescue data suggest that the YxxL motif located within F13L may possess L domain activity. Phenotypic differences in plaques created by the recombinant mutants and plaques produced by way of the rescue assay may be due to the transient nature of F13L expression associated with the rescue assay. It is likely that the mutant PCR products were successful in the rescue of viral release but that the particles formed were defective in that they contained the more abundant F13L-deleted genome.
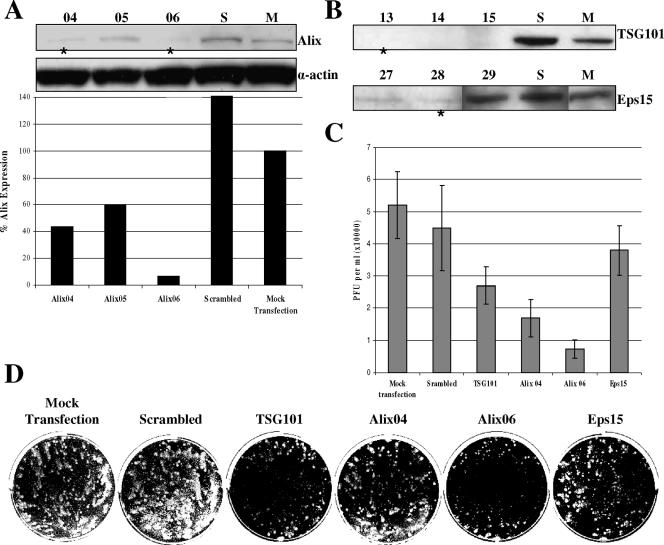
To confirm the apparent function of the F13L YxxL motif in EEV release, the roles of several endogenous trafficking components were examined. AIP1/Alix (hereafter referred to as Alix) was first identified as a novel mouse protein that undergoes calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein (29). Alix has also been shown to play a role in the formation of late endosomes and in endocytic membrane trafficking (31). Studies involving equine infectious anemia virus (EIAV), which contains the most extensively characterized viral YxxL-type L domain, have demonstrated that a lack of Alix expression results in a dramatic decrease in the production of EIAV virions (27, 38). The YxxL-Alix relationship has also been established in HIV-1 and murine leukemia virus, although to a lesser extent due to the presence of a second L domain motif, PTAP and PPxY, respectively (27, 35). Furthermore, Alix has been recognized as a binding partner of the EIAV p9 protein (38). Thus, if the F13L YxxL motif functions as an L domain, Alix may also play a role in the formation of VV EEV. We also sought to determine whether TSG101, a known binding partner of viral PTAP L domains (12, 40), had any effect on EEV release. While it is true that VV does not contain the PTAP motif in any of its known envelope proteins, the C-terminal proline-rich domain of Alix does contain a PSAP motif and has been shown to bind to TSG101 (27, 38, 41). Finally, it has been proposed that F13L is recycled from the plasma membrane via an association with clathrin complexes which lead to the formation of early endosomes (23). Blocking the F13L endocytic retrieval pathway using a dominant-negative form of the accessory protein Eps15 (2) demonstrated a decreased quantity of EEV released from cells (24). To compare the effects of each of these trafficking factors on VV EEV release, an analysis of VV comet formation following the depletion of Alix, TSG101, or Eps15 via RNA interference (RNAi) transfection was performed. Additionally, the amount of EEV released into the medium was quantified by plaque assay. Commercially available sets of RNAi oligonucleotides targeting Alix, TSG101, and Eps15 as well as a scrambled control were purchased from Invitrogen (Carlsbad, CA). Prior to this study, the knockdown efficiency of each oligo set was qualitatively evaluated by immunoblot analysis, and Alix expression was further analyzed by densitometry (Fig. 3A and B). The best set was chosen for each target, with the exception of Alix, in which case, two sets of differing efficiencies were chosen in an attempt to demonstrate a correlation between Alix expression and EEV release. BSC40 cells underwent liposome-mediated transfection with 200 pmol of each oligo set. Fresh growth medium was added at 24 h posttransfection. At 48 h posttransfection, the growth medium was replaced with medium containing 50 PFU/well of VV strain IHDJ for 1 h at 37°C. IHDJ was chosen for this assay since it possesses a mutation in the A34R protein that allows it to release up to 40 times more EEV than other strains of VV, resulting in the formation of comet-like plaques (4). The viral inoculum was then replaced with growth medium and incubated at 37°C for an additional 30 h, at which time the supernatant was collected and subjected to IMV depletion via a 1-h incubation with anti-A27L antisera. Viral titers were then determined using BSC40 cells. The most dramatic effect on EEV release was associated with the depletion of Alix, followed by the depletion of TSG101 and to a lesser extent the depletion of Eps15 (Fig. 3C). Cell monolayers were fixed and stained and revealed an inhibition of comet formation that correlated with the results obtained for the EEV quantification (Fig. 3D), suggesting that VV may interact with Alix in a way similar to that which has been described for EIAV. Moreover, these results also imply that TSG101 and Eps15 may be involved in EEV egress as well. IMV formation does not appear to be affected by the depletion of any of the three host factors or the scrambled control as all four wells treated with RNAi demonstrated primary plaques of similar size and quantity relative to those of the untreated control.
