Abstract
The replication and pathogenicity of influenza A virus (FLUAV) are controlled in part by the alpha/beta interferon (IFN-α/β) system. This virus-host interplay is dependent on the production of IFN-α/β and on the capacity of the viral nonstructural protein NS1 to counteract the IFN system. Two different mechanisms have been described for NS1, namely, blocking the activation of IFN regulatory factor 3 (IRF3) and blocking posttranscriptional processing of cellular mRNAs. Here we directly compare the abilities of NS1 gene products from three different human FLUAV (H1N1) strains to counteract the antiviral host response. We found that A/PR/8/34 NS1 has a strong capacity to inhibit IRF3 and activation of the IFN-β promoter but is unable to suppress expression of other cellular genes. In contrast, the NS1 proteins of A/Tx/36/91 and of A/BM/1/18, the virus that caused the Spanish influenza pandemic, caused suppression of additional cellular gene expression. Thus, these NS1 proteins prevented the establishment of an IFN-induced antiviral state, allowing virus replication even in the presence of IFN. Interestingly, the block in gene expression was dependent on a newly described NS1 domain that is important for interaction with the cleavage and polyadenylation specificity factor (CPSF) component of the cellular pre-mRNA processing machinery but is not functional in A/PR/8/34 NS1. We identified the Phe-103 and Met-106 residues in NS1 as being critical for CPSF binding, together with the previously described C-terminal binding domain. Our results demonstrate the capacity of FLUAV NS1 to suppress the antiviral host defense at multiple levels and the existence of strain-specific differences that may modulate virus pathogenicity.
The genome of influenza A virus (FLUAV) consists of eight RNA segments that encode nine structural proteins and two nonstructural proteins, called NS1 (41) and PB1-F2 (11). NS1 is a virulence factor of FLUAV by virtue of conferring resistance to the antiviral effects of the host interferon (IFN) system (21, 40, 63). Previous studies with recombinant FLUAV carrying deletions in the NS1 gene (delNS1) showed a strong attenuation in IFN-competent systems, whereas the NS1-deleted virus replicated to levels similar to those of wild-type virus in cell culture and in mice with a defect in the IFN system (16, 23, 39).
The expression of type I IFNs (IFN-α/β) is induced in response to viral infection. Viral single-stranded and double-stranded RNAs (dsRNAs) with phosphorylated 5′ ends are among the viral products that induce IFN-α/β (32, 42, 55). These viral RNA molecules activate a variety of cellular signaling pathways, resulting in the activation of transcription factors, such as the IFN regulatory factors (IRFs) and the stress-induced transcription factors NF-κB and c-Jun/ATF2 (34, 60, 64). Upon activation, these latent transcription factors move from the cytoplasm into the nucleus and initiate the expression of type I IFNs. IFN-α/β subtypes bind to a common type I IFN receptor, thus activating the JAK-STAT signaling pathway. This results in the expression of more than 300 IFN-stimulated genes (13, 59), including some that encode antiviral proteins. The best-studied IFN-stimulated genes with antiviral activity against FLUAV replication are the Mx GTPases (28), protein kinase R (PKR) (6, 30), and the 2′-5′ oligoadenylate synthetase/RNase L system (48). In order to replicate despite this efficient antiviral program, many viruses evolved mechanisms to escape the IFN system (22, 29), with the NS1 protein of FLUAV being the first and one of the best-studied examples of a viral IFN antagonist among negative-stranded RNA viruses (21).
FLUAV NS1 is a multifunctional protein with regulatory activities that affect a variety of host cell functions, as follows. (i) It binds dsRNA (15, 71) and thus prevents activation of the 2′-5′ oligoadenylate synthetase/RNase L system (48). In addition, sequestration of dsRNA and/or interaction with the RNA helicase RIG-I (26, 47, 55) is likely to prevent activation of the transcription factors IRF3, IRF7, NF-κB, and c-Jun/ATF2 by NS1 (46, 66, 68, 73). (ii) NS1 suppresses the induction of RNA interference, most likely by its capacity to sequestrate small interfering RNAs (8, 44). (iii) It directly binds to PKR and blocks PKR activation (43), leading to stimulation of host protein synthesis in NS1-expressing cells (58). (iv) NS1 inhibits host cell mRNA processing (45) and blocks nuclear export of polyadenylated cellular transcripts (20, 57, 61). (v) In addition, the NS1 proteins of some virus strains have been shown to suppress the antiviral function of IFNs and tumor necrosis factor alpha (24, 31, 63). (vi) NS1 has a direct enhancing effect on FLUAV replication by binding and activation of phosphatidylinositol 3-kinase (27). (vii) Finally, it forms a trimeric complex consisting of the eukaryotic translation initiation factor eIF4GI and poly(A)-binding protein I (PABI) to enhance the translational initiation of viral mRNAs (10).
The NS1 protein can be divided into two major domains, the N-terminal RNA-binding domain (amino acids 1 to 73) and the C-terminal effector domain (amino acids 74 to 230) (see Fig. 2A) (40). The RNA-binding domain is sufficient to prevent IFN induction provided that it is linked to a dimerization domain that reconstitutes a functional RNA-binding site (72). The effector domain binds to distinct cellular proteins and affects their function. NS1 interacts with the 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF) (49, 51) and with PABII (12), leading to a block of cellular mRNA processing in FLUAV-infected cells. A recent large sequence analysis identified a potential PDZ domain binding motif consisting of the last four C-terminal amino acids of NS1 that might influence the activity of PDZ domain-containing proteins, which are often involved in cellular signal transduction pathways (52).
FIG. 2.
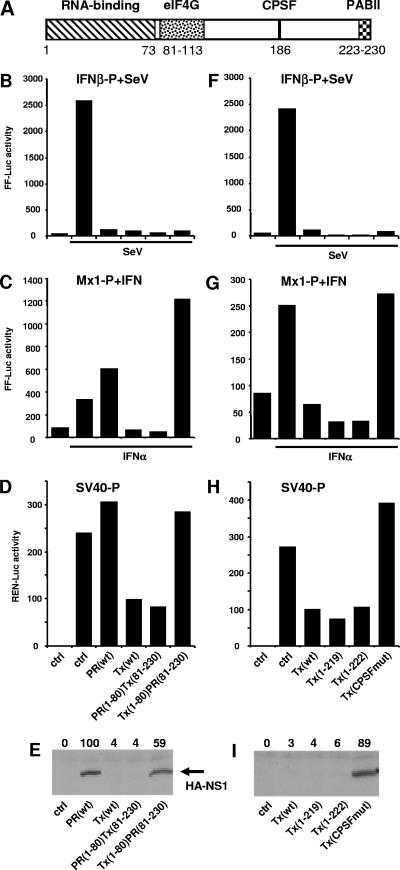
NS1 of A/Tx/36/91 mediates a general inhibition of gene expression. (A) Schematic representation of functional domains in the NS1 protein, showing the RNA-binding domain (positions 1 to 73) (40), the eIF4G-binding domain (positions 81 to 113) (1), the CPSF interaction domain (around position 184) (51), and the PABII-binding domain (positions 223 to 230) (12). Numbers indicate amino acid positions. 293T (B, E, F, and I) and Vero (C, D, G, and H) cells were cotransfected with reporter plasmids carrying the FF-Luc gene under the control of the IFN-β promoter (B and F) or the Mx1 promoter (C and G) and REN-Luc under the control of the constitutive SV40 promoter (D and H), together with expression plasmids encoding HA-tagged versions of NS1, an NS1 chimera of A/PR/8/34 and A/Tx/36/91, or C-terminally truncated Tx NS1 constructs or with an empty plasmid (ctrl). At 16 h posttransfection, the cells were infected with SeV (B and F) or treated with 200 U/ml of IFN-α2a (C and G) for 16 h and then analyzed for reporter gene expression. (E and I) Lysates of 293T cells were analyzed for NS1 protein expression by Western blotting using an HA-specific antiserum. The numbers indicate the relative intensities of the NS1 protein bands, with PR NS1 activity defined as 100. The data from one representative experiment are shown.
