Abstract
Molecular analyses of the protease-resistant prion protein (PrPres) from a few natural scrapie isolates showed by Western blotting some partial similarities with those observed in experimental ovine bovine spongiform encephalopathy (BSE). They showed a low apparent molecular mass of unglycosylated PrPres, although diglycosylated PrPres was less abundant than in ovine BSE. The prototype of such cases is the CH1641 experimental scrapie isolate. We analyzed PrPres molecular features from three French natural “CH1641-like” isolates, in comparison with CH1641 and BSE, after transmission of the disease in ovine transgenic mice (TgOvPrP4). One of these isolates (TR316211) behaved like the CH1641 isolate, with PrPres features in mice similar to those in the sheep brain. From two other isolates (O100 and O104), two distinct PrPres phenotypes were identified in mouse brains, with either high (h-type) or low (l-type) apparent molecular masses of unglycosylated PrPres, the latter being similar to that observed with CH1641, TR316211, or BSE. Both phenotypes could be found in variable proportions in the brains of the individual mice. In contrast with BSE, l-type PrPres from “CH1641-like” isolates showed lower levels of diglycosylated PrPres. From one of these cases (O104), a second passage in mice was performed for two mice with distinct PrPres profiles. This showed a partial selection of the l-type phenotype in mice infected with a mouse brain with predominant l-type PrPres, and it was accompanied by a significant increase in the proportions of the diglycosylated band. These results are discussed in relation to the diversity of scrapie and BSE strains.
Prion diseases, such as Creutzfeldt-Jakob disease in humans, scrapie in sheep and goats, and bovine spongiform encephalopathy (BSE) in cattle, are closely associated with the accumulation in infected tissues of an abnormal form of a host-encoded cellular prion protein (PrP C) (34). This disease-associated form of the protein (PrPd) differs from the normal form in its biochemical properties, including insolubility in nondenaturing detergents and partial resistance to degradation by proteases. Whereas the normal protein is fully sensitive to proteases (PrPsen), the abnormal prion protein is only partly degraded (PrPres), with its amino-terminal end being removed. The exact relationship between the infectious agent allowing the transmission of the disease and the prion protein remains controversial (13, 31, 34), but the expression of the prion protein is required for the development of the clinical disease, as demonstrated by transmission studies in mice devoid of the prnp gene encoding the mouse prion protein (12). A major hallmark of these diseases is the existence of different strains essentially defined by different features of the disease, including differences in the incubation periods and in the distributions of brain lesions, as shown by transmission experiments in mice of different genotypes (21). This has been related to some extent with the biochemical and biophysical properties of the disease-associated PrP protein, which might be related to the conformation of the protein (6, 7, 14, 30, 35-37, 40, 41).
Until recently, in contrast with the diversity of experimental strains originating from scrapie, the infectious agent involved in BSE in cattle was thought to be extremely uniform and stable following transmission to other species and to be linked to the infection by a single major strain (9, 10). A typical molecular signature of the BSE agent was identified by PrPres Western blot analysis (3, 14, 23, 30, 38, 42). These were the first experimental data to suggest a relationship between the variant form of Creutzfeldt-Jakob disease in humans and the BSE agent infecting cattle (14), which was later confirmed by the analysis of the pathogenic behavior of the infectious agent following transmission of the disease in wild-type mice (11). Such an approach has also made it possible to search for the possible transmission of the BSE agent under natural conditions in animals, such as sheep and goats (3, 22, 23, 33, 38, 39, 42). This recently led to the identification of the first, and so far unique, case of prion disease in a goat with molecular and biological properties similar to that found in BSE (18). Although still unconfirmed by bioassay, another case in a goat with molecular properties indistinguishable from BSE was recently reported in the United Kingdom (26).
However, a few cases of transmissible spongiform encephalopathies (TSEs) that showed partial similarities to experimental ovine BSE were described in sheep. This was first demonstrated in the CH1641 experimental scrapie isolate, originally isolated from a British scrapie case and then maintained by serial transmissions in sheep (20, 24). This experimental isolate was characterized by a lower apparent molecular mass of unglycosylated PrPres, very close to that found in ovine BSE (3, 24, 38). Similarly, we previously reported two cases (O100 and O104) with similar properties within a French scrapie-infected flock, although detailed analysis by both Western blotting and immunohistochemistry also revealed features different from those of ovine BSE in these cases (32). In both experimental (CH1641) and natural (O100 and O104) situations, molecular characterization using an immunohistochemical approach has been helpful for discrimination of these isolates from BSE (25, 32).
The usefulness of transgenic mice expressing the ovine prion protein for the detection and characterization of the infectious agent involved in sheep prion diseases has been emphasized by several studies in recent years (2, 16, 17, 43). The transmission in TgOvPrP4 ovine transgenic mice of the CH1641 isolate, which had previously consistently failed to transmit to wild-type mice, was previously reported (24). We now report the detailed molecular characterization of PrPres proteins of three similar French natural TSE isolates (O100, O104, and TR316211) following the transmission of the disease in TgOvPrP4 mice, in comparison with CH1641, all of them sharing some molecular similarities with ovine BSE regarding the low apparent molecular mass of unglycosylated PrPres.
MATERIALS AND METHODS
TSE sheep isolates.
TSE sheep isolates included the CH1641 experimental scrapie isolate (kindly provided by N. Hunter, Institute for Animal Health, Edinburgh, United Kingdom) and three unusual French natural TSE isolates. Molecular analyses of CH1641 transmitted to TgOvPrP4 ovine transgenic mice have been previously described (2). The three natural isolates were two V136R154Q171 homozygous sheep diagnosed with TSE in 1996 and 1997 within a scrapie-infected flock (32) in Pyrénées-Atlantiques and a case identified in 2003 in an A136R154Q171 homozygous sheep in another region of France (Allier). The last isolate also showed, as previously described in experimental ovine BSE (2), a low apparent molecular mass of unglycosylated PrPres (0.1 to 0.4 kDa lower than in cattle BSE) and reduced PrPres labeling with P4 monoclonal antibody.
Mouse lines and experimental infections.
The ovine transgenic mouse line TgOvPrP4 expressing the ovine prion protein (A136R154Q171 sequence) under the control of the neuron-specific enolase promoter on a murine prnp null background has been previously described (17). The mice were cared for and housed according to the guidelines of the French Ethical Committee (decree 87-848) and European Community Directive 86/609/EEC. Experiments were performed in the biohazard prevention area (A3) of the authors' institution with the approval of the Rhône-Alpes Ethical Committee for Animal Experiments.
Four- to 6-week-old female mice were inoculated intracerebrally with 10% (wt/vol) brain homogenates in 5% glucose in distilled water (20 μl per animal), and the brains were sampled at the terminal stage of the disease or after death of the animal for intercurrent disease or ageing. The whole brain of every second mouse was frozen and stored at −80°C before Western blot analysis, and the other brains were fixed in 4% paraformaldehyde for histopathological studies.
Extraction of PrPres.
PrPres was obtained following concentration by ultracentrifugation, as previously described (4). Dissociation of half of the mouse brain after sagittal section was performed in 5% glucose in distilled water (10% [wt/vol]), using disposable blenders, and complete homogenization was obtained by forcing the brain suspension through a 0.4-mm-diameter needle. A 330-μl volume was brought to 1.2 ml in 5% glucose before incubation with proteinase K (10 μg/100 mg brain tissue; Roche) for 1 h at 37°C. N-Lauroyl sarcosyl (30%; 600 μl; Sigma) was added. After incubation at room temperature for 15 min, samples were centrifuged at 100,000 × g for 2 h on a 10% sucrose cushion in a Beckman TL100 ultracentrifuge. The pellets were resuspended and heated for 5 min at 100°C in 50 μl TD4215 denaturing buffer (4% sodium dodecyl sulfate, 2% β-mercaptoethanol, 192 mM glycine, 25 mM Tris, 5% sucrose).
In some experiments, deglycosylation was performed using peptide:N-glycosidase F (PNGase) (kit P07043; BioLabs). Denatured samples of PrPres in TD4215 buffer (1 to 2 μl) were mixed with denaturing buffer from the PNGase kit, G7 buffer, NP-40, and PNGase according to the manufacturer's instructions. After incubation at 37°C for 1 h, samples were ready for Western blot analysis, following appropriate dilution in TD4215 buffer.
Western blot analysis.
Samples were run in 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted to nitrocellulose membranes in transfer buffer (25 mM Tris, 192 mM glycine, 10% isopropanol) at a constant 400 mA for 1 h. The membranes were blocked for 1 h with 3% bovine serum albumin in phosphate-buffered saline-Tween 20 (0.1%) (PBST). After two washes in PBST, the membranes were incubated (1 h at room temperature) with the monoclonal antibody Bar233 (1/5,000; kindly provided by J. Grassi, C. E. A.-Saclay) or P4 (0.2 μg/ml; r-biopharm, Germany) against the ovine PrP sequences 144-FGNDYEDRYYRE-155 and 89-GGGGWGQGGSHSQWNK-104 PrP, respectively (32). After three washes in PBST, the membranes were incubated (30 min at room temperature) with peroxidase-labeled conjugate anti-mouse immunoglobulin G (heavy and light chains; 1/2,500 in PBST) (reference 1010-05; Clinisciences). Streptavidin (5 ng/ml; S5512) was added to the conjugate solution. After three washes in PBST, bound antibodies were detected either on films after exposure of the membranes to Kodak Biomax MR films (Sigma) using ECL (Amersham) chemiluminescent substrate or, for quantitative studies, by direct capture with the Versa Doc (Bio-Rad) analysis system using a Supersignal (Pierce) chemiluminescent substrate. Quantitative studies were performed using Quantity One (Bio-Rad) software following at least three independent runs of the samples. Glycoform ratios were expressed as mean percentages (± standard deviations) of the total signal for the three PrPres glycoforms, and the apparent molecular masses were evaluated by comparison of the positions of the PrPres bands with a biotinylated marker (B2787; Sigma). The intensities of PrPres signals obtained with Bar233 or P4 antibody were compared by quantification of total PrPres signals obtained with both antibodies.
RESULTS
Two distinct molecular behaviors of “CH1641-like” isolates in TgOvPrP4 ovine transgenic mice.
We transmitted the disease by intracerebral inoculation in TgOvPrP4 ovine transgenic mice of three natural sheep TSE isolates (O100, O104, and TR316211) and one from the CH1641 experimental scrapie source. These four isolates showed a lower apparent molecular mass of unglycosylated PrPres in sheep brain than in cattle BSE and reduced PrPres labeling by P4 monoclonal antibody, as found in experimental ovine BSE. The transmission results, with mean survival periods and detection of PrPd assessed by Western blotting or immunohistochemistry, are given in Table 1, showing that these four isolates were efficiently transmitted to ovine transgenic mice.
TABLE 1.
TSE sources transmitted to TgOvPrP4 ovine transgenic micea
| TSE source | Breed | PrP genotypeb | Survival period (days p.i. [mean ± SD]) | PrPd-positive micec |
|---|---|---|---|---|
| Sheep isolates | ||||
| CH1641 | Cheviot | AxQ/AxQ | 245 ± 17 (p1) | 12/12 |
| 220 ± 31 (p2) | 11/11 | |||
| O100 | Manech tête rousse | VRQ/VRQ | 364 ± 61 | 12/12 |
| O104 | Manech tête rousse | VRQ/VRQ | 248 ± 50 (p1) | 10/10 |
| 204 ± 6 (p2 from a PrPres l-type mouse) | 13/13 | |||
| 238 ± 29 (p2 from a PrPres h-type mouse) | 13/13 | |||
| TR316211 | Unknown | ARQ/ARQ | 235 ± 26 | 8/8 |
| Experimental strainsd | ||||
| Chandler | 396 ± 100 | 9/9 | ||
| C506M3 | 333 ± 26 | 11/11 | ||
| 87V | 258 ± 44 | 10/10 | ||
| 79A | 342 ± 20 | 12/12 | ||
| BSE | 375 ± 51 | 12/12 |
Incubation periods of the disease and results of detection of disease-associated PrP by Western blotting or immunohistochemistry are shown.
Amino acids 136, 154, and 171.
Number of positive mice/number of mice examined by Western blotting or immunohistochemistry.
Second passage in TgOvPrP4 mice from previously reported primary passage (2).
We then examined the PrPres Western blot profile from each of the positive mice available for Western blot analysis. The results (Fig. 1) showed two distinct behaviors depending on the isolate. For both TR316211 natural and CH1641 experimental TSE isolates, all the mice showed similar PrPres patterns, with Bar 233 antibody directed against the 144-to-155 ovine PrP sequence, characterized by a low apparent molecular mass of unglycosylated PrPres (∼18.5 to 19.0 kDa) (Fig. 1A and B); in comparison with a PrPres control from TgOvPrP4 mice infected with the C506M3 scrapie strain, the apparent molecular mass of the unglycosylated PrPres was 0.5 to 1 kDa lower in mice infected with the TR316211 isolate. This was associated with strongly reduced labeling by P4 antibody, directed against the 93-to-99 ovine PrP sequence, compared to that observed with Bar233 antibody.
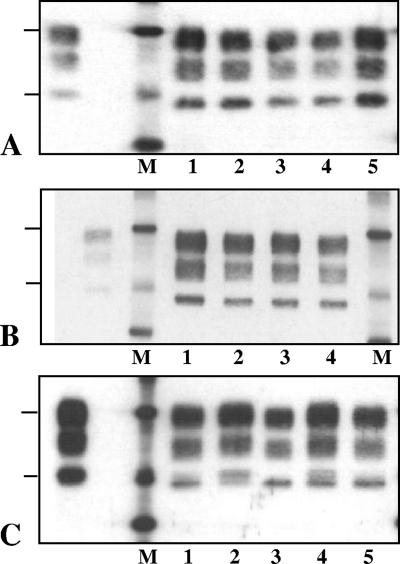
FIG. 1.
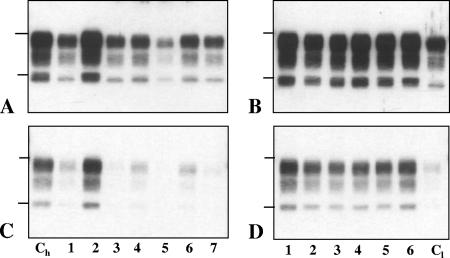
Western blot features of PrPres detected with Bar233 antibody in individual TgOvPrP4 mice infected with CH1641 (A), TR316211 (B), or O104 (C) sheep isolates, shown in lanes at the right of the molecular mass marker (M). PrPres from TgOvPrP4 mice infected with C506M3 is shown in the lane to the left of the molecular mass marker in each panel. The bars on the left of the panel indicate the 29.0- and 20.1-kDa marker positions.
In contrast, not all of the mice infected with the O100 or O104 isolate (Fig. 1C) showed the same molecular pattern with Bar 233 antibody. More precisely, Western blot analyses showed the possible presence of two PrPres species, characterized by a higher (h-type) or a lower (l-type) apparent molecular mass of unglycosylated PrPres, in variable proportions depending on the individual mice. In some mice, both types could be detected (Fig. 1C, lane 4), while in other mice a marked preponderance of one of the two types was observed (Fig. 1C, lanes 2 and 3). While l-type PrPres had an apparent molecular mass similar to that found in CH1641-infected mice, h-type PrPres was similar to that found in C506M3-infected mice. Such differences between individual mice were also detected between the different mice after deglycosylation by PNGase treatment (Fig. 2A). This was also associated with reduced labeling by P4 monoclonal antibody in mice with l-type PrPres, including after PNGase treatment (Fig. 2B). A P4-labeled PrPres population, however, could be detected in all of the mice following sufficient exposure of the blots, but contrary to what was observed with Bar 233 antibody in those mice that showed a mixture of the two PrPres types, the unglycosylated PrPres always appeared as a single h-type band, as observed in the C506M3 scrapie strain control. These data are consistent with the preferential detection of h-type PrPres by P4 antibody.
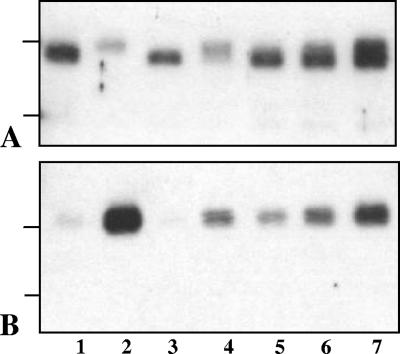
FIG. 2.
Western blot features of PrPres after PNGase deglycosylation in TgOvPrP4 mice infected with BSE (lane 1), C506M3 (lane 2), and O104 at first passage (lanes 3 to 7). The O104-infected samples in lanes 3 to 7 are from the same mice as those in Fig. 1C, lanes 3, 2, 5, 1, and 4, respectively. PrPres was detected with Bar233 (A) and P4 (B) monoclonal antibodies. The bars on the left of the panels indicate the 20.1- and 14.3-kDa marker positions.
Using Bar 233 and P4 antibody, l-type PrPres from mice infected with the three natural TSE isolates or with experimental scrapie CH1641 behaved similarly to PrPres in mice infected with experimental ovine BSE on the bases of the apparent molecular masses of unglycosylated PrPres and reduced PrPres labeling by P4 monoclonal antibody (Fig. 3). Analysis of the glycoform proportions, however, showed, as in sheep isolates, lower proportions of the diglycosylated band in mice infected with the three natural TSE isolates and with CH1641 (Fig. 4B) than in BSE-infected mice (Fig. 4A).
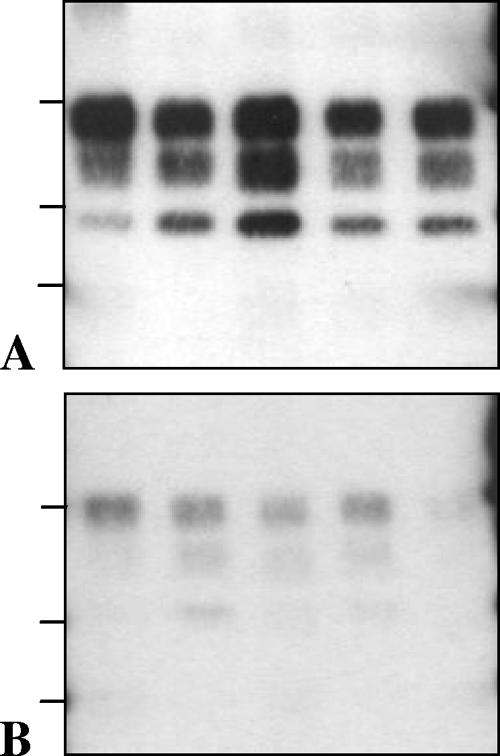
FIG. 3.
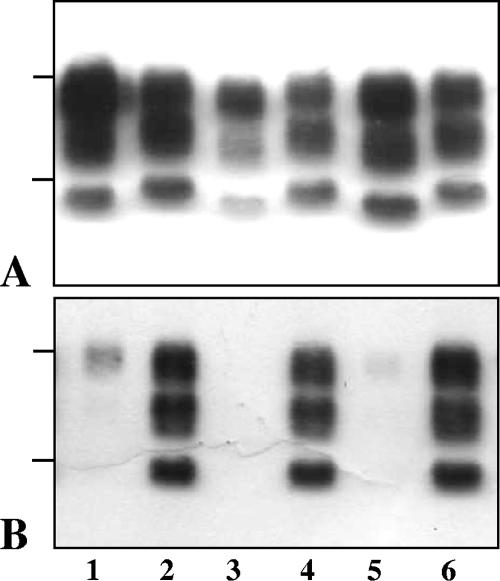
Western blot features of PrPres in TgOvPrP4 mice infected with, from the left to the right lanes in each panel, BSE, CH1641, TR316211, O104, and O100. In the last two cases, the samples were from one of the inoculated mice showing predominant l-type PrPres. PrPres was detected by Bar233 (A) and P4 (B) antibodies. The bars on the left of the panels indicate the 29.0-, 20.1-, and 14.3-kDa marker positions.
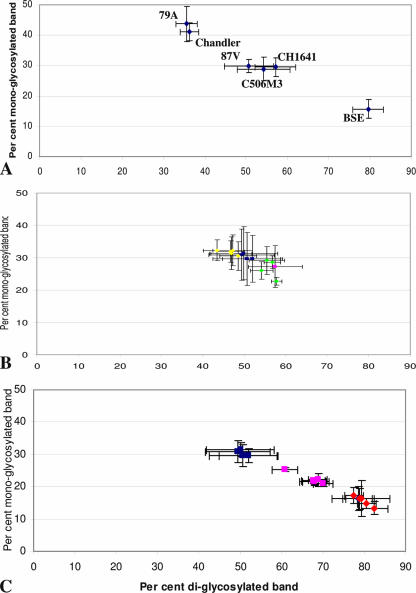
FIG. 4.
Glycoform ratios (means ± standard deviations) of PrPres detected in TgOvPrP4 mouse brains with Bar 233 antibody. (A) Means obtained from all of the Western blot-positive mice (five or six) of the experimental groups at the second passage for experimental scrapie or BSE strains derived from mouse-adapted strains and for CH1641. (B) Means obtained from each of the individual mice with l-type PrPres in the experimental groups at the first passage of CH1641 (yellow diamonds), TR316211 (green diamonds), O100 (pink square), and O104 (blue squares). (C) Means obtained from each of the individual mice with l-type PrPres of the experimental groups infected at the first (blue squares) or second (pink squares) passage from isolate O104 or at the second passage from mouse-adapted BSE (red diamonds).
Partial selection of the two PrPres phenotypes following a second passage in ovine transgenic mice.
We then studied, in comparison with the CH1641 isolate, the behavior at the second passage in TgOvPrP4 mice of one of the natural isolates (O104), which showed variable PrPres Western blot profiles among the different infected mice at the first passage. Two different mice from the first passage of O104 were chosen, one with only l-type PrPres detectable (“l-type mouse”) (Fig. 1C, lane 3) and the other one (“h-type mouse”) with predominant h-type PrPres (Fig. 1C, lane 2). As summarized in Table 1, transmission results showed a slightly shortened incubation period at the second passage (10 to 44 days difference in the mean incubation periods) and, for the O104 isolate, also indicated that the incubation period was significantly shorter (P = 0.0002) and less variable in mice inoculated with the first passage l-type mouse (204 ± 6 days postinoculation [p.i.]) than with the first passage h-type mouse (238 ± 29 days p.i.). These incubations are in the same range as those observed with the CH1641 isolate (220 ± 31 days p.i. at the second passage).
As for analysis of the PrPres profiles with Bar233 antibody, all but one (six of seven) of the mice inoculated with the l-type mouse (group 1) showed a single band corresponding to l-type PrPres, and all of the mice (six of six) (group 2) inoculated with predominant h-type PrPres showed a mixture of the two PrPres types. This was shown by the analysis of the apparent molecular masses using Bar 233 antibody (Fig. 5A and B) and by P4 labeling, which showed recognition of the single h-type PrPres band (Fig. 5C and D). All the mice inoculated with CH1641 at the second passage, as at the first passage, had l-type PrPres.
FIG. 5.
Western blot features of PrPres detected with Bar233 (A and B) or P4 (C and D) antibody in TgOvPrP4 mice infected with O104 isolate at the second passage. (A and C) PrPres from the seven mice inoculated with an l-type PrPres mouse at the first passage; the left lanes (Ch) show a PrPres control from one of the other six mice inoculated with predominant h-type. (B and D) PrPres from the six mice inoculated with predominant h-type PrPres; the right lanes (Cl) show l-type PrPres from one of the other seven mice inoculated with l-type PrPres. The bars on the left of the panels indicate the 29.0- and 20.1-kDa marker positions.
Molecular comparisons of l-type PrPres in mice infected with “CH1641-like” natural isolates or with experimental scrapie and BSE sources.
Finally, we compared the PrPres electrophoretic patterns in mice with l-type PrPres with that found in a panel of experimental scrapie or BSE sources originating from scrapie or BSE strains previously adapted in wild-type mice and then transmitted to TgOvPrP4 ovine transgenic mice. Analyses were performed at the second passage in TgOvPrP4 mice in order to reproduce the absence of a species barrier during transmission of sheep isolates to ovine transgenic mice. The transmission results from these experiments are summarized in Table 1.
In each of these experimental groups, PrPres profiles were similar in all the individual mice, and for each strain, they were similar at this second passage in TgOvPrP4 mice to those previously described at the first passage. This implies that the 87V scrapie strain and BSE shared an l-type PrPres, as shown with Bar 233 antibody (Fig. 6A), that was poorly reactive with P4 antibody (Fig. 6B), which thus appears comparable, regarding the PrPres cleavage, to the situation in some (O100 and O104) or all (TR316211 and CH1641) of the mice infected with “CH1641-like” sheep TSE samples. In contrast, mice infected with the C506M3, Chandler, and 79A scrapie strains showed higher apparent molecular masses of unglycosylated PrPres and strong PrPres labeling by P4 antibody.
FIG. 6.
Western blot features of PrPres in TgOvPrP4 mice infected with CH1641 isolate (lane 1) and with Chandler, 87V, 79A, BSE, and C506 strains (lanes 2 to 6, respectively) at the second passage in TgOvPrP4 mice. PrPres was detected by Bar233 (A) and P4 (B) antibodies. The bars on the left of the panels indicate the 29.0- and 20.1-kDa marker positions.
As for glycoform ratios, BSE was heavily diglycosylated and Chandler and 79A were poorly diglycosylated, while 87V, C506M3, and CH1641 shared close and intermediate levels of diglycosylated PrPres (Fig. 4A). Glycoform ratios in mice with l-type PrPres infected with “CH1641-like” isolates were similar to those observed with the 87V, C506M3, and CH1641 scrapie sources (Fig. 4B). However, a major change occurred at the second passage performed from the O104-inoculated mice with l-type PrPres (Fig. 4C). This led to a shift toward significantly higher levels (P < 0.0001) of the diglycosylated band, although these proportions remained significantly lower (P < 0.0001) than those found in BSE-infected mice (Fig. 6C).
DISCUSSION
We report the analysis in ovine transgenic mice (TgOvPrP4) of several natural TSE isolates identified in France, the study of which had been triggered by the observation of similarities with experimental ovine BSE in regard to some of the molecular features of PrPres. The situation of these isolates was not unprecedented, since a well-known scrapie isolate (CH1641), identified decades ago in the United Kingdom and then used for experimental studies of scrapie, had been shown by Western blot analysis to present similar features (24). While the last experimental source had always failed to be transmitted to wild-type mice, the associated strain of TSE agent could not be studied by such methods (20, 24). We previously reported that TgOvPrP4 mice allowed efficient transmission of CH1641 and maintained the PrPres features initially described in the sheep brain (2). By Western blotting, only some subtle differences from ovine BSE were found, the most obvious being the proportions of the different PrPres glycoforms. As previously described, at different passages during serial transmission of CH1641 in sheep (25), proximity between CH1641 and BSE in the apparent molecular masses of PrPres bands and in PrPres labeling by P4 antibody were demonstrated during two successive passages in TgOvPrP4 mice. All of our previous experiments with TgOvPrP4 mice had shown, and the present study confirms, that these mice faithfully reproduce the PrPres molecular features of the inoculated sources (2, 16), even following cross-species transmissions, such as scrapie and BSE from wild-type mice (2) or BSE from cattle (15). We have now extended our initial observations with the CH1641 source, and we report that a natural sheep TSE isolate (TR316211) showed the same molecular behavior in TgOvPrP4 mice.
Two other French natural TSE isolates (O100 and O104) from the same scrapie-infected flock were previously shown to exhibit molecular features comparable to those of the CH1641 isolate (32). These two isolates, however, showed a distinct and puzzling behavior after transmission in TgOvPrP4 mice with the presence of two PrPres types, either “CH1641-like,” i.e., with a low apparent molecular mass of unglycosylated PrPres (l-type), or with a higher apparent molecular mass of unglycosylated PrPres (h-type) and thus comparable to most classical scrapie cases (3, 23, 33, 38, 42). Whereas both molecular types could be identified in variable proportions depending on the individual mouse, the l-type phenotype could be more clearly separated by a second passage in ovine transgenic mice. Although this could be suggested by our data, it is presently unknown whether these two molecular phenotypes represent two distinct prion strains, a concept which has been based on the analysis of the phenotype of the disease (incubation periods, distribution and nature of brain lesions, and molecular features of PrPres) following transmission in wild-type mouse lines. While further studies are being considered to analyze the histopathological features in these ovine transgenic mice compared to scrapie and BSE strains, it is nevertheless not possible to rely on a large background of knowledge of prion strains, as in wild-type mice. Interestingly, as previously described for the CH1641 isolate, we failed in our attempts to transmit the disease from one of these three natural isolates (O100) in both RIII and C57BL/6 mice. It also cannot be established from our data whether the strains associated with l-type PrPres are identical in the three French natural isolates and in CH1641, although in TgOvPrP4 mice, their associated molecular features are quite similar and their incubation periods were in the same range (∼200 to 250 mean incubation periods), at least at the second passage.
The possible existence of a mixture of strains from a single scrapie case, which can only be separated by biological cloning, has been documented following bioassay in mice or hamsters (9, 10, 27). The best-studied example, however, has been that originating from transmissible mink encephalopathy, which allowed the isolation of two strains (HY and DY) characterized by major differences in clinical signs and incubation periods of the disease in a hamster model (8). This was found to be associated with distinct biochemical properties of PrPres, including a 1- to 2-kDa-faster migration of DY PrPres (∼19 kDa) than of HY PrPres (∼21 kDa), identified by Western blotting, associated with different cleavage sites for proteinase K and different β-sheet conformations (6, 7, 13). In some experiments, a mixture of the ∼19- and ∼21-kDa PrPres proteins could be identified, associated with a mixture of clinical signs (5). These data strongly resemble our molecular data in TgOvPrP4 mice following inoculation of the two unusual sheep isolates, O100 and O104. A major difference with transmissible mink encephalopathy studies, however, lies in the fact that our experiments did not involve any species barrier associated with a different prnp sequence of the experimental model. This favors the hypothesis that the two phenotypes found in mice could be the result of the presence of two strains in sheep as well, rather than of a possible change associated with the experimental model.
Detailed analysis of the two sheep brains for which such puzzling observations were made (O100 and O104) dismissed the idea that such sheep could represent BSE in sheep (32). In addition to the subtle differences observed by Western blotting, such as the lower proportions of diglycosylated PrPres and more intense PrPres labeling by P4 monoclonal antibody, the most obvious differences were revealed by epitope mapping of PrPd by immunohistochemistry using different antibodies against different PrP regions. Unlike ovine BSE, strong intraneuronal P4 labeling was indeed observed in some brain stem nuclei, but not in others. Given the results obtained in ovine transgenic mice, it is tempting to speculate that this could have been the result of the presence of the two PrPres phenotypes in sheep brains as well, which might possibly be present in different brain stem nuclei. This would be consistent with the intermediate PrPres P4 labeling observed by Western blotting. Importantly, during the second passage in TgOvPrP4 mice, which is not affected by variations in infectious titers in the sheep brain, this incubation period was significantly shorter than that found following BSE transmission in the mouse model. These data suggest a major biological difference between l-type-associated strains and BSE in these experiments. Although we cannot fully exclude the possibility that the BSE strain properties might be affected by passage in sheep, as was recently demonstrated (19), incubation periods at the second passage from ovine BSE were in the same range (more than 300 days p.i.) as those here shown for BSE initially derived from mouse-adapted BSE. It should be noted, however, that, intriguingly, after a second passage in TgOvPrP4 mice from an l-type mouse brain infected with the O104 isolate, an increase in the proportions of the diglycosylated PrPres occurred, and molecular differences from BSE, although still statistically significant, were less important. It is unknown whether this reflects the selection process of l-type PrPres, allowing accurate identification of the intrinsic properties of this PrPres form, or a genuine molecular change. Such a phenomenon, however, was not observed in scrapie sources derived from mouse-adapted experimental scrapie strains. This reinforces the need to initiate studies to characterize the infectious agent present in these mice infected with “CH1641-like” isolates at different passages in comparison with BSE and CH1641. The availability of TgOvPrP4 mouse brain tissues with different molecular features offers some opportunities for further attempts to transmit and characterize the involved strain(s) of infectious agent.
Together with the observation that 87V led to an incubation period shorter than those of other strains and in the same range as the “CH1641-like” isolates, further examination of possible similarities between these strains is required. It is indeed puzzling that 87V is the only scrapie strain that showed some molecular similarities with BSE in wild-type mice (29, 30, 36) that were also reproduced in TgOvPrP4 mice (2). 87V transmits to wild-type mice, but in C57BL mice with the sinc s7 allele, incubation periods are very long (>700 days p.i.) and some mice do not develop the disease during their life spans (9). We previously emphasized the intriguing common behaviors of 87V, CH1641, and BSE regarding their unusual interactions with the genetic background of the host (1). The CH1641 isolate has been considered in different studies to be a prototype of a particular group of scrapie strains (“C” strains) (20, 24).
Although isolates with such molecular features have rarely been reported (24, 32), the precise prevalence of this molecular phenotype in sheep is presently unknown. Furthermore, our data question the possibility that they might coexist within a mixture of strains that could in some cases prevent their identification by current discriminatory molecular methods. The identification and characterization of this particular phenotype could only be achieved by initiating transmission studies in a transgenic mouse model. Interestingly, a recent study has shown the existence of two distinct PrPres phenotypes in cattle that had been intracerebrally inoculated with a British scrapie brain pool collected in the 1990s (28). These two phenotypes, which differed between animals or, in one case, in the brain from a single animal, clearly recall the results obtained here in TgOvPrP4 mice. In a second experiment with a scrapie brain pool prepared from 11 sheep collected prior to 1975, all inoculated cattle showed PrPres with low apparent molecular masses, close to that of BSE. Experiments with the post-1990 brain pool showing five of seven animals with high apparent molecular masses of unglycosylated PrPres, however, demonstrated that such molecular changes were not necessarily associated with sheep-cattle transmission and with the presence of the bovine prnp gene of the experimental host. Such a possibility could have been considered following the finding of l-type PrPres after transmission, with long incubation periods, of three scrapie isolates in bovine transgenic mice (19). Since experiments in cattle were performed with a scrapie brain pool prepared from a limited number of sheep and since both scrapie pools showed a Western blot PrPres pattern like those of most scrapie cases (“h-type”), these data could suggest that TSE sources associated with l-type PrPres might be more frequent in sheep than expected and/or highly pathogenic for cattle.
Acknowledgments
We acknowledge Eric Morignat for statistical analysis of the data; Jérémy Verchère, Dominique Canal, and Johann Vulin for the Western blot experiments; and Emilie Antier and Clément Lavigne for the follow-up of animal experiments.
This work was supported by grants from GIS “Infections à prions.”
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Baron, T. 2002. Identification of inter-species transmission of prion strains. J. Neuropathol. Exp. Neurol. 61:377-383. [DOI] [PubMed] [Google Scholar]
- 2.Baron, T., C. Crozet, A.-G. Biacabe, S. Philippe, V. J. A. Bencsik, J.-Y. Madec, D. Calavas, and J. Samarut. 2004. Molecular analysis of the protease-resistant prion protein in scrapie and bovine spongiform encephalopathy transmitted to ovine transgenic and wild-type mice. J. Virol. 78:6243-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, T., J.-Y. Madec, D. Calavas, Y. Richard, and F. Barillet. 2000. Comparison of French natural scrapie isolates with bovine spongiform encephalopathy and experimental scrapie infected sheep. Neurosci. Lett. 284:175-178. [DOI] [PubMed] [Google Scholar]
- 4.Baron, T. G. M., and A.-G. Biacabe. 2001. Molecular analysis of the abnormal prion protein during coinfection of mice by bovine spongiform encephalopathy and a scrapie agent. J. Virol. 75:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartz, J. C., R. A. Bessen, D. McKenzie, R. F. Marsh, and J. M. Aiken. 2000. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J. Virol. 74:5542-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessen, R. A., and R. F. Marsh. 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 66:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessen, R. A., and R. F. Marsh. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68:7859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen, R. A., and R. F. Marsh. 1992. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 73:329-334. [DOI] [PubMed] [Google Scholar]
- 9.Bruce, M. 1996. Strain typing studies of scrapie and BSE, p. 223-236. In H. Baker and R. M. Ridley (ed.), Prion diseases. Humana Press, Totowa, NJ.
- 10.Bruce, M. E., A. Boyle, S. Cousens, I. McConnell, J. Foster, W. Goldmann, and H. Fraser. 2002. Strain characterization of natural sheep scrapie and comparaison with BSE. J. Gen. Virol. 83:695-704. [DOI] [PubMed] [Google Scholar]
- 11.Bruce, M. E., R. G. Will, J. W. Ironside, I. McConnell, D. Drummond, A. Suttle, L. McCardle, A. Chree, J. Hope, C. Birkett, S. Cousens, H. Fraser, and C. J. Bostock. 1997. Transmission to mice indicates that “new variant” CJD is caused by the BSE agent. Nature 389:498-501. [DOI] [PubMed] [Google Scholar]
- 12.Büeler, H., A. Aguzzi, A. Sailer, R.-A. Greiner, P. Autenried, M. Aguet, and C. Weissmann. 1993. Mice devoid of PrP are resistant to scrapie. Cell 73:1339-1347. [DOI] [PubMed] [Google Scholar]
- 13.Chesebro, B. 1999. Prion protein and the transmissible spongiform encephalopathy diseases. Neuron 24:503-506. [DOI] [PubMed] [Google Scholar]
- 14.Collinge, J., K. C. L. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383:685-690. [DOI] [PubMed] [Google Scholar]
- 15.Cordier, C., A. Bencsik, S. Philippe, D. Bétemps, F. Ronzon, D. Calavas, C. Crozet, and T. Baron. 2006. Transmission and characterization of BSE sources in two ovine transgenic mouse lines (TgOvPrP4 and TgOvPrP59). J. Gen. Virol. 87:3763-3771. [DOI] [PubMed] [Google Scholar]
- 16.Crozet, C., A. Bencsik, F. Flamant, S. Lezmi, J. Samarut, and T. Baron. 2001. Florid plaques in ovine PrP transgenic mice infected with an experimental ovine BSE. EMBO Rep. 21:952-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crozet, C., F. Flamant, A. Bencsik, D. Aubert, J. Samarut, and T. Baron. 2001. Efficient transmission of two different sheep scrapie isolates in transgenic mice expressing the ovine PrP gene. J. Virol. 75:5328-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eloit, M., K. Adjou, M. Coulpier, J.-J. Fontaine, R. Hamel, T. Lilin, S. Messiaen, O. Andreoletti, T. Baron, A. Bencsik, A.-G. Biacabe, V. Beringue, H. Laude, A. Le Dur, J. L. Vilotte, E. Comoy, J.-P. Deslys, J. Grassi, S. Simon, F. Lantier, and P. Sarradin. 2005. BSE agent signatures in a goat. Vet. Rec. 156:523-524. [DOI] [PubMed] [Google Scholar]
- 19.Espinosa, J. C., O. Andréoletti, J. Castilla, M. E. Herva, M. Morales, E. Alamillo, F. D. San-Segundo, C. Lacroux, S. Lugan, F. J. Salguero, J. P. M. Langeveld, and J. M. Torres. 2006. Sheep-passaged BSE agent exhibits altered pathobiological properties in bovine-PrP transgenic mice. J. Virol. doi: 10.1128/JVI.01356-06. [DOI] [PMC free article] [PubMed]
- 20.Foster, J. D., and A. G. Dickinson. 1988. The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet. Rec. 123:5-8. [DOI] [PubMed] [Google Scholar]
- 21.Fraser, H., and A. G. Dickinson. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol. 78:301-311. [DOI] [PubMed] [Google Scholar]
- 22.Gretzschel, A., A. Buschmann, M. Eiden, U. Ziegler, G. Luhken, G. Erhardt, and M. Groschup. 2005. Strain typing of German transmissible spongiform encephalopathies: field cases in small ruminants by biochemical methods. J. Vet. Med. B 52:55-63. [DOI] [PubMed] [Google Scholar]
- 23.Hill, A. F., K. C. L. Sidle, S. Joiner, P. Keyes, T. C. Martin, M. Dawson, and J. Collinge. 1998. Molecular screening of sheep for bovine spongiform encephalopathy. Neurosci. Lett. 255:159-162. [DOI] [PubMed] [Google Scholar]
- 24.Hope, J., S. C. E. R. Wood, C. R. Birkett, A. Chong, M. E. Bruce, D. Cairns, W. Goldmann, N. Hunter, and C. J. Bostock. 1999. Molecular analysis of ovine prion protein identifies similarities between BSE and an experimental isolate of natural scrapie, CH1641. J. Gen. Virol. 80:1-4. [DOI] [PubMed] [Google Scholar]
- 25.Jeffrey, M., L. Gonzalez, A. Chong, D. Foster, W. Goldmann, N. Hunter, and S. Martin. 2006. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J. Comp. Pathol. 134:17-29. [DOI] [PubMed] [Google Scholar]
- 26.Jeffrey, M., S. Martin, L. Gonzalez, J. Foster, J. P. M. Langeveld, F. G. Zijderveld, J. Grassi, and N. Hunter. 2006. Immunohistochemical features of PrP(d) accumulation in natural and experimental goat transmissible spongiform encephalopathies. J. Comp. Pathol. 134:171-181. [DOI] [PubMed] [Google Scholar]
- 27.Kimberlin, R. H., and C. A. Walker. 1978. Evidence that the transmission of one source of scrapie agent to hamster involves separation of agent strains from a mixture. J. Gen. Virol. 39:487-496. [DOI] [PubMed] [Google Scholar]
- 28.Konold, T., Y. H. Lee, M. J. Stack, C. Horrocks, R. B. Green, M. Chaplin, M. M. Simmons, S. A. C. Hawkins, R. Lockey, J. Spiropoulos, J. W. Wilesmith, and G. A. H. Wells. 2006. Different prion disease phenotypes result from inoculation with two temporally separated sources of sheep scrapie from Great Britain. BMC Vet. Res. doi: 10.1186/1746-6148-2-31. [DOI] [PMC free article] [PubMed]
- 29.Kuczius, T., and M. H. Groschup. 1999. Differences in proteinase K resistance and neuronal deposition of abnormal prion proteins characterize bovine spongiform encephalopathy (BSE) and scrapie strains. Mol. Med. 5:406-418. [PMC free article] [PubMed] [Google Scholar]
- 30.Kuczius, T., I. Haist, and M. H. Groschup. 1998. Molecular analysis of bovine spongiform encephalopathy and scrapie strain variation. J. Infect. Dis. 178:693-699. [DOI] [PubMed] [Google Scholar]
- 31.Lasmézas, C. I., J.-P. Deslys, O. Robain, A. Jaegly, V. Beringue, J.-M. Peyrin, J.-G. Fournier, J.-J. Hauw, J. Rossier, and D. Dormont. 1997. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 275:402-405. [DOI] [PubMed] [Google Scholar]
- 32.Lezmi, S., S. Martin, S. Simon, E. Comoy, A. Bencsik, J.-P. Deslys, J. Grassi, M. Jeffrey, and T. Baron. 2004. Comparative molecular analysis of the abnormal prion protein in field scrapie cases and experimental BSE in sheep using Western blot and immunohistochemical methods. J. Virol. 78:3654-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonno, R., E. Esposito, G. Vaccari, M. Conte, S. Marcon, M. Di Bari, C. Ligios, G. Di Guardo, and U. Agrimi. 2003. Molecular analysis of cases of Italian sheep scrapie and comparison with cases of bovine spongiform encephalopathy (BSE) and experimental BSE in sheep. J. Clin. Microbiol. 41:4127-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safar, J., H. Wille, V. Itrri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 4:1157-1165. [DOI] [PubMed] [Google Scholar]
- 36.Somerville, R. A., A. Chong, O. U. Mulqueen, C. R. Birkett, S. C. E. R. Wood, and J. Hope. 1997. Biochemical typing of scrapie strains. Nature 386:564. [DOI] [PubMed] [Google Scholar]
- 37.Somerville, R. A., R. C. Oberthür, U. Havekost, MacDonald, F., D. M. Taylor, and A. G. Dickinson. 2002. Characterization of thermodynamic diversity between transmissible agent strains and its theoretical implications. J. Biol. Chem. 277:11084-11089. [DOI] [PubMed] [Google Scholar]
- 38.Stack, J., M. J. Chaplin, and J. Clark. 2002. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases, and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 104:279-286. [DOI] [PubMed] [Google Scholar]
- 39.Sweeney, T., T. Kuczius, M. McElroy, M. G. Parada, and M. H. Groschup. 2000. Molecular analysis of Irish sheep scrapie cases. J. Gen. Virol. 81:1621-1627. [DOI] [PubMed] [Google Scholar]
- 40.Telling, C. G., P. Parchi, S. J. De Armond, P. Cortelli, P. Montagna, R. Gabizon, J. Mastrianni, E. Lugaresi, P. Gambetti, and S. B. Prusiner. 1996. Evidence for the conformation of the pathologic isoform of the prion enciphering and propagating prion diversity. Science 274:2079-2082. [DOI] [PubMed] [Google Scholar]
- 41.Thomzig, A., S. Spassov, M. Friedrich, D. Naumann, and M. Beekes. 2004. Discriminating scrapie and bovine spongifrom encephalopathy isolates by infrared spectroscopy of pathological prion protein. J. Biol. Chem. 279:33847-33854. [DOI] [PubMed] [Google Scholar]
- 42.Thuring, C. M., J. H. F. Erkens, J. G. Jacobs, J. G. Bossers, L. J. M. Van Keulen, G. J. Garssen, F. G. Van Zijderveld, S. J. Ryder, M. H. Groschup, M. H. Sweeney, and J. P. M. Langeveld. 2004. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size immunoreactivity and glycoprofile of prion protein. J. Clin. Microbiol. 42:972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilotte, J. L., S. Soulier, R. Essalmani, M. G. Stinnakre, D. Vaiman, L. Lepourry, J. C. DaSilva, N. Besnard, M. Dawson, A. Buschmann, M. Groschup, S. Petit, M. F. Madelaine, S. Rakatobe, A. LeDur, D. Vilette, and H. Laude. 2001. Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. J. Virol. 75:5977-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]