FIG. 3.
Depletion of host trafficking factors by RNAi. Six-well plates containing BSC40 cells seeded at a density of 1.5 × 105 cells/well were transfected with RNAi oligonucleotides overnight and then incubated for an additional 24 h in growth medium. At 48 h posttransfection, cells were harvested, lysed, and subjected to immunoblot analysis using commercially available monoclonal antibodies purchased from BD Biosciences Pharmingen (Philadelphia, PA). (A) Determination of Alix inhibition. The amount of protein loaded into each well was normalized to the actin-loading control by densitometry analysis. *, oligo sets selected for use in subsequent assays. (B) Determination of TSG101 and Eps15 inhibition. *, oligo set selected for use in subsequent assays. (C) EEV quantification. Six-well plates containing BSC40 cells seeded at a density of 1.5 × 105 cells/well were transfected with RNAi oligonucleotides overnight and then incubated for an additional 24 h in growth medium. At 48 h posttransfection, cells were infected with VV strain IHDJ for 1 h, at which time the viral inoculum was replaced with fresh growth medium. Cells were then incubated for an additional 29 h. At 30 h postinfection, the supernatant was collected and treated with anti-A27L antisera for 1 h. Titers were determined via plaque assay. Bars and vertical lines represent the mean PFUs and standard deviations for each RNAi treatment. (D) Comet inhibition. Cells were fixed, and comets were visualized by staining with 0.1% crystal violet.
The identification and characterization of viral L domains have provided a more solid understanding regarding the way in which enveloped virions are produced and released into the extracellular environment. By encoding sequence domains that mimic those of the host cell, nascent viral particles are able to usurp established pathways for their own dissemination, and VV, with its highly conserved L domain-like motif, is likely no exception (Table 1). In the manuscript, we report on the analysis of the YxxL domain identified within the VV F13L envelope protein. A panel of F13L-GFP fusion constructs containing mutations to the YxxL motif was generated (Fig. 1A) and used in conjunction with a recombinant F13L-deleted mutant VV in a trans complementation assay. The results obtained indicate that the ability of the deletion virus to form plaques may be restored by the introduction of a functional F13L ORF (Fig. 1B). However, in order for rescue to proceed, certain minimum requirements within the YxxL motif were established: (i) single alanine substitutions at each residue restored the plaque-forming ability, albeit to various degrees, but constructs lacking both the Y and the L residues were not functional, indicating that these residues work in tandem to direct EEV release, and (ii) the YxxL motif is absolutely required for plaque formation and cannot be deleted or replaced with other known L domains, as was demonstrated by a lack of complementation for the AAAA construct, the YxxL deletion construct, and the PTAP substitution construct. This suggests that VV EEV utilizes a very specific process to achieve budding. Recombinant YxxL motif mutant viruses supported these observations in that isolates containing alanine substitutions for either the Y or the L residue demonstrated an extremely small, asymmetrical plaque phenotype, while alanine substitutions for either of the P residues produced much larger, circular plaques, similar to those observed for the parental virus strains (Fig. 2). At no time were isolates containing an AxxA or an AAAA mutation, a YxxL deletion, or a YxxL substitution obtained, implying that the result of this type of recombination may be detrimental to the formation of mature virions. RNAi experiments involving the depletion of known host protein trafficking components also demonstrated measurable effects on EEV release (Fig. 3C and D). A reduction in the expression of Alix, an established binding partner of the YxxL L domain of EIAV (38), resulted in a 3.5- to 4.5-fold reduction of EEV release. Data from these experiments also support a less significant role for TSG101 in VV EEV release. Since VV lacks a PTAP domain, TSG101 is not thought to participate in the direct binding of VV proteins but may instead act as a trafficking regulator through its association with Alix. These data also support previous assertions that EEV release is somewhat dependent upon the recycling of F13L from the plasma membrane (23, 24), although from these experiments, it appears as though endocytosis is not as crucial for EEV release as Alix and possibly TSG101.
The role of the VV F13L YxxL motif and potential host binding factors in the production and release of EEV particles is of great interest. The highly conserved nature of the motif coupled with its sensitivity to mutation suggests that the YxxL motif located within the F13L protein may conduct late domain function in order to subvert host protein sorting machinery and facilitate EEV release, making it the subject of future research. One possible role for the YxxL motif may be in the wrapping of IMV particles to form IEV, or perhaps the motif is responsible for the recruitment of cellular factors to the site of IEV particle release. Recent studies indicate that Alix is involved in intracellular membrane metabolism through interactions with endophilins (6) and with 2,2′-dioleoyllysobisphosphatidic acid (LBPA) (25, 28). As a result of this relationship, the recruitment of Alix by viral L domains may be necessary to facilitate the release of enveloped VV particles from the plasma membrane. Experiments to decipher how the YxxL motif functions in extracellular enveloped particle production are under way. If the YxxL motif is recognized as a bona-fide L domain, it will be of interest to identify potential antiviral targets common to both enveloped DNA and enveloped RNA viruses with the intention of preventing a systemic viral infection.
Acknowledgments
This work was supported by NIH grant 5R44AI056409-06.
The authors thank Sean Amberg for a contributing role in the conception of the project and Chelsea Byrd for critical analysis of the manuscript.
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.Ansarah-Sobrinho, C., and B. Moss. 2004. Role of the I7L protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 78:6335-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benmerah, A., M. Bayrou, N. Cerf-Bensussan, and A. Dautry-Varsat. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J. Cell Sci. 112:1303-1311. [DOI] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2002. The vaccinia virus I7L gene product is the core protein protease. J. Virol. 76:8973-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatellard-Causse, C., B. Blot, N. Cristina, S. Torch, M. Missotten, and R. Sadoul. 2002. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J. Biol. Chem. 277:29108-29115. [DOI] [PubMed] [Google Scholar]
- 7.Chen, T. F., J. D. Yoder, and D. E. Hruby. 2004. Mass spectrometry analysis of synthetically myristoylated peptides. Eur. J. Mass Spectrom. 10:501-508. [DOI] [PubMed] [Google Scholar]
- 8.Ciancanelli, M. J., and C. F. Basler. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J. Virol. 80:12070-12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. L., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Côté, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. TSG101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 14.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosenbach, D. W., D. O. Ulaeto, and D. E. Hruby. 1997. Palmitylation of the vaccinia virus 37-kDa major envelope protein. J. Biol. Chem. 272:1956-1964. [DOI] [PubMed] [Google Scholar]
- 16.Grosenbach, D. W., and D. E. Hruby. 1998. Biology of vaccinia virus acylproteins. Front. Biosci. 3:d354-364. [DOI] [PubMed] [Google Scholar]
- 17.Grosenbach, D. W., S. G. Hansen, and D. E. Hruby. 2000. Identification and analysis of vaccinia virus palmitylproteins. Virology 275:193-206. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, S. G., D. W. Grosenbach, and D. E. Hruby. 1999. Analysis of the site occupancy constraints of primary amino acid sequences in the motif directing palmitylation of the vaccinia virus 37 kDa envelope protein. Virology 254:124-137. [DOI] [PubMed] [Google Scholar]
- 19.Hiller, G., H. Eibl, and K. Weber. 1981. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J. Virol. 39:903-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honeychurch, K. M., C. M. Byrd, and D. E. Hruby. 2006. Mutational analysis of the potential catalytic residues of the VV G1L metalloproteinase. Virol. J. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husain, M., and B. Moss. 2003. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J. Virol. 77:9008-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husain, M., and B. Moss. 2005. Role of receptor-mediated endocytosis in the formation of vaccinia virus extracellular enveloped particles. J. Virol. 79:4080-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, T., M. Beuchat, J. Chevallier, A. Makino, N. Mayran, J. Escola, C. Lebrand, P. Cosson, T. Kobayashi, and J. Gruenberg. 2002. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 35:32157-32164. [DOI] [PubMed] [Google Scholar]
- 26.Martin, K. H., C. A. Franke, and D. E. Hruby. 1999. Novel acylation of poxvirus A-type inclusion proteins. Virus Res. 60:147-157. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuo, H., J. Chevallier, N. Mayran, I. Le Blanc, C. Ferguson, J. Faure, N. S. Blanc, S. Matile, J. Dubochet, R. Sadoul, R. G. Parton, F. Vilbois, and J. Gruenberg. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531-534. [DOI] [PubMed] [Google Scholar]
- 29.Missotten, M., A. Nichols, K. Rieger, and R. Sadoul. 1999. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene-2 (ALG-2) protein. Cell Death Differ. 6:124-129. [DOI] [PubMed] [Google Scholar]
- 30.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 31.Odorizzi, G. 2006. The multiple personalities of Alix. J. Cell Sci. 119:3025-3032. [DOI] [PubMed] [Google Scholar]
- 32.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakaguchi, T., A. Kato, F. Sugahara, Y. Shimazu, M. Inoue, K. Kiyotani, Y. Nagai, and T. Yoshida. 2005. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79:8933-8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segura-Morales, C., C. Pescia, C. Chatellard-Causses, R. Sadoul, E. Bertrand, and E. Basyuk. 2005. TSG101 and Alix interact with murine leukemia virus gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280:27004-27012. [DOI] [PubMed] [Google Scholar]
- 36.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 37.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 38.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Göttlinger. 2003. AIP1/Alix is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 39.Strecker, T., R. Eichler, J. ter Meulen, W. Weissenhorn, H. Dieter Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. TSG101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 42.Yoder, J. D., T. Chen, and D. E. Hruby. 2004. Sequence-independent acylation of the vaccinia virus A-type inclusion protein. Biochemistry 43:8297-8302. [DOI] [PubMed] [Google Scholar]