There is an ongoing debate about the contributions of the different functional domains of NS1 to its anti-IFN effects. Experimental data support the importance of the dsRNA-binding property of NS1 to prevent the activation of IRF3 and thereby the induction of IFN expression (21). However, work by others showed a suppression of IFN production by a block of pre-mRNA processing, in spite of fully activated IRF3. This inhibitory effect was mediated by the NS1 effector domain and its influence on the various cellular NS1-binding partners (40). To investigate this discrepancy, we directly compared the activities of NS1 gene products from three different human FLUAV (H1N1) strains and found significant differences in their functions. Based on our results, we speculate that strains of FLUAV have evolved two fundamentally different strategies to circumvent the establishment of the IFN-induced antiviral host defense, namely, prevention of IFN induction by the amino-terminal dsRNA-binding domain and suppression of cellular gene expression, most likely by a block of nuclear pre-mRNA processing. These apparently redundant strategies are likely to synergize to mediate efficient evasion of the IFN-α/β response.
MATERIALS AND METHODS
Cells and viruses.
293, 293T, A549, BSR-T7 (9), and Vero cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For infection studies, virus stocks were diluted in phosphate-buffered saline supplemented with 0.3% bovine serum albumin. Stocks of Sendai virus (SeV) strain Cantell (5) and Newcastle disease virus coding for green fluorescent protein (NDV-GFP) (54) were grown in 10-day-old embryonated chicken eggs.
Recombinant FLUAV was generated on the basis of the rescue plasmids of A/WSN/33, as described previously (19). For A/WSN/33 carrying the NS gene segments of A/PR/8/34 (62) or A/Tx/36/91 (69), the respective cDNA constructs were used. A recombinant mutant A/WSN/33 virus, WSN(Mut), carrying mutations in the NS1 gene that lead to amino acid exchanges of lysine 38 and arginine 41 with alanines was described previously (15).
Plasmids.
Expression constructs for NS1 under the control of the chicken β-actin promoter (pCAGGS) have been described for A/PR/8/34 (68) and A/BM/1/18 (4). cDNAs encoding the NS1 protein of A/Tx/36/91 (3) was cloned into pCAGGS by using the EcoRI and XhoI restriction sites, creating pCAGGS-NS1(Tx/91). The NS1-encoding cDNAs contained silent mutations in the splice acceptor site, resulting in splice acceptor mutants (SAM), to avoid splicing events and expression of the NS2/NEP.
To generate N-terminally hemagglutinin (HA)-tagged full-length NS1 expression constructs of PR/34 and Tx/91, the open reading frames encoding NS1(SAM) were amplified using appropriate primers. The PCR products were cloned into the pCAGGS-HA-NH2 expression plasmid downstream of the sequence encoding the HA tag, using SmaI and XhoI restriction sites. pCAGGS-HA-NH2 is a modified version of the original vector pCAGGS.MCS (50) that contains an HA tag sequence followed by a multiple cloning site to make N-terminally HA-tagged versions of the expressed cDNAs. All HA-tagged NS1 cDNAs were further subcloned into pcDNA3 by using EcoRI and XhoI restriction sites. HA-tagged NS1 chimeras or C-terminal truncations of the NS1 protein were constructed by using the unique NcoI restriction site or a PCR-based approach (primer sequences are available upon request), and the cDNAs were cloned into pCAGGS-HA-NH2. Mutation of the CPSF interaction site (amino acid positions 184 to 188) from GLEWN to RFLRY was performed as described previously (51). Single amino acid exchanges (positions 103 and/or 106) were introduced into PR NS1 and Tx NS1 by site-directed mutagenesis and PCR with appropriate primer pairs. The plasmid encoding the V protein of simian virus 5 (SV5), pCAGGS-V(SV5), was kindly provided by Megan Shaw, Mount Sinai School of Medicine, New York, NY. Plasmid pCAGGS-eGFP was generated by cloning the enhanced GFP open reading frame (peGFP-C1; Clontech) into the pCAGGS expression vector.
A reporter plasmid carrying the firefly luciferase (FF-Luc) gene under the control of the Mx1 promoter was created by insertion of a 2,300-bp EcoRI/SmaI fragment of the mouse Mx1 promoter (33) into pGL3 (Promega, Mannheim, Germany), creating pGL3-Mx1P-Luc (35). We also used reporter plasmids carrying the FF-Luc gene under the control of the IFN-β promoter (p125Luc; kindly provided by Takashi Fujita [75]), the T7 RNA polymerase promoter (pTM1-FF-Luc; kindly provided by Friedemann Weber [74]), an IRF1 promoter (pIRF-1-Luc; kindly provided by Stephen Goodbourn [36]), and the NF-κB-responsive element (pNF-κB-Luc; Stratagene, Amsterdam, The Netherlands). For activation of the NF-κB-responsive element, the cells were cotransfected with p288.1-LMP1, encoding the late membrane protein of Epstein-Barr virus (kindly provided by Wolfgang Hammerschmidt [2]). The reporter plasmid pRL-SV40, carrying the Renilla luciferase gene (REN-Luc) under the control of the constitutive SV40 promoter, was purchased from Promega.
Human CPSF was cloned by PCR, using a primer pair specific for the human cDNA (GenBank accession no. NM_006693) (primers target nucleotide positions 37 to 57 and 843 to 823). The product was cloned into a modified pCAGGS expression plasmid containing a C-terminal Flag tag by using the SacI and SmaI restriction enzymes.
Reporter gene assays.
For activation of the IFN-β promoter, 293T cells were transfected with 1 μg of p125Luc and 4 μg of the indicated NS1 expression plasmids, using the CaPO4 method of a mammalian transfection kit (Stratagene). After 16 h, the cells were infected with 1 PFU/cell of the SeV stock for 16 h. To measure activation of the IRF1 promoter, 293T cells were transfected with pIRF-1-Luc and treated with 100 U/ml of IFN-γ (a gift from Peter Staeheli) for 16 h. To detect the effect of NS1 on an NF-κB-driven promoter, the cells were transfected with pNF-κB-Luc and p288.1-LMP1 for 24 h. To measure activation of the Mx promoter, Vero cells were transfected with 1 μg of pGL3-Mx1P-Luc and 2 μg of the indicated NS1 expression plasmids, using Lipofectamine 2000 as described by the manufacturer (Invitrogen, Karlsruhe, Germany). At 16 h posttransfection, cells were treated with 200 U/ml recombinant IFN-α2a (Roche, Mannheim, Germany) for 16 h. At 32 h posttransfection, cells were harvested and lysed in 200 μl of passive lysis buffer (Promega, Mannheim, Germany). To measure gene expression under the control of the constitutive SV40 promoter, Vero cells were transfected with 10 ng of pRL-SV40 and 2 μg of the indicated NS1 constructs, using Lipofectamine 2000, for 24 h. An aliquot of 20 μl was used to measure FF-Luc and REN-Luc activities as described by the manufacturer (dual-luciferase reporter assay system; Promega). For T7-driven expression, 0.3 μg of pTM1-FF-Luc together with 2 μg of the NS1 expression plasmids was transfected into BSR-T7 cells, using FuGene (Roche). Expression of FF-Luc activity was measured after 24 h. All experiments were repeated independently and yielded similar results.
Detection of IFN-β induction by reverse transcription-PCR (RT-PCR).
A549 cells were infected with 0.5 PFU/cell for 10 h, and total RNA was isolated and reverse transcribed. PCRs (30 cycles) were performed using primer pairs specific for the cDNAs of human IFN-β (GenBank accession no. V00534) (primers target nucleotide positions 580 to 599 and 876 to 855), NP of A/PR/8/34 (accession no. NC_004522) (primers target positions 580 to 600 and 1120 to 1104), and human γ-actin (accession no. BC021036) (primers target positions 1652 to 1672 and 2057 to 2036).
