Abstract
MicroRNAs (miRNAs) are increasingly being recognized as major regulators of gene expression in many organisms, including viruses. Among viruses, members of the family Herpesviridae account for the majority of the currently known virus-encoded miRNAs. The highly oncogenic Marek's disease virus type 1 (MDV-1), an avian herpesvirus, has recently been shown to encode eight miRNAs clustered in the MEQ and LAT regions of the viral genome. The genus Mardivirus, to which MDV-1 belongs, also includes the nononcogenic but antigenically related MDV-2. As MDV-1 and MDV-2 are evolutionarily very close, we sought to determine if MDV-2 also encodes miRNAs. For this, we cloned, sequenced, and analyzed a library of small RNAs from the lymphoblastoid cell line MSB-1, previously shown to be coinfected with both MDV-1 and MDV-2. Among the 5,099 small RNA sequences determined from the library, we identified 17 novel MDV-2-specific miRNAs. Out of these, 16 were clustered in a 4.2-kb long repeat region that encodes R-LORF2 to R-LORF5. The single miRNA outside the cluster was located in the short repeat region, within the C-terminal region of the ICP4 homolog. The expression of these miRNAs in MSB-1 cells and infected chicken embryo fibroblasts was further confirmed by Northern blotting analysis. The identification of miRNA clusters within the repeat regions of MDV-2 demonstrates conservation of the relative genomic positions of miRNA clusters in MDV-1 and MDV-2, despite the lack of sequence homology among the miRNAs of the two viruses. The identification of these novel miRNAs adds to the growing list of virus-encoded miRNAs.
Marek's disease represents a highly contagious T-cell lymphoid neoplasia of chickens associated with huge economic losses to the poultry industry. The causative agent of the disease is an avian herpesvirus, Marek's disease virus (MDV), belonging to the genus Mardivirus, a member of the Alphaherpesvirinae subfamily (16). In addition to herpesvirus of turkeys (Meleagrid herpesvirus 1), the genus Mardivirus includes two distinct MDV species, MDV type 1 (MDV-1) (Gallid herpesvirