Abstract
The Epstein-Barr virus transcription factor Zta (encoded by BZLF1) is a bZIP protein containing an α-helical coiled-coil homodimerization motif (zipper). The Zta zipper forms less-stable dimers than other bZIP proteins, and an adjacent region (CT) interacts with the zipper to form a novel structure that is proposed to strengthen the dimer. Here we question the role of the CT region for Zta function. Cross-linking experiments demonstrate that the entire CT region lies adjacent to the zipper. Detailed analyses of Zta truncation mutations identify an involvement of the proximal CT region (221 to 230) in dimer formation with a further contribution from the distal region (236 to 243). Biophysical analyses reveal that residues 221 to 230 enhance the stability of the coiled coil. The ability of the Zta truncation mutants to interact with three Zta-binding sites also requires the proximal CT region. Fine mapping of DNA-binding requirements highlighted the contribution of these amino acids for Zta function. Thus, the proximal part of the CT region is required to aid the dimerization of Zta and thereby its DNA-binding ability. In contrast, although the distal part of the CT region aids dimerization, it promotes only a modest increase in DNA binding. To probe this further, we defined the contribution from the CT region for Zta to transactivate a promoter embedded within the viral genome. From this we conclude that the proximal part of the CT region is absolutely required, whereas the distal part is dispensable.
Epstein-Barr Virus (EBV), a lymphocryptovirus, is responsible for severe human malignancies, such as Hodgkin's disease, Burkitt's lymphomas, nasopharyngeal carcinoma, and gastric carcinoma, and is the causative agent of infectious mononucleosis (8, 19). EBV replication is tightly regulated, with only a few latency-associated genes expressed in the majority of infected cells. The product of the BZLF1 gene, the lytic switch transactivator Zta (ZEBRA, EB1, and Z), mediates the disruption of EBV from latency (6, 7, 32). Indeed, a virus lacking the BZLF1 gene is unable to trigger the viral replicative cycle of EBV (10). The cascade of lytic EBV gene expression is initiated by Zta, which initially autoregulates its own promoter through binding to two Z-responsive elements (ZREs) (12, 33). The expression of Zta then induces activation of the BRLF1 and BMLF1 genes through the ZREs in their promoters. Together the BMRF1 and BRLF1 gene products activate the expression of approximately 100 viral genes (9, 15, 31). Zta is also an essential factor for EBV replication, since it can directly bind to the origin of lytic replication (25, 26) in addition to promoting cell cycle arrest (3, 4).
Zta is a member of the bZIP family of transcription factors (26-30). The general structure of bZIP family members, including Zta, comprises a transactivation domain, a DNA contact region (basic), and a dimerization region (zipper) (17). Early work from several groups revealed that Zta binds to DNA as a multimer and mutations within the zipper prevent this binding (5, 11, 16, 18).
The basic region of Zta conforms to the bZIP model; however, the zipper region does not. The zipper regions of other bZIP family members contain a repeat of hydrophobic leucine residues every seven amino acids and form amphipathic α-helical structures that dimerize through their hydrophobic surfaces to form coiled coils. In contrast, Zta lacks the usual heptad repeats of leucine seen in canonical bZIP members, with other hydrophobic residues taking their place (5, 11, 16, 18). This led us to question whether Zta dimerizes through a coiled-coil motif. Biophysical evidence indicates that the dimerization domain of Zta forms a weak coiled coil (14). This prompted us to ask whether additional regions outside the zipper aid dimer stability and unmasked a contribution from the C-terminal or CT region, which is adjacent to the zipper (13). The recently solved crystal structure of Zta, containing part of the CT region, revealed complex interactions between the zipper and CT regions, which are proposed to stabilize dimer formation (20, 23). The CT region of Zta is highly conserved in all EBV isolates and EBV-related viruses but has no homology with proteins from other species (14). Preliminary analysis of the role of this region revealed that the CT region is necessary for Zta to interact with a ZRE and to transactivate a synthetic ZRE-dependent reporter construct (13).
Here we investigate the involvement of the CT region in Zta structure and function and demonstrate that the entire CT region lies in close proximity to the zipper. The contribution of the CT region to dimerization, DNA-binding, and transactivation of Zta is established using a variety of in vitro assays and an in vivo assay.
MATERIALS AND METHODS
Cell culture.
Cells of the 293-BZLF1-KO epithelial cell line, which contains the B95-8 strain lacking the BZLF1 gene (10), were maintained in RPMI 1640 medium supplemented with 10% (vol/vol) of fetal bovine serum, 100 U/ml of penicillin, 100 μl/ml of streptomycin, and 2 mM of l-glutamine in a humidified 5%-CO2 environment at 37°C. The medium was also supplemented with 100 μg/ml of hygromycin for maintenance of the recombinant virus. All reagents were from Invitrogen.
Transfection.
293-BZLF1-KO cells were transfected using the Effectene transfection reagent kit (QIAGEN). In a six-well plate, 4 × 105 cells were seeded prior to transfection with 0.8 μg of DNA.
Expression vectors and mutagenesis.
Termination codons were introduced into the coding sequence of pSP64Zta (24) by site-directed mutagenesis, replacing a termination codon (TAA) for the indicated amino acid codon. Thus, for D228ter the codon encoding D is replaced with a termination codon and the terminal residue within the resulting protein is V227. Some mutants have been described previously (13). The following oligonucleotides were used to generate the mutations: D228 Ter, AGCCTGGATGTTTAAGACTCCATTATCC; I231 Ter, ATGTTGACTCCATTTAAATCCCCCGGACACC; T234 Ter, TCCATTATCCCCCGGTAAACACCAGATGTTTTAC; V237 Ter, CGGACACCAGATTAAGTTTTACACGAG; and L243 Ter, GTTTTACACGAGGATCTCTAGTTAAATTTCTAA.
The C-terminal truncation mutants and wild-type Zta were cloned into the pBABE puro vector using the BamHI and EcoRI sites for in vivo expression (21).
Zta1 to -3 were generated by sequential rounds of mutagenesis, starting by substituting H239-D241 for alanine (Zta1), additionally substituting R233 and D236 for alanine (Zta2), and finally additionally substituting D226 and D228 for alanine (Zta3). The following primers were used: Zta1, CCCCGGACACCAGATGTTTTAGCCGCGGCTCTCTTAAATTTCTAAC; Zta2, GACTCCATTATCCCCGCGACACCAGCTGTTTTAGCCGCGGCTCTC; and Zta3, GATGTGCCCAAGCCTGGCTGTTGCCTCCATTATCCCCGCGACACC.
Dimerization assay.
Zta proteins were generated in a wheat germ translation system incorporating [35S]methionine (Promega). Four microliters of the translated protein was mixed with 9 μl of buffer (10 mM KPO4, 10 mM dithiothreitol) at 37°C. Cross-links were generated following the addition of glutaraldehyde to a final concentration of 0.05% (vol/vol) and a 30-min incubation at 37°C. Excess glutaraldehyde was quenched by the addition of 100 mM glycine and the cross-linked proteins analyzed on a 10% bis-Tris gel in morpholinepropanesulfonic acid buffer (Novex) and detected by phosphorimaging (STORM; Amersham).
EMSA.
One oligonucleotide (10 pmol) was labeled at the 5′ end with [γ-33P]ATP (25 μCi) using polynucleotide kinase (Roche), and 20 pmol of the complementary oligonucleotide strand was added to the labeled single strand and incubated for 2 min at 95°C, 10 min at 65°C, and 30 min at 37°C to anneal the strands. The 5′ oligonucleotide sequences of the ZIIIB and AP1 probes used are as follows: ZIIIB, 5′-GTACATTAGCAATGCCTG-3′; and AP-1, 5′-GATCCATGACTCAGAGGAAAACATACG-3′. The ZRE1 probe was described elsewhere (2). The electrophoretic mobility shift assay (EMSA) was performed as previously described (1).
CD analysis.
Circular dichroism (CD) spectroscopy analysis was performed on two synthetic peptides, M221pep (acetyl-LLQHYREVAAAKSSENDRLRLLLKQ-amide) and I231pep (acetyl-LLQHYREVAAAKSSENDRLRLLLKQCPSLDVDSI-amide), which were synthesized, purified by high-pressure liquid chromatography, and verified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Alta Biosciences). Millimolar stock solutions of these peptides were reconstituted in 25 mM of potassium phosphate (pH 7.0), 100 mM NaCl, and 1 mM dithiothreitol. Concentrations were determined by the absorbance at 280 nm, assuming a molar extinction coefficient of 1,490 M−1 cm−1 for both peptides. CD spectroscopy was carried out with a Jasco J-715 spectropolarimeter fitted with a Peltier temperature controller. For thermal unfolding experiments, the signal at 222 nm was measured as the temperature was increased in a stepwise manner at a rate of 1°C/min over the range from 2°C to 90°C. The midpoint temperature of unfolding was taken as the maximum of the first derivative of the melting curves obtained.
Analytical ultracentrifugation.
Sedimentation equilibrium studies were conducted at 5°C with a Beckman Optima XL-I analytical ultracentrifuge fitted with an An-60 Ti rotor as described elsewhere (34). A 100-μl sample of peptide (at a starting concentration of 500 μM peptide [pH 7]) was used with a 110-μl sample of matched buffer as the reference. The sample was equilibrated for 24 to 72 h at rotor speeds of 25,000, 30,000, 35,000, 40,000, 50,000, 55,000, and 60,000 rpm. Equilibrium curves were recorded at 280 nm. The resulting data sets were fitted simultaneously with routines from Ultrascan II, version 8.0 (Borries Demeler). Two fitting methods were used: the first assumed a single ideal species, and the second assumed a monomer-dimer (M221pep) or dimer-tetramer (I231pep) equilibrium. The molecular mass, molar extinction coefficient, and partial specific volume at 20°C were calculated from the amino acid composition of each peptide: these were 2,991.9 Da, 1,490 cm−1 M−1, and 0.7448 cm3 g−1, respectively, for M221pep and 4,050 Da, 1,490 cm−1 M−1, and 0.7401 cm3 g−1, respectively, for I231pep; the solvent density at 20°C calculated from the buffer composition was 1.000529 g cm−3.
Gene expression analysis using real-time PCR.
Following transfection of the 293-BZLF1-KO cells with various pBABE-BZLF1 CT mutants, cells were lysed and total RNA was isolated using the RNeasy kit (QIAGEN), followed by the synthesis of cDNA using the ImProm-II reverse transcription system (Promega). Quantitative real-time PCR was performed using SYBER green master mix (QIAGEN) mixed with 0.4 μM of each primer and 1 μl of DNA to a final volume of 25 μl. The sequences of the primers used in determining gene expression are as follows: L32 (housekeeping gene), 5′-CAACATTGGTTATGCAAGCAACA-3′ and 5′-TGACGTTGTGGACCAGGAACT-3′; Zta (for transfection efficiency), 5′-CTATCAGGACCTGGGAGGGC-3′ and 5′-CACAGCACACAAGGCAAAGG-3′; and BMRF1, 5′-AGAATGGCCCTACAAGTCG-3′ and 5′-CACACGTGACTGCCGAAGTG-3′. The Real time PCR cycling condition was as follows: step 1, one cycle at 50°C for 2 min, followed by 95°C for 10 min; step 2, 40 cycles at 95°C for 15 s and 60°C for 1 min.
Cross-linking and mass spectrometry analysis.
pRSETbZIPCT was generated by amplifying the coding region from amino acid 134 to 245 of Zta and subcloning the resulting fragment into the BamHI and EcoRI sites of pRSETA using the following oligonucleotides: 5′-GGGAATTCTTAGAAATTTAAGAGATCCTCG-3′ and 5′-CCGGATCCATGGGGGCTAACCAAGGACAAC-3′.
His-bZIPCT Zta was produced from pRSETbZIPCT in bacteria and purified. Elution from the nickel beads was performed with phosphate-buffered saline supplemented with 150 mM imidazole and 0.5% (vol/vol) NP-40. The protein was buffer exchanged into 0.1 M morpholineethanesulfonic acid (pH 4.8) using a Slide-a-lyzer dialysis system (Pierce). To cross-link neighboring residues within the protein, 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) was added to a final concentration of 2 mg/ml and incubated at 20°C for 2 h. Where appropriate, the protein was subsequently cleaved with cyanogen bromide in an acidic environment (0.1 M HCl). Following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualization, proteins were excised from the gel and subjected to trypsin digestion followed by mass spectrometry (University of Sussex proteomics service).
RESULTS
Cross talk between the CT region and the zipper region extends to the basic region.
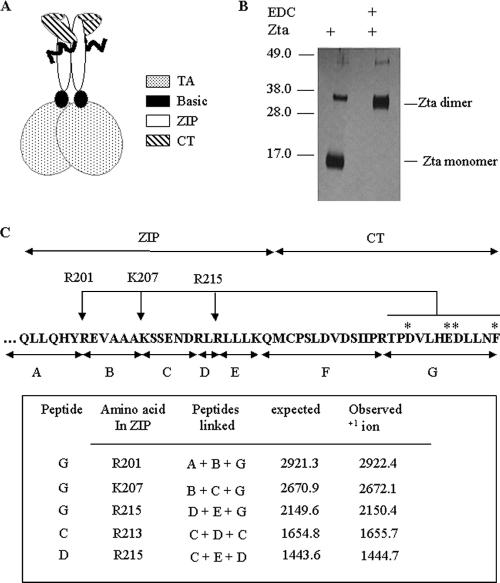
The recently published crystal structure of Zta contained only half of the CT region (23). In order to probe the structure of the CT region further, we sought to determine its proximity to the zipper using a zero-spaced cross-linking agent, EDC. A polyhistidine-tagged version of the bZIP and CT regions of Zta was expressed and purified from Escherichia coli and cross-linked (Fig. 1A and B). As anticipated, the molecular weight of the protein changed from that of a monomer to that of a dimer. Therefore, at least one cross-link per molecule was formed. It was anticipated that cross-links would be formed between the two zipper regions and potentially between the CT region and the zipper. To question whether these links were formed, we treated the protein with cyanogen bromide, which cleaves after methionine residues. Only one methionine is present in the zipper and CT regions, at the boundary between them (M221), so cleavage by cyanogen bromide should separate these regions if they are not cross-linked together. The size of the cleaved protein was consistent with two zippers and two CT regions cross-linked together, demonstrating that the CT region lies in close proximity to the zipper region and suggesting that the links between the CT region and zipper occurred in the majority of molecules (data not shown).
FIG. 1.
The distal CT region closely interacts with the zipper region of Zta. (A) Schematic diagram of Zta showing interaction of part of the CT region with the zipper as determined by Petosa and colleagues (23). The wavy line represents the part of the CT region of unknown structure. (B) Migration of the bZIPCT protein in SDS-PAGE before and after cross-linking with EDC. (C) Following excision and trypsin digestion, resulting peptides were identified by mass spectrometry. Potential tryptic peptides are indicated beneath the sequence, and asterisks indicate acidic residues in peptide G. Peptides A, B, and D cross-linked to the extreme CT region (peptide G) and peptides C and D linked to the zipper were identified, allowing identification of specific cross-linked residues in the ZIP region.
To further map the location of the interactions, we subjected the protein to analysis by mass spectrometry both in its native state and following cross-linking. Trypsin digestion of the native protein generated specific peptide fragments of Zta, shown in Fig. 1C. This analysis cannot be comprehensive, since some cross-linked species would be of too great a mass to resolve accurately; however, following cross-linking, two cross-links within the zipper and three cross-links involving the “G” peptide, which encompasses the distal part of the CT region and the zipper, were identified. As EDC forms peptide bonds between amine and carboxyl groups, a trypsin cleavage site is removed when cross-linking occurs, so a cross-link between peptide “G” and residue R201 removes the trypsin recognition site at the boundary of peptides “A” and “B” and results in a mass spectrometry signal for “A + B + G,” seen in Fig. 1. All of the cross-linked species identified in the analysis are shown in Fig. 1; none of these were detected in the non-cross-linked sample analyzed in parallel. This shows that one of the four acidic groups in peptide “G” is cross-linked to R201, one to K207, and one to R215. The contacts may occur in either an intra- or interchain manner and could potentially stabilize the structure. From the crystal structure of Zta, it is known that the proximal part of the CT region runs antiparallel to the zipper (23); these results indicate that the CT region continues in that direction, maintaining close contact with the zipper region throughout its length.
Dimerization of Zta requires residues in the proximal and distal parts of the CT region.
Zta truncation mutants (Fig. 2A) and wild-type Zta were translated in vitro and labeled with [35S]methionine (Fig. 2B). The name of each mutation reflects the amino acid residue that was converted to a termination codon, e.g., H199ter. Labeled proteins were incubated with glutaraldehyde for 30 min and then analyzed for dimer formation (Fig. 2C). Preliminary experiments demonstrated that the assay is linear over a 90-min period (data not shown). We found that K219ter and H199ter formed negligible dimers under these conditions and M221ter, L225ter, and D228ter formed 10% as many dimers as full-length Zta, but the addition of residues to I230 allowed efficient dimer formation (75% of that observed for Zta). In addition, residues in the distal part of the CT region, to L243, aided dimer formation further.
FIG. 2.
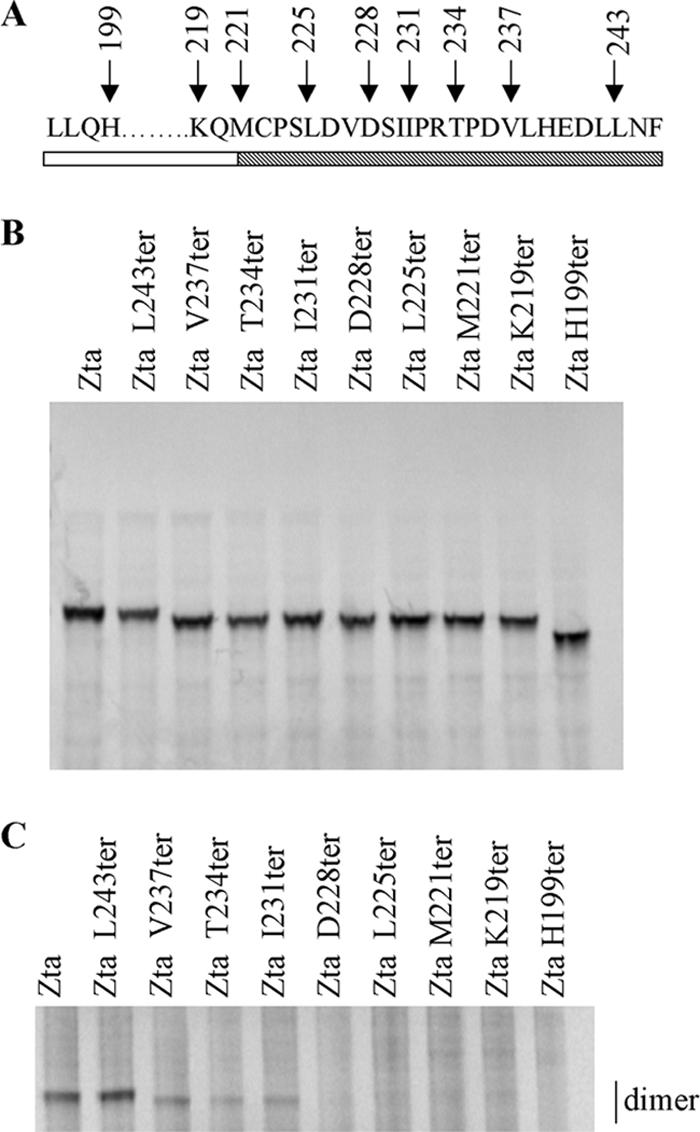
Both the proximal and distal parts of the CT region contribute to dimerization. (A) Schematic diagram showing sequence of the truncation mutants spanning the zipper and CT regions. The zipper region is shown as an open box and the CT region as a cross-hatched box. (B) Amounts of translation product generated from wheat germ extract for Zta and Zta truncated mutants used in the dimerization assay were assessed by SDS-PAGE and quantitated by phosphorimaging. (C) Equivalent amounts of each protein were incubated at 37°C and cross-linked by addition of 0.05% glutaraldehyde for 30 min. The reaction was quenched by the addition of glycine, and dimeric proteins were separated on a 10% bis-Tris gel and visualized using phosphorimaging. A representative gel is shown.
The proximal CT region contributes to the stability of the cooperatively folded helical dimer.
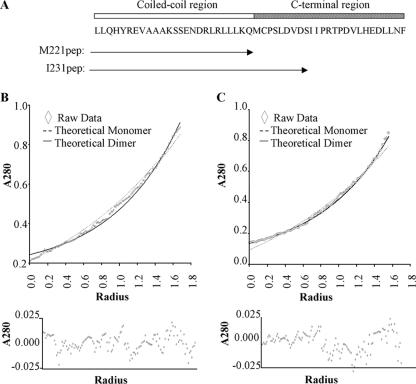
To analyze the contribution of the proximal CT region to dimer formation further, we used short synthetic peptides, containing either the zipper alone (M221pep) or the zipper plus the proximal CT region (I231pep), to undertake quantitative biophysical analyses (Fig. 3A). To probe the oligomerization state of the two peptides directly and quantitatively, sedimentation equilibrium experiments were performed with the analytical ultracentrifuge. When analyzed assuming a single ideal species in solution, the data for the longer peptide, I231pep, gave a molecular mass of 8,417 Da (Fig. 3C). This is consistent with the peptide forming a dimer (expected Mr for the dimer = 8,101.4). Analysis of the data assuming a monomer-dimer model failed to return consistent values for the dissociation constant, indicating that no monomer was present in the assay. In contrast, the data for M221pep (Fig. 3B) suggested that this peptide was mainly monomeric at 5°C, with a small proportion of dimer formed. This is consistent with the data from Fig. 2 showing that M221ter is poor at dimerization.
FIG. 3.
Stability of dimers revealed by analytical ultracentrifugation. (A) Schematic representation of residues contained within the two synthetic peptides and their locations compared to Zta structure. Representative analytical ultracentrifuge data sets for M221pep (B) and I231pep (C), recorded at 50,000 rpm, are shown and provide curves typical of sedimented and equilibrated species. Protein concentration (measured as A280) versus distance (r) from the center of the centrifuge rotor. The starting protein concentration was 500 μM. Experimental data points are shown for sedimentation of the peptide (diamonds). Also shown are simulated curves calculated assuming a completely monomeric (broken line) or dimeric (solid line) protein. The residual signals, calculated as the difference between the experimental and fitted data for a single ideal species, show no systematic errors, indicating that the fits are robust (lower panels).
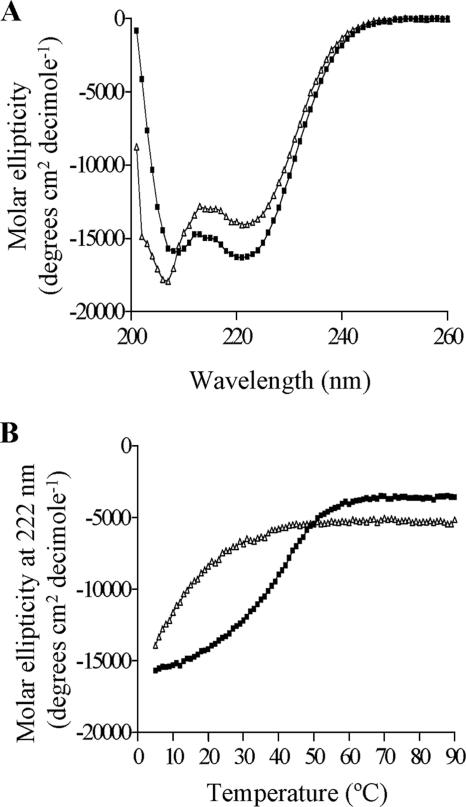
The secondary structures of both peptides were then probed using CD spectroscopy. Both displayed spectra indicating an α-helical structure, with minima at 208 and 222 nm (Fig. 4A). Consistently, as judged by the intensity of the signal at 222 nm, I231pep displayed a higher degree of helical structure in solution over a range of peptide concentrations (Fig. 4A; also data not shown). Thermal denaturation experiments revealed that the unfolding characteristics of the M221pep and I231pep peptides were also very different (Fig. 4B). The sigmoidal curve obtained for I231pep is consistent with a cooperative structure. In contrast, the shorter peptide, M221pep, at best showed the tail end of a sigmoidal (unfolding) curve indicative of much-reduced stability. Furthermore, the Tm values for I231pep increased with increasing peptide concentration, which is consistent with oligomer (i.e., possible coiled-coil dimer) formation (data not shown).
FIG. 4.
CD spectroscopy reveals that the C-terminal region of Zta contributes to stability of the coiled-coil dimer. (A) CD analysis determining the secondary structure of the M221pep and I231pep (at a concentration of 100 mM), shown by white triangles and black squares, respectively. (B) The melting curve was obtained by measuring a signal obtained at the wavelength of 222 nm for M221pep or I231pep through a range of temperature (from 2°C to 90°C).
Taken together, the biophysical data show that both peptides have a tendency to form helical structures in solution. This is consistent with the leucine zipper model. However, the ability to form stably folded dimers is markedly greater for the longer peptide, I231pep, which includes part of the CT region, than for the shorter peptide, M221pep, which encompasses the zipper region only. Thus, these experiments establish the importance of the proximal part of the CT region for Zta folding and explain the relative instability of the zipper region of Zta reported hitherto.
The proximal CT region is critical for Zta to bind to ZREs.
To address the ability of the Zta truncation mutants to bind to DNA in vitro, EMSAs were carried out using three different ZREs (AP-1, ZRE1, and ZIIIB) (Fig. 5). As expected, H199ter, the mutant lacking the zipper and CT regions, showed negligible DNA binding to any site. Analysis of the binding sites revealed that K219ter, M221ter, and L225ter were unable to interact with the AP1, ZIIIB, and ZRE1 sites. In contrast, I231ter and T234ter were able to interact almost as well as full-length Zta, and V237ter and L243ter were able to interact at least as well as full-length Zta. Thus, residues to I231ter are required for interaction with DNA.
FIG. 5.
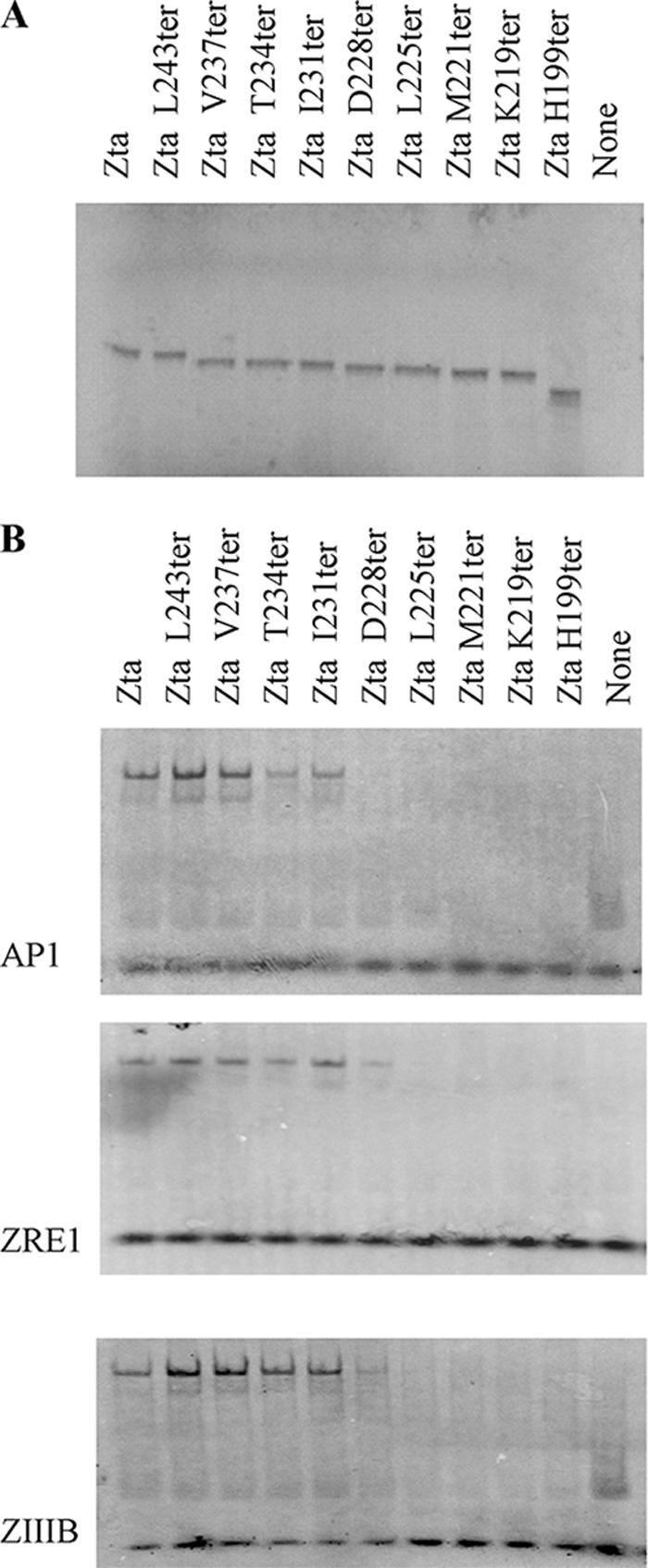
Requirement of the proximal CT region for interaction with three ZREs. (A) Proteins were assessed by SOS-PAGE. Abilities of indicated Zta truncation mutants to bind to ZREs were determined by EMSA analysis. In all cases the probe was in excess. Following separation of free and bound ZREs by electrophoresis, locations of the complexes were determined using phosphorimaging, with subsequent quantitation. Duplicate experiments were undertaken, and representative images are shown (B).
A further series of mutants, sequentially replacing all charged amino acids in the CT region with alanine residues, was generated (Zta1 to -3) (Fig. 6). The effect of these substitutions was determined by comparing the abilities of Zta and Zta1, -2, and -3 to interact with the ZIIIB and AP1 sites (Fig. 6). No contribution was observed for residues in the distal CT, but further mutation of both D226 and D228 in Zta3 dramatically reduced the ability of Zta to interact with both sites. Thus, mutation of D226 and D228, either alone or in conjunction with the other charged residues in the CT region, severely compromises DNA binding. Zta3 is also compromised in its ability to form dimers compared to Zta (Fig. 6). Overall, this shows that the proximal CT region contributes to the ability of Zta to interact with DNA and that the region required correlates with the region that aids dimerization.
FIG. 6.
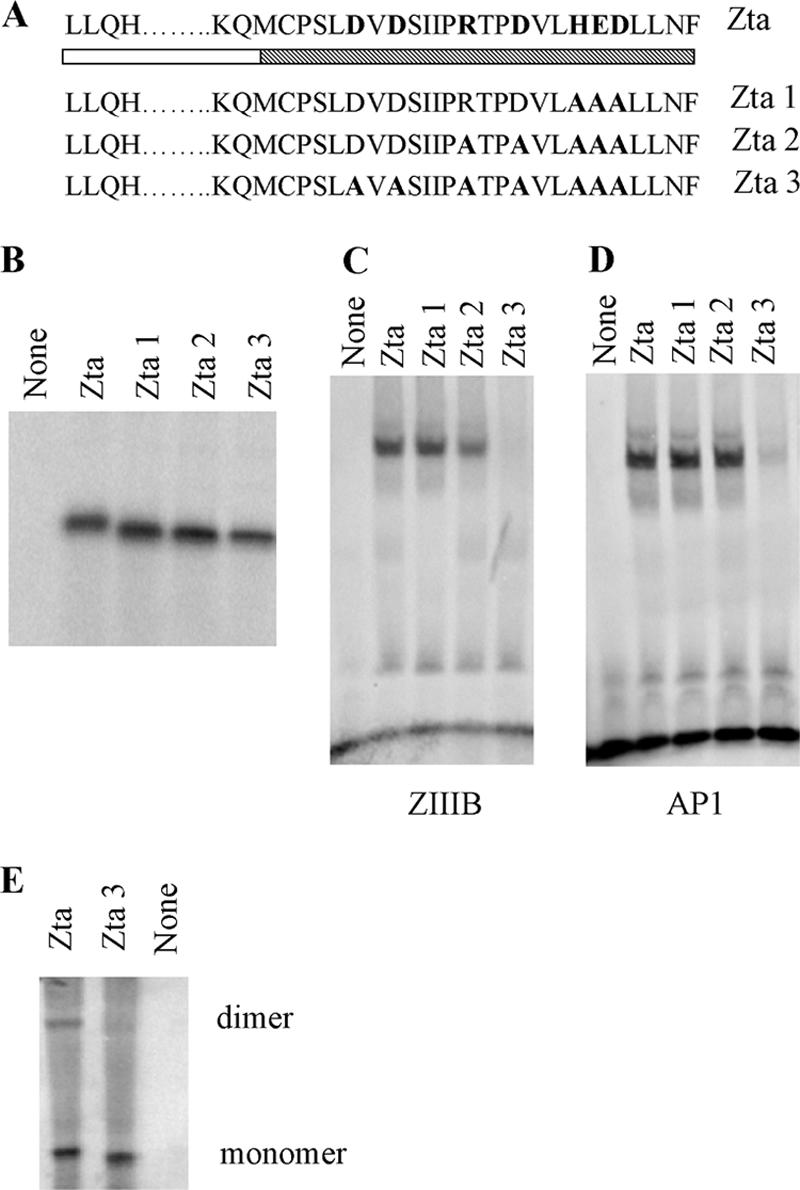
Importance of residues 226 and 228 for DNA-binding function of Zta. (A) Schematic representation of mutations of Zta1, Zta2, and Zta3, with the shading as described in Fig. 2. (B) Amounts of translation product generated for Zta and Zta truncated mutants used in the dimerization assay were assessed by SDS-PAGE and quantitated by phosphorimaging. (C and D) Equivalent amounts of protein were analyzed for their ability to bind DNA by EMSA analysis. Following separation of free and bound ZREs by electrophoresis, the locations of the complexes were detected using phosphorimaging, with subsequent quantitation. Duplicate experiments were undertaken, and representative images are shown for ZIIIB (C) and AP1 (D). (E) Equivalent amounts of protein were assessed for their abilities to form dimers as described in the legend to Fig. 2. Proteins of both monomer and dimer sizes are shown.
The distal CT region is not required for transactivation function of Zta in vivo.
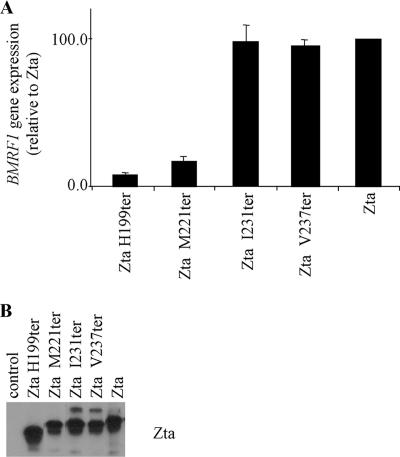
Since both the proximal and distal parts of the CT region influence dimerization but only the proximal CT region appears to be required for DNA binding in vitro, we sought to further question whether the distal CT region contributes anything to in vivo functions of Zta. A comparison of the abilities of truncation mutants of Zta to activate the transcription of a Zta-responsive gene embedded in the EBV genome revealed that I231ter was able to transactivate BMRF1 as well as wild-type Zta (Fig. 7), whereas M221ter was not. Therefore, the proximal CT region is required for this in vivo function, whereas the distal CT region can be dispensed with. It is therefore unlikely that the modest contribution of the distal CT region to dimerization and DNA binding contributes to DNA-binding-related functions in vivo.
FIG. 7.
The CT region is required for transactivation in vivo. (A) Expression vectors for indicated Zta truncation mutants were introduced into 293-BZLF1-KO cells, and expression of a viral gene, embedded in the genome, was detected using quantitative real-time PCR. Expression of Zta RNA was also detected and was used as a measure of transfection efficiency. The level of expression of the housekeeping gene, L32, was used to standardize signals. Experiments were undertaken in duplicate, and relative expression of BMRF1 was determined. These levels were then expressed relative to the level seen following expression of Zta. (B) Total protein extracts from cells were analyzed for expression of Zta using Western blot analysis.
DISCUSSION
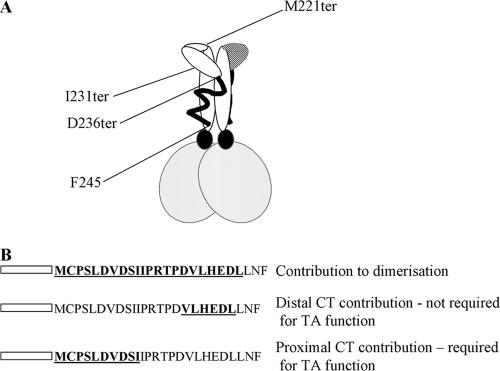
A contribution from the CT region to the dimerization ability of Zta has long been suspected (5, 11, 22). The recent resolution of the crystal structure of Zta, containing the basic region, the zipper region, and part of the CT region (to residue 236), revealed interactions between the amino-terminal part of the CT region and the zipper region that could potentially stabilize dimer formation (23). Here we show that the entire CT region lies in close contact with the zipper region, extending almost as far as the basic region (Fig. 8A). This provides a large potential interface over which stabilizing inter- or intramolecular interactions could occur.
FIG. 8.
Schematic diagram of structure of Zta and requirement of the CT region for function. (A) The schematic diagram of the structure of Zta is based on the crystal structure of Zta, which ends at D236 (23). The additional information that the distal part of the CT region (ending at F245) contacts the zipper and locations of the termination mutants I231ter and M221 ter are shown. (B) Locations of amino acids that contribute to dimerization are shown in bold and underlined. Each line represents mutants that support the conclusion shown on the right-hand side. Transactivation in vivo is represented as TA.
In order to address the mechanisms by which the CT region contributes to Zta function, we sought to determine whether the CT region impacts on the core properties of Zta: to fold as a dimer and to bind to DNA. Analysis of the ability of the Zta termination mutants to form dimers revealed that the zipper and residues to I230 were required and that the dimer is strengthened by the inclusion of further amino acids to L242 (Fig. 8B). The contribution of residues 221 to 230 in the proximal CT region to dimer formation was further probed using biophysical approaches. The results revealed that although the zipper region is able to fold as an α-helix in the absence of the CT region, the propensity of the peptides to fold into an α-helix and their ability to form dimers are increased considerably by the addition of 10 amino acids from the proximal CT region. The crystal structure of Zta shows that residues 221 to 224 form a hairpin loop immediately adjacent to the zipper region of Zta and residues 225 to 230 form part of the four-helix bundle (23). The hairpin loop is considered to be important to align the CT region and the zipper region into the four-helix bundle. The biophysical data support the role of the proximal CT region in stabilizing the structure of the zipper. Mutation of charged amino acids in the CT region highlights the contribution of D226 and D228 to DNA binding (Fig. 8B) and provides support for the proposal that an intramolecular salt bridge between D228 and R215 may stabilize the zipper (23).
An analysis of the relevance of the entire CT region for the transactivation function of Zta revealed that I231ter was able to transactivate a viral gene embedded in the genome as well as full-length Zta. This demonstrates that the ability of the proximal CT region to promote the dimerization and DNA-binding functions of Zta enables it to transactivate in vivo. In contrast, the more modest contribution to dimer stability promoted by the distal CT region is not required either for DNA-binding stability or for transactivation function in vivo. It remains to be determined whether the enhanced dimer stability observed when the entire CT region is included in the protein has any impact on functions of Zta that do not require DNA binding, such as interaction with host cell signaling proteins.
Acknowledgments
We thank Henri-Jaques Delecluse for 293-BZLF1-KO cells, the University of Sussex Proteomics Service for mass spectrometry, and Sarah Valencia for molecular biology assistance.
This work was supported by grants from the Medical Research Council and the Wellcome Trust.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Al Mehairi, S., E. Cerasoli, and A. J. Sinclair. 2005. Investigation of the multimerization region of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) protein K-bZIP: the proposed leucine zipper region encodes a multimerization domain with an unusual structure. J. Virol. 79:7905-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099-1104. [DOI] [PubMed] [Google Scholar]
- 3.Cayrol, C., and E. Flemington. 1996. G(0)/G(1), growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. [DOI] [PubMed] [Google Scholar]
- 4.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G(0)/G(1) cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. N., D. L. Y. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr-Virus Zta transactivator—a member of the bZip family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford, D. H. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. London B 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell, P., D. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemington, E., and S. Speck. 1990. Evidence of coiled-coil dimer formation by an Epstein-barr virus transactivator that lacks a heptad repeat of leucine residues. Proc. Natl. Acad. Sci. USA 87:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr Virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks, M. R., S. S. Al-Mehairi, and A. J. Sinclair. 2003. The zipper region of Epstein-Barr virus bZIP transcription factor Zta is necessary but not sufficient to direct DNA binding. J. Virol. 77:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks, M. R., S. Balesaria, C. Medina-Palazon, M. J. Pandya, D. N. Woolfson, and A. J. Sinclair. 2001. Biophysical analysis of natural variants of the multimerization region of Epstein-Barr virus lytic-switch protein BZLF1. J. Virol. 75:5381-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenney, S., J. Kamine, E. Holley-Guthrie, J. C. Lin, E. C. Mar, and J. Pagano. 1989. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 17.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macsween, K. F., and D. H. Crawford. 2003. Epstein-Barr virus—recent advances. Lancet Infect. Dis. 3:131-140. [DOI] [PubMed] [Google Scholar]
- 20.Morand, P., M. Budayova-Spano, M. Perrissin, C. W. Muller, and C. Petosa. 2006. Expression, purification, crystallization and preliminary X-ray analysis of a C-terminal fragment of the Epstein-Barr virus ZEBRA protein. Acta Crystallogr. Sect. F Struct. Biol. Crystalliz. Commun. 62:210-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packham, G., A. Economou, C. M. Rooney, D. T. Rowe, and P. J. Farrell. 1990. Structure and function of the Epstein-Barr virus BZLF1 protein. J. Virol. 64:2110-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petosa, C., P. Morand, F. Baudin, M. Moulin, J. B. Artero, and C. W. Muller. 2006. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol. Cell 21:565-572. [DOI] [PubMed] [Google Scholar]
- 24.Rooney, C. M., D. T. Roew, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 26.Schwarzmann, F., M. Jager, N. Prang, and H. Wolf. 1998. The control of lytic replication of Epstein-Barr virus in B lymphocytes. Int. J. Mol. Med. 1:137-142. [DOI] [PubMed] [Google Scholar]
- 27.Sinclair, A. J. 2003. bZIP proteins of human gamma herpesviruses. J. Gen. Virol. 84:1941-1949. [DOI] [PubMed] [Google Scholar]
- 28.Sinclair, A. J. 2006. Unexpected structure of Epstein-Barr virus lytic cycle activator Zta. Trends Microbiol. 14:289-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinclair, A. J., and P. J. Farrell. 1992. Epstein-Barr-virus transcription factors. Cell Growth Differ. 3:557-563. [PubMed] [Google Scholar]
- 30.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 31.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West, M. J., H. M. Webb, A. J. Sinclair, and D. N. Woolfson. 2004. Biophysical and mutational analysis of the putative bZIP domain of Epstein-Barr virus EBNA 3C. J. Virol. 78:9431-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]