NS1/CPSF coimmunoprecipitation.
To monitor the interaction of the different NS1 constructs with Flag-CPSF, HA-tagged NS1 cDNAs in pcDNA3 were expressed using a TNT transcription/translation kit (Promega) in the presence of 35S-labeled methionine. The cDNA coding for Flag-CPSF was expressed by transfection into 293T cells. At 48 h posttransfection, cells were lysed in 50 mM Tris (pH 7.5), 200 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, and 10% glycerol. The cleared cell lysates were incubated with the radiolabeled NS1 proteins in the presence of 1 μg of a monoclonal anti-Flag antibody (Sigma) and 25 μl of protein A-Sepharose beads (Amersham) for 2 h at 4°C. The beads were then washed three times in lysis buffer and incubated in sodium dodecyl sulfate (SDS) sample buffer for 5 min at 95°C. The proteins were subjected to 12% SDS-polyacrylamide gel electrophoresis (12% SDS-PAGE) and transferred to membranes by Western blotting. The 35S-labeled proteins were detected by autoradiography, and the precipitated Flag-CPSF was detected using a polyclonal Flag-specific rabbit antiserum (Sigma).
NS1/RIG-I coimmunoprecipitation.
To investigate binding of NS1 to RIG-I, 293T cells were transfected with pCAGGS-Flag-RIG-I, encoding human RIG-I N-terminally fused to a Flag tag. At 48 h posttransfection, the cells were infected with 2 PFU/cell of the recombinant viruses for an additional 8 h. The lysates were then prepared as described previously (47) and were used for immunoprecipitation of Flag-RIG-I, using a monoclonal anti-Flag antibody and protein A-Sepharose beads. The precipitated proteins were analyzed by Western blotting using a polyclonal rabbit anti-NS1 antibody (67) and a polyclonal Flag-specific rabbit antiserum (Sigma).
Immunofluorescence and Western blot analysis.
For immunofluorescence of the recombinant NS1 proteins, MDCK cells were transfected in suspension with 2 μg of the NS1 expression plasmids, using Lipofectamine 2000, fixed in 3% paraformaldehyde after 48 h, and permeabilized with 0.5% Triton X-100. Cells were stained using a polyclonal rabbit antiserum directed against the HA tag (sc-805; Santa Cruz) and a fluorophore (Cy2)-conjugated secondary donkey anti-mouse antibody (Dianova, Hamburg, Germany). To detect the expression levels of the NS1 and NP proteins, transfected or infected cells were lysed in passive lysis buffer in the presence of protease inhibitors (Complete; Roche), and the proteins were subjected to 12% SDS-PAGE followed by Western blotting. NP and NS1 proteins were detected using rabbit polyclonal antisera directed against NP and NS1 (67). The HA-tagged proteins were detected using a polyclonal rabbit antiserum directed against the HA tag (sc-805; Santa Cruz). To detect the production of endogenous MxA, A549 cells were treated with 400 U/ml of IFN-α2a for 20 h. The cells were then lysed, and MxA expression was analyzed by Western blotting using mouse monoclonal antibody M143 directed against human MxA (18). The intensities of the bands were quantified using Quantify One software (Bio-Rad). The “relative intensity” of the PR NS1 protein was set to 100.
IRF3 dimerization.
To detect IRF3 dimerization, 293 cells were infected with 1 PFU/cell of the recombinant viruses for 12 h and then superinfected with 1 PFU/cell of SeV stock for an additional 6 h. The cells were then lysed, and IRF3 monomers and dimers were separated by 7.5% nondenaturing PAGE in the presence of 1% deoxycholate as described previously (34). IRF3 was detected by Western blotting using polyclonal rabbit antiserum FL-425 (Santa Cruz).
NDV-GFP complementation assay.
The NDV-GFP complementation assay was performed similarly to a previously described method (65). A549 or Vero cells were transfected with 2 μg of the NS1 expression plasmids, using 2 μl of Lipofectamine 2000. At 16 h posttransfection, the cells were treated with 150 U/ml of IFN-α2a for 12 h and then infected with 1 PFU per cell of NDV-GFP. The replication of the virus was analyzed after 24 h by the expression of GFP, using a fluorescence microscope. To determine the efficiency of transfection, we used in parallel a pCAGGS expression plasmid encoding GFP (pCAGGS-eGFP) and determined the percentage of green fluorescence-positive cells at 16 h posttransfection.
RESULTS
NS1 is a potent suppressor of IFN-β induction and IRF3 activation.
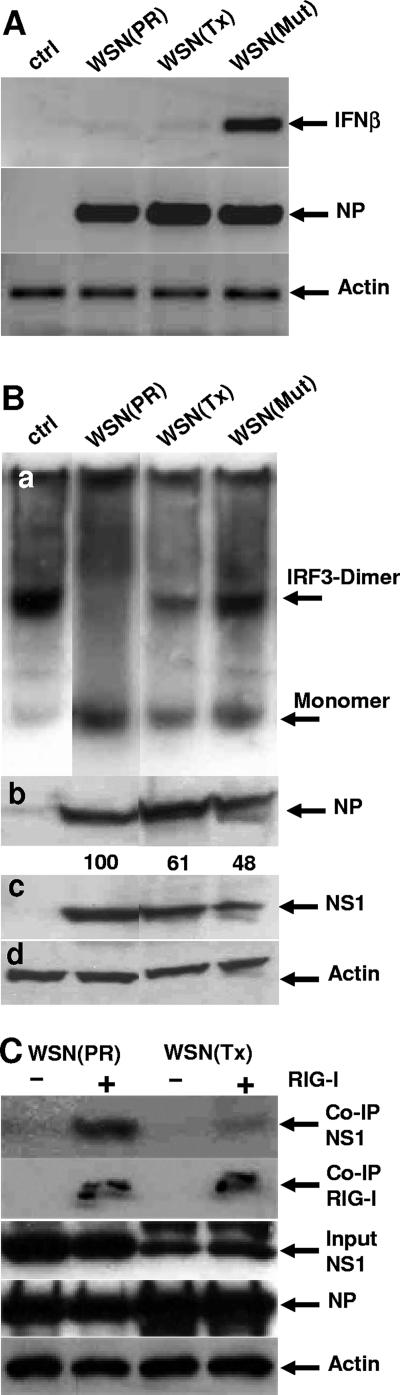
Two general models of the IFN-antagonistic function of NS1 have been proposed. NS1 mainly works by preventing activation of IRF3, a transcription factor involved in IFN-β induction. Alternatively, NS1 blocks the expression of cellular genes at the posttranscriptional level and, thereby, IFN gene expression. To compare these two activities of NS1, we generated recombinant viruses on the basis of A/WSN/33 that carried the NS genomic segments from two different (H1N1) FLUAV isolates, namely, A/PR/8/34 and A/Tx/36/91. Furthermore, we cloned NS1 cDNA expression constructs of A/PR/8/34 and A/Tx/36/91 under the control of the chicken β-actin promoter. Using these tools, we first studied the influence of NS1 on IFN-β induction by the recombinant viruses, i.e., WSN(PR) and WSN(Tx), respectively. Human lung epithelial A549 cells were infected with 0.5 PFU/cell for 10 h. A recombinant virus with inactivating mutations in NS1 (R38A/K41A), called WSN(Mut), which was shown to be a strong IFN inducer (15), was used as a positive control. WSN(Mut) strongly induced IFN-β expression, whereas infection with the viruses carrying the functional NS gene segments, i.e., WSN(PR) and WSN(Tx), showed only weak induction, as assessed by RT-PCR analysis (Fig. 1A). Analysis of the NS gene segments of A/PR/8/34 and A/Tx/36/91 showed specific differences in the amino acid sequences of the NS1 and NS2/NEP gene products. However, the viruses carrying the respective NS segments infected the cells with comparable efficiencies, as detected by RT-PCR analysis of viral NP transcripts (Fig. 1A), indicating that the differences in the NS2/NEP protein do not influence the replication of the recombinant viruses. These results indicate that the two NS1 proteins both have the potency to control IFN induction, leading to an inefficient production of IFN-β in infected cells.
FIG. 1.
Control of IFN-β induction and IRF3 activation by NS1. (A) Induction of IFN-β by recombinant A/WSN/33 carrying the NS gene segment of A/PR/8/34 or A/Tx/36/91 or an inactive NS1 mutant. A549 cells were infected with 0.5 PFU/cell of the recombinant viruses or were left uninfected (ctrl). At 10 h postinfection, total RNA was isolated, and transcripts for IFN-β, viral NP, and β-actin were detected by RT-PCR. (B) IRF3 dimerization assay. 293 cells were infected with the recombinant viruses (1 PFU/cell) or were left uninfected (ctrl). After 12 h, the cells were superinfected with 1 PFU/cell of SeV for an additional 6 h. The cells were then lysed and analyzed for the presence of IRF3 monomers and dimers by nondenaturing gel electrophoresis followed by Western blotting using an IRF3-specific antibody (a). In parallel, the lysates were analyzed by denaturing gel electrophoresis and Western blotting for the accumulation of viral NP (b), NS1 (c), and β-actin (d) as an internal standard. The lanes were cut out from the same gel. The numbers in panel c indicate the relative intensities of the NS1 protein bands normalized to the expression of NP and actin, with PR NS1 activity defined as 100. (C) NS1 association with RIG-I. 293T cells were transfected with a Flag-RIG-I expression construct or empty vector and were infected with the recombinant viruses (2 PFU/cell) 48 h later. At 8 h postinfection, lysates of the cells were subjected to immunoprecipitation (co-IP) using an anti-Flag antibody. The precipitated proteins were analyzed by Western blotting using polyclonal antibodies directed against NS1 and the Flag peptide. Infection of the cells was monitored by the accumulation of viral NP and NS1 in the cell lysates (input). β-Actin was used as a loading control.
To further analyze the effect of NS1 on IFN induction, we monitored IRF3 activation in infected cells. Upon activation, IRF3 forms homodimers that are necessary for transcriptional activation (34). To test the effects of the two NS1 proteins, 293 cells were infected with a recombinant virus carrying the NS genomic segment of A/PR/8/34 or A/Tx/36/91 or with WSN(Mut). To ensure full activation of IRF3, the cells were superinfected with SeV 12 h after FLUAV infection for an additional 6 h. The cell lysates were then analyzed for the accumulation of IRF3 dimers by native gel electrophoresis. As expected, untreated SeV-infected cells showed the formation of IRF3 dimers, whereas expression of PR NS1 completely prevented IRF3 dimerization (Fig. 1B, panel a). In contrast, cells infected with the virus encoding Tx NS1 showed partial dimerization of IRF3. However, compared with the stronger activation of IRF3 in WSN(Mut)-infected cells, Tx NS1 showed a significant suppression of IRF3 dimerization (Fig. 1B, panel a). The detection of viral NP suggests comparable infection rates by the three viruses (Fig. 1B, panels b to d). Furthermore, we studied the binding of the NS1 proteins to RIG-I, an interaction that was recently reported using NS1 of A/PR/8/34 (47, 55). Using Flag-RIG-I-transfected cells that were infected with the recombinant viruses, we tested the two NS1 proteins for coprecipitation with RIG-I. Consistent with their ability to inhibit IRF3 dimerization, both PR NS1 and Tx NS1 were found to bind to RIG-I in virus-infected cells (Fig. 1C). These results demonstrate that both NS1 variants are able to strongly suppress IFN-β induction by virus infection, interfere with IRF3 activation, and associate with RIG-I. Although in later assays PR NS1 appeared to be more effective than Tx NS1, this might have been due to the slightly lower expression levels of Tx NS1 (Fig. 1B, panel c, and C).
Using HA-tagged NS1 expression constructs, we examined the IFN-antagonistic activity independent of the virus context. To this end, we cotransfected FF-Luc cDNA under the control of the IFN-β promoter, together with the different NS1 expression plasmids, into 293T cells. To stimulate the IFN-β promoter, cells were infected with SeV. The infection strongly induced reporter gene expression compared to that in the uninfected control (Fig. 2B). Coexpression of PR NS1 and Tx NS1 suppressed the induction of SeV-induced reporter gene expression (Fig. 2B). When we analyzed the expression levels of the HA-tagged cDNA constructs used in the reporter assays, we found a positive Western blot signal for PR NS1 but no detectable signal for Tx NS1 (Fig. 2E). To follow up on this profound difference between the levels of expression of the two NS1 proteins, we tested their effects on the levels of expression of other reporter constructs. We analyzed reporter gene expression under the control of the IFN-induced Mx1 promoter or the constitutively active SV40 promoter. Cells were cotransfected with the luciferase reporter plasmids together with the NS1 expression constructs. After treatment with 200 U/ml of IFN-α2a, Mx1 promoter-controlled reporter gene expression was stimulated sixfold (Fig. 2C). Cotransfection of PR NS1 slightly increased reporter gene expression, whereas cotransfection of Tx NS1 strongly suppressed the effect of IFN (Fig. 2C). Analysis of SV40 promoter-controlled gene expression showed a similar but weaker effect (Fig. 2D), indicating that Tx NS1 but not PR NS1 leads to the suppression of reporter gene expression. This block in gene expression by Tx NS1 is most likely also responsible for the reduction of its own gene expression in transfected cells, explaining the lack of detectable Tx NS1 protein in the Western blot analysis (Fig. 2E).
Characterization of domains responsible for blocking IFN action.
The NS1 protein consists of an N-terminal RNA-binding domain (positions 1 to 73) and a C-terminal effector domain (positions 74 to 230) (Fig. 2A). To analyze the contributions of these two domains to the differential PR NS1 and Tx NS1 effects, we used a unique NcoI restriction site present at position 241 of the NS1 cDNA that divided the NS1 open reading frame into an N-terminal domain from amino acids 1 to 80 and a C-terminal domain from amino acids 81 to 230. Using this strategy, we generated HA-tagged chimeras encoding the N-terminal domain of PR NS1 fused to the C terminus of Tx NS1 [PR(1-80)Tx(81-230)] and vice versa [Tx(1-80)PR(81-230)]. Expression of the chimeric cDNAs suppressed SeV-induced IFN-β promoter-controlled gene expression as efficiently as did that of the wild-type constructs (Fig. 2B). However, luciferase gene expression from the IFN-stimulated Mx1 promoter or the SV40 promoter was reduced by PR(1-80)Tx(80-239) but not by Tx(1-80)PR(81-230) (Fig. 2C and D), indicating that the C-terminal region of Tx NS1, but not that of PR NS1, leads to a block in gene expression. Western blot analysis indicated the same effect. Only Tx(1-80)PR(81-230)-NS1 could be detected by the HA antibody (Fig. 2E), suggesting that the C terminus of Tx NS1 strongly suppresses its own expression in transfected cells.
Several functional motifs have been identified in the C-terminal effector domain of NS1. A CPSF interaction site was mapped around the glutamic acid at position 186 (51), a PABII-binding site has been reported for amino acids 223 to 237 of NS1 of A/Ud/72 (12), and a putative PDZ interaction motif consisting of amino acids 227 to 230 (52) has been described (Fig. 2A). To test the contributions of these functional motifs to the effects of NS1 in our reporter assay systems, we transfected cells with two Tx NS1 constructs that were deleted in the last 8 and 11 C-terminal amino acids, namely, Tx(1-222) and Tx(1-219), respectively, and with a Tx NS1 construct that was mutated in the CPSF-binding motif as described previously (51) [Tx(CPSFmut)]. All four expression constructs showed comparable suppressive effects on IFN-β promoter activation by SeV (Fig. 2F). However, the expression of reporter genes under the control of the Mx1 promoter or SV40 promoter was impaired only by wild-type Tx NS1 and the C-terminally truncated constructs, whereas Tx NS1 with the inactivating mutation in the CPSF interaction site lost its suppressive activity (Fig. 2G and H). The Western blot analysis confirmed this result, with apparently suppressed expression of the Tx NS1 and truncated fragments but a clear signal for Tx(CPSFmut) (Fig. 2I), indicating a minor role of the PABII and PDZ interactions but a dominant function of CPSF interaction for the suppressing activity of Tx NS1 on gene expression.
Dissection of the functional domains of FLUAV NS1.
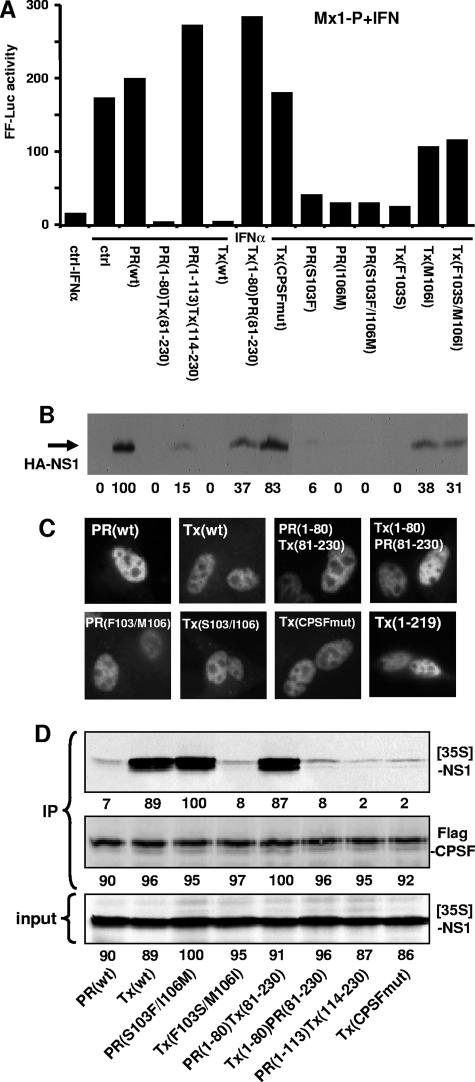
Surprisingly, screening of the amino acid sequences of PR NS1 and Tx NS1 did not show profound sequence differences around the CPSF-binding motif. Because the middle region between amino acids 81 and 113 was also identified as critical for some of the functions associated with NS1 (1) (Fig. 2A), we constructed a chimera consisting of the first 113 amino acids of PR NS1 fused to the C terminus of Tx NS1 [PR(1-113)Tx(114-230)]. We compared the activity of this construct with those of wild-type Tx NS1 and PR(1-80)Tx(81-230) in the Mx1 and SV40 promoter-driven reporter assays and found a reversal of the inhibitory activity, very similar to the effect of Tx(CPSFmut) (Fig. 3A and data not shown). This reversion of the inhibitory effect of Tx NS1 on gene expression also correlated with the detection of the chimeric NS1 proteins by Western blot analysis (Fig. 3B). To investigate whether differences in subcellular distribution may account for the differential effects of the NS1 proteins, a subset of wild-type, chimeric, and mutated constructs were also analyzed for their subcellular localization. Although the detection of the NS1 proteins by Western blot analysis differed significantly, we were able to detect accumulations of the recombinant NS1 proteins by immunofluorescence analysis. All NS1 proteins showed a mainly nuclear localization (Fig. 3C), consistent with the presence of intact nuclear localization signals.
FIG. 3.
Identification of NS1 domains responsible for inhibition of gene expression and CPSF binding. (A) Vero cells were cotransfected with reporter plasmids carrying the FF-Luc gene under the control of the Mx1 promoter and the REN-Luc gene under the control of the SV40 promoter, together with expression plasmids encoding HA-tagged versions of NS1 of A/PR/8/34 and A/Tx/36/91 or with empty plasmid (ctrl). At 16 h posttransfection, the cells were treated with 200 U/ml of IFN-α2a for 16 h and then analyzed for reporter gene expression. (B) Lysates of transfected cells were analyzed for NS1 protein expression by Western blotting using an HA-specific antiserum. The numbers indicate the relative intensities of the NS1 protein bands, with PR NS1 activity defined as 100. (C) Analysis of subcellular accumulation of NS1 proteins. MDCK cells were transfected with the NS1 expression plasmids for 48 h. The cells were then fixed and analyzed by immunofluorescence, using a polyclonal antibody directed against the HA tag. (D) Analysis of CPSF binding. The HA-tagged NS1 constructs were expressed by in vitro translation in the presence of [35S]methionine (input). Flag-tagged CPSF was expressed in transfected 293T cells. The cell lysate was mixed with the 35S-labeled NS1 proteins and subjected to coimmunoprecipitation (IP) using a monoclonal anti-Flag antibody coupled to protein A-Sepharose. The beads were washed three times, and the precipitated proteins were analyzed for the presence of 35S-labeled NS1 by autoradiography and for Flag-CPSF by Western blot analysis using a Flag-specific rabbit antiserum. The numbers indicate the relative intensities of the protein bands, with the strongest signal defined as 100.
Next, we tested the chimeric constructs for interaction with CPSF in a coimmunoprecipitation assay. The HA-NS1 constructs were expressed in the presence of [35S]methionine by in vitro transcription and translation. The labeled NS1 proteins were mixed with Flag-tagged CPSF and subjected to immunoprecipitation using a Flag-specific antibody. We found almost no CPSF binding for PR NS1 but a strong association of Tx NS1 that was abolished by mutation of the CPSF-binding site in Tx NS1 [Tx(CPSFmut)] (Fig. 3D). Analysis of the chimeric constructs demonstrated the importance of the domain of Tx NS1 from amino acids 81 to 113 for interaction with CPSF (Fig. 3D).
In the middle domain, the PR NS1 sequence differs at only two amino acids from most other NS1 sequences, including that of Tx NS1. Therefore, we exchanged these two positions in PR NS1 with the sequence of Tx NS1 [PR(S103F/I106M)] and vice versa [Tx(F103S/M106I)]. Exchange of only these two amino acids converted PR NS1 into a CPSF-interacting protein, whereas CPSF binding of the Tx NS1 mutant was abolished (Fig. 3D). The two mutants were tested for effects on Mx1 and SV40 promoter-driven reporter gene expression. The PR NS1 mutant blocked reporter gene expression in both test systems, whereas mutation of Tx NS1 reverted the inhibitory effect significantly (Fig. 3A and data not shown), indicating that the middle domain of NS1 (at least the critical phenylalanine 103 and methionine 106) is involved in CPSF interaction, in addition to the classical CPSF-binding region around position 186. The same interactions between CPSF and the PR8 and TxNS1 mutants were found using a glutathione S-transferase fusion construct of the F2/F3 domain of CPSF (data not shown).
FLUAV NS1 blocks IFN action.
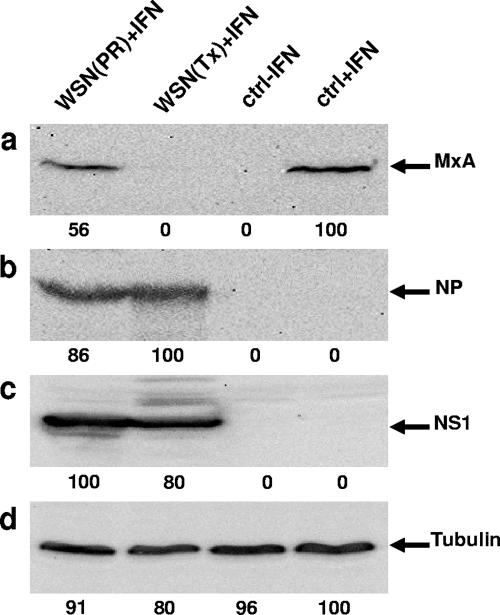
The reporter assays using transfected expression plasmids suggested that Tx NS1 has the capacity to suppress not only IFN production but also IFN-induced gene expression. We thought to test this assumption in a natural situation, i.e., with a recombinant virus and an endogenous IFN-stimulated gene, such as the human MxA gene. A549 cells were infected with recombinant A/WSN/33 viruses carrying segment 8 of A/PR/8/34 or A/Tx/36/91 and were additionally treated with 400 U/ml of human IFN-α to induce MxA synthesis. The expression of MxA was determined by Western blot analysis, which showed a strong induction in IFN-treated cells but not in untreated cells (Fig. 4a). However, the accumulation of MxA was completely absent in cells infected with WSN(Tx), whereas infection with WSN(PR) had only a minor effect on MxA induction. The detection of the viral nucleoprotein, the NS1 protein, and β-tubulin suggested comparable infections with the recombinant viruses and loading of comparable amounts of cell lysates (Fig. 4b to d). Thus, recombinant FLUAV encoding Tx NS1 but not the virus coding for PR NS1 is able to prevent the induction of IFN-stimulated genes in infected cells.
FIG. 4.
NS1 of A/Tx/36/91, but not that of A/PR/8/34, blocks IFN action. A549 cells were infected with recombinant viruses (1 PFU/cell) encoding NS gene segments of A/PR/8/34 and A/Tx/36/91 or were left uninfected (ctrl). The cells were then treated with 400 U/ml of IFN-α2a for 20 h or were left untreated (ctrl-IFN). Cell lysates were analyzed for the induction of MxA (a) and the accumulation of viral NP (b), NS1 (c), and β-tubulin (d) by Western blot analysis. The numbers indicate the relative intensities of the protein bands, with the strongest signal defined as 100.
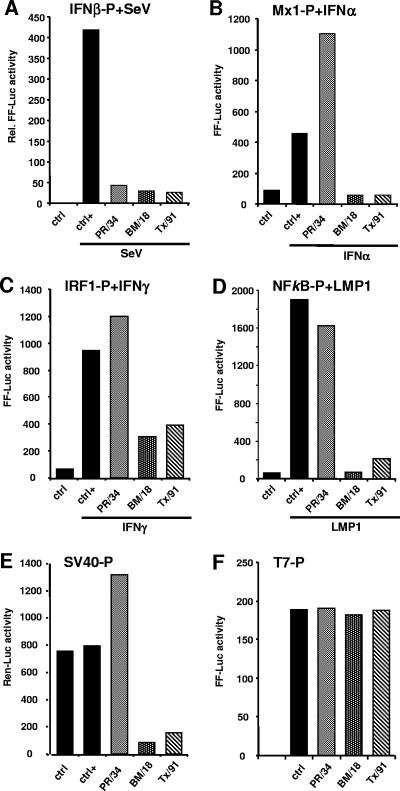
NS1 of A/BM/1/18 blocks the activity of IFN.
Recently, the NS1 gene of the pandemic FLUAV A/BM/1/18 was cloned (4) and showed an efficient suppression of IFN-stimulated gene expression in cDNA microarray analysis (24, 37). We included the NS1 cDNA of A/BM/1/18 in our study and compared its activity with those of PR NS1 and Tx NS1. Using the Mx1 and SV40 promoter constructs, we found a strong suppression of reporter gene expression by BM NS1, similar to that by Tx NS1, whereas cotransfection of PR NS1 had an enhancing effect (Fig. 5B and E). In addition, BM NS1 and Tx NS1 were able to suppress reporter gene expression induced by IFN-γ or NF-κB (Fig. 5C and D). Independent of these differences, all three NS1 constructs efficiently suppressed reporter gene expression under the control of the IFN-β promoter (Fig. 5A). To test whether this is a more general effect on gene expression or specific for transcripts produced by cellular RNA polymerase II, we used reporter gene expression under the control of the bacteriophage T7 RNA polymerase promoter. For this purpose, BSR-T7 cells that constitutively express the T7 RNA polymerase (9) were cotransfected with the NS1-encoding expression plasmids and a T7-driven reporter construct. Under these conditions, the NS1 constructs showed no effect on reporter gene expression (Fig. 5F), indicating that only nuclear transcripts produced by the cellular RNA polymerase II were targets of the NS1 inhibitory effect. Our results indicate that BM NS1 and Tx NS1 follow the same strategy, which differs from that of PR NS1, by suppressing the action of type I IFNs.
FIG. 5.
NS1 of A/BM/1/18 inhibits RNA polymerase II-driven gene expression. 293T (A, C, and D), Vero (B and E), and BSR-T7 (F) cells were cotransfected with a reporter plasmid carrying the Luc gene under the control of the IFN-β promoter (A), the Mx1 promoter (B), the IRF1 promoter (C), an NF-κB-driven promoter (D), the constitutive SV40 promoter (E), or the T7 promoter (F) together with expression plasmids encoding NS1 proteins or with empty plasmid (ctrl). At 16 h posttransfection, the cells were infected with SeV (A) or were treated with 200 U/ml of IFN-α2a (B) or 100 U/ml of IFN-γ (C) for 16 h and then analyzed for reporter gene expression. Two independent experiments yielded similar results. The data from one experiment are shown.
NS1 proteins of A/BM/1/18 and A/Tx/36/91 prevent IFN-induced antiviral action.
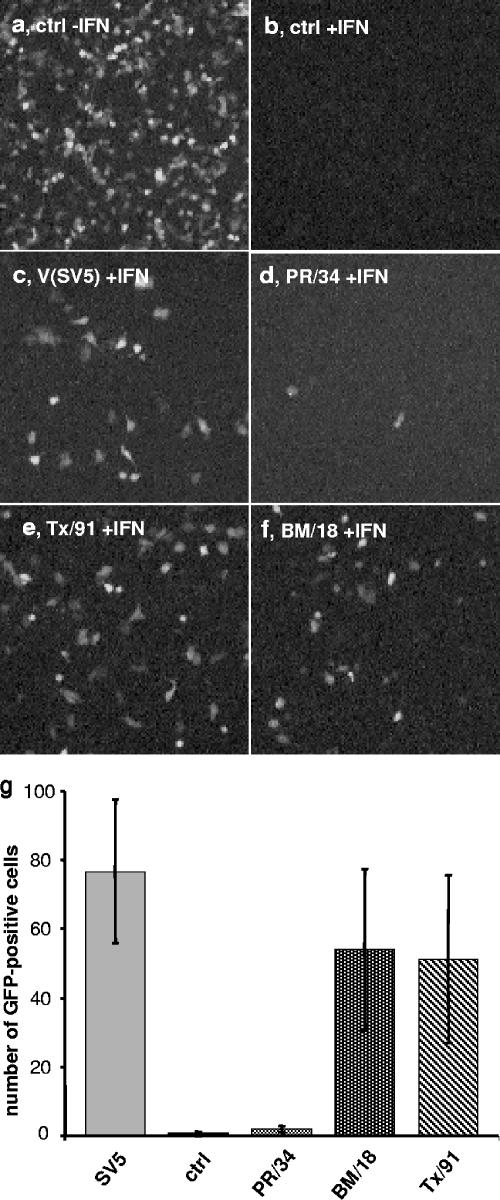
To show a potential effect of NS1 on the antiviral activity of type I IFNs, we used an NDV-GFP complementation assay (65). This assay is based on the high sensitivity of recombinant NDV-GFP to the antiviral activity of IFN-α. A549 cells were transfected with the NS1 expression plasmids. The cells were treated with 150 U/ml of IFN-α and then infected with NDV-GFP. In untreated cells, NDV was able to replicate, resulting in the expression of GFP in the infected cells, whereas pretreatment of the cells with IFN-α prevented virus-encoded GFP expression (Fig. 6a and b). When a viral IFN antagonist like the V protein of SV5, which is known to interfere with IFN signaling (14), was expressed in the cells, the replication of NDV was rescued, as evidenced by GFP expression in the cells despite the pretreatment with IFN-α (Fig. 6c). Transfection of PR NS1 did not influence the inhibitory effect of IFN-α (Fig. 6d), whereas transfection of Tx NS1 and BM NS1 rescued NDV-GFP expression with efficiencies similar to that of SV5 V (Fig. 6e and f). A quantitative analysis of this experiment demonstrated that the efficiencies of Tx NS1 and BM NS1 at reconstituting NDV replication are in a comparable range to that of SV5 V (Fig. 6g). The same effects of the different NS1 cDNAs were observed in IFN-treated Vero cells (not shown). These results are in line with the reporter assay results and suggest that the NS1 proteins of A/BM/1/18 and A/Tx/36/91 suppress the antiviral action of IFN-α by a block of IFN-stimulated gene expression.
FIG. 6.
NS1 proteins of A/Tx/36/91 and A/BM/1/18 suppress the antiviral effect of IFN-α. A549 cells were transfected with empty vector (a and b), with an expression plasmid encoding the V protein of SV5 (c), or with NS1 expression plasmids (d to f). After 16 h, the cells were treated with 150 U/ml of IFN-α2a (b to f) for 12 h or were left untreated (a) and then infected with 1 PFU/cell of NDV-GFP. At 24 h postinfection, GFP expression was analyzed by fluorescence microscopy. (g) The numbers of GFP-positive cells for a given area were determined for three independent experiments.
DISCUSSION
The viral NS1 gene product of FLUAV mediates resistance to the IFN system so that the virus is able to replicate even in the presence of a functional IFN system. In the present study, we compared the capacities of NS1 proteins of various FLUAV strains to confer an IFN-suppressive property and found remarkable differences. Whereas the NS1 protein of A/PR/8/34 most strongly blocked induction of IFN by suppression of IRF3 activation via its N-terminal RNA-binding domain, the NS1 proteins of A/BM/1/18 and A/Tx/36/91 induced a shutoff of host gene expression via the C-terminal effector domain, allowing virus growth even in IFN-treated cells. This activity that was absent in PR NS1 was found to be dependent on the association of Tx NS1 with the CPSF component of the cellular pre-mRNA processing system. Mutational analysis of Tx NS1 identified a new, as yet undescribed domain contributing to CPSF interaction located in the middle region of NS1, which proved to be responsible for the block of gene expression and seems to be required together with the previously described C-terminal CPSF interaction site (51). Therefore, our data indicate that the multifunctional NS1 protein of FLUAV evolved with at least two strategies to prevent the induction of the IFN-induced antiviral host defense. These results have potential implications on the virulence and pathogenicity associated with specific strains of FLUAV.
We used cDNA constructs carrying segment 8 of three different FLUAV H1N1 strains. The NS1 cDNA from A/BM/1/18 was derived from frozen tissue samples from a patient with a fatal case of the 1918 FLUAV “Spanish influenza” pandemic (4). A/PR/8/34 is a mouse-adapted strain of one of the first human FLUAV isolates considered to be descendants of the 1918 pandemic FLUAV strain that circulated in the human population until 1957 (53). A/Tx/36/91 was introduced in this study as the prototype of a recently isolated human H1N1 virus (3). A comparison of the predicted amino acid sequences of the three NS1 gene products did not show great differences. The NS1 protein of A/PR/8/34 is very similar (95%) to that of A/BM/1/18, from which it originated. The NS1 protein of A/Tx/36/91 shows around 90% amino acid identity to those of the two other H1N1 strains.
Both PR NS1 and Tx NS1, in the context of the same background (recombinant A/WSN/33 virus) or expressed from cDNA plasmids, showed efficient suppression of endogenous IFN-β gene expression and activation of the IFN-β promoter. When we further analyzed the effects of these two NS1 variants on IRF3 activation, both reduced IRF3 dimerization compared to that of the control, consistent with earlier observations for PR NS1 (68). NS1 chimeric constructs, C-terminal deletion constructs, and point mutations had a predominantly nuclear localization and were active in blocking virus-induced IFN-β promoter activation, indicating that this effect is highly conserved and is performed by the N-terminal RNA-binding domain, as also described in recent studies (15, 26, 31). Moreover, it has been shown that fusion of only the N-terminal region of NS1 to a heterologous dimerization domain could functionally replace the C-terminal effector domain, emphasizing the importance of the N-terminal RNA-binding domain for suppression of IFN induction (72).
In addition to the block of IFN-β induction, we found that the NS1 proteins of A/Tx/36/91 and A/BM/1/18, but not NS1 of A/PR/8/34, strongly suppressed (i) IFN-induced gene expression of endogenous MxA, (ii) reporter gene expression under the control of a variety of promoters, and (iii) establishment of an antiviral state in the cells. These activities are due to an inhibition of cellular pre-mRNA polyadenylation and splicing by NS1 based on complex formation of NS1 with the 30-kDa subunit of CPSF (49) and with PABII (12). FLUAV mRNAs escape this inhibition because they do not require CPSF for polyadenylation, which is achieved by stuttering of the viral polymerase (56, 76). In addition, NS1 was recently documented to interact with components of the nuclear pore machinery, resulting in a general inhibition of cellular mRNA export (61). By using C-terminally truncated constructs and mutational analysis of the published CPSF interaction motif in Tx NS1, we found that interaction of NS1 with CPSF but not with PABII is critical for the general block of gene expression observed in our study. In addition, the analysis did not show an effect by the putative C-terminal PDZ interaction motif (52) or the PABII-binding motif. Therefore, we would exclude an importance of the potential PABII and PDZ interaction domains for the anti-IFN effects measured in our study, which is in agreement with recently published data by others (25, 31).
The amino acid sequences of PR NS1 and Tx NS1 do not differ significantly around the CPSF interaction site (glutamic acid at position 186). However, the two proteins differ markedly in their potencies in interacting with CPSF or with the F2F3 zinc finger region of CPSF that was characterized as the NS1-binding site (70). Surprisingly, our analysis identified a second domain in the middle region of NS1 (amino acids 81 to 113) that is important for CPSF binding and that has previously been characterized as an interaction site for the cellular translational activator eIF4GI (1). By coprecipitation experiments combined with mutational analysis, we found that this site, especially amino acids 103 and 106, is critical for CPSF interaction in combination with the C-terminal CPSF interaction site around position 186. Single amino acid exchanges in the middle region could convert PR NS1 into a CPSF-associated molecule that gained the ability to suppress IFN-induced Mx1 promoter activation and gene expression under the control of the constitutive SV40 promoter, indicating the critical importance of this region for NS1 function. A BLAST search analysis of different NS1 sequences revealed conservation of the Phe-103 and Met-106 residues that are present in the NS1 proteins of A/Tx/36/91 and A/BM/1/18 and in most other NS1 sequences. The corresponding amino acid residues in the middle region of A/PR/8/34 NS1 are found in some few other FLUAV isolates, indicating an alternative evolution of A/PR/8/34, at least in its NS1-based strategy to suppress the innate immune response. In addition, we also found that the NS1 protein from the swine virus A/sw/Tx/4199-2/98, like PR NS1, is unable to inhibit general gene expression (A. Sólorzano and A. García-Sastre, unpublished observation). The three-dimensional structure of the NS1 effector domain (7) reveals that Glu-186 is located in the same area of the monomeric molecule as the amino acid residues at positions 103 and 106, supporting our assumption that both regions are cooperatively involved in direct interaction of NS1 with CPSF.
A recent publication proved the importance of this middle region of NS1 for suppression of an IFN-α-, IFN-γ-, and tumor necrosis factor alpha-induced antiviral state in porcine lung epithelial cells (63). This function could be transferred to a A/PR/8/34-based recombinant virus by introducing the eighth segment of A/HK/156/97 or by a single amino acid exchange from aspartic acid to glutamic acid at position 92 of PR NS1 (63). However, all virus strains used in the present study lacked a glutamic acid at position 92, indicating an additional strategy of avian influenza viruses to limit the antiviral potency of the host.
In conclusion, our direct functional comparison of three different NS1 molecules demonstrates two major anti-IFN strategies. A/PR/8/34 NS1 might serve as a prototype for a viral IFN antagonist that specifically blocks IFN induction and activation of PKR and 2′-5′ oligoadenylate synthetase via its N-terminal RNA-binding domain. The anti-IFN capacity of A/Tx/36/91 NS1 is, in addition, based on a more general block of host pre-mRNA processing mediated by two distinct domains in the C-terminal effector domain, namely, the well-characterized region around amino acid 186 (70) and the newly identified middle region around positions 103 and 106 (this study). These regions are required for binding to the host factor CPSF, resulting in an inhibition of cellular mRNA processing. Recently, it was also shown that NS1 can inhibit host gene expression by interacting with components of the nuclear pore and blocking mRNA export (61). Future experiments are required to investigate whether the same NS1 region is also involved in this activity. Inhibition of host gene expression is a strategy also adopted by the NS1 protein of A/BM/1/18 and might account for the suppression of IFN-stimulated gene expression (24; this study) and, at least partially, for the highly virulent phenotype of this pandemic virus (38, 69). In addition, it seems conceivable that the existence of two different mechanisms to inhibit the IFN response by the NS1 protein might increase the capacity of the virus to adapt to new environments and hosts, a hallmark of FLUAV evolution that is shaped by the ability of specific viral strains and/or genes to occasionally jump from birds to mammals and humans. It will also be interesting to investigate whether the multifunctional activities of NS1 might account for its recently described ability to inhibit dendritic cell function (17). Finally, the presented results hopefully help to clarify the long-lasting discussion about the molecular action of the FLUAV NS1 IFN antagonist.
Acknowledgments
This work was partly supported by grants from the Deutsche Forschungsgemeinschaft (Ko 1579/4-1/2) to G.K. and from the NIH (R01AI46954, P01AI58113, U19AI62623/CIVIA, and U01AI70469) to A.G.-S.
We thank Otto Haller and Peter Palese for helpful discussions and constant support and Simone Gruber and Richard Cádagan for excellent technical assistance. We are grateful to Randy Albrecht, Christopher Basler, Washington Cardenas, Karl-Klaus Conzelmann, Takashi Fujita, Stephen Goodbourn, Wolfgang Hammerschmidt, Man-Seong Park, Megan Shaw, Alicia Sólorzano, Peter Staeheli, Silke Stertz, and Friedemann Weber for expression constructs, cell lines, reporter plasmids, antibodies, and technical advice.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Aragon, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortin, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 3.Baskin, C. R., A. Garcia-Sastre, T. M. Tumpey, H. Bielefeldt-Ohmann, V. S. Carter, E. Nistal-Villan, and M. G. Katze. 2004. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina). J. Virol. 78:10420-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. [DOI] [PubMed] [Google Scholar]
- 8.Bucher, E., H. Hemmes, P. de Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85:983-991. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgui, I., T. Aragon, J. Ortin, and A. Nieto. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 14.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Sesma, A., S. Marukian, B. J. Ebersole, D. Kaminski, M. S. Park, T. Yuen, S. C. Sealfon, A. Garcia-Sastre, and T. M. Moran. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 80:6295-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flohr, F., S. Schneider-Schaulies, O. Haller, and G. Kochs. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463:24-28. [DOI] [PubMed] [Google Scholar]
- 19.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govorkova, E. A., J. E. Rehg, S. Krauss, H. L. Yen, Y. Guan, M. Peiris, T. D. Nguyen, T. H. Hanh, P. Puthavathana, H. T. Long, C. Buranathai, W. Lim, R. G. Webster, and E. Hoffmann. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 79:2191-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo, Z., L. M. Chen, H. Zeng, J. A. Gomez, J. Plowden, T. Fujita, J. M. Katz, R. O. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell. Mol. Biol. 36:263-269. [DOI] [PubMed] [Google Scholar]
- 27.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. USA 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haller, O., and G. Kochs. 2002. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 29.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayman, A., S. Comely, A. Lackenby, S. Murphy, J. McCauley, S. Goodbourn, and W. Barclay. 2006. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology 347:52-64. [DOI] [PubMed] [Google Scholar]
- 32.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 33.Hug, H., M. Costas, P. Staeheli, M. Aebi, and C. Weissmann. 1988. Organization of the murine Mx gene and characterization of its interferon- and virus-inducible promoter. Mol. Cell. Biol. 8:3065-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 35.Jorns, C., D. Holzinger, R. Thimme, H. C. Spangenberg, M. Weidmann, J. Rasenack, H. E. Blum, O. Haller, and G. Kochs. 2006. Rapid and simple detection of IFN-neutralizing antibodies in chronic hepatitis C non-responsive to IFN-alpha. J. Med. Virol. 78:74-82. [DOI] [PubMed] [Google Scholar]
- 36.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 37.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319-323. [DOI] [PubMed] [Google Scholar]
- 38.Kobasa, D., A. Takada, K. Shinya, M. Hatta, P. Halfmann, S. Theriault, H. Suzuki, H. Nishimura, K. Mitamura, N. Sugaya, T. Usui, T. Murata, Y. Maeda, S. Watanabe, M. Suresh, T. Suzuki, Y. Suzuki, H. Feldmann, and Y. Kawaoka. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703-707. [DOI] [PubMed] [Google Scholar]
- 39.Kochs, G., I. Koerner, L. Thiel, S. Kothlow, B. Kaspers, N. Ruggli, A. Summerfield, J. Pavlovic, J. Stech, and P. Staeheli. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chicken. J. Gen. Virol. 88:1403-1409. [DOI] [PubMed] [Google Scholar]
- 40.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 41.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 42.Levy, D. E., and I. J. Marie. 2004. RIGging an antiviral defense—it's in the CARDs. Nat. Immunol. 5:699-701. [DOI] [PubMed] [Google Scholar]
- 43.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 44.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 50.Niwa, H., K. Yamamura, and J. Miyazali. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 51.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 52.Obenauer, J. C., J. Denson, P. K. Mehta, X. Su, S. Mukatira, D. B. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. M. Rakestraw, R. G. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. W. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576-1580. [DOI] [PubMed] [Google Scholar]
- 53.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10:S82-S87. [DOI] [PubMed] [Google Scholar]
- 54.Park, M.-S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and E. S. C. Reis. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′ phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 56.Poon, L. L., D. C. Pritlove, E. Fodor, and G. G. Brownlee. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvath, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. Garcia-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuel, C. E. 2001. Antiviral action of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 61.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 104:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schickli, J. H., A. Flandorfer, T. Nakaya, L. Martinez-Sobrido, A. Garcia-Sastre, and P. Palese. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B 356:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine response. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 64.Servant, M. J., N. Grandvaux, and J. Hiscott. 2002. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem. Pharmacol. 64:985-992. [DOI] [PubMed] [Google Scholar]
- 65.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 67.Solorzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 70.Twu, K. Y., D. L. Noah, P. Rao, R. L. Kuo, and R. M. Krug. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 80:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, X., C. F. Basler, B. R. Williams, R. H. Silverman, P. Palese, and A. Garcia-Sastre. 2002. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NF-κB and induction of type I interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 75.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the IFN-alpha/beta system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng, H., H. A. Lee, P. Palese, and A. Garcia-Sastre. 1999. Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J. Virol. 73:5240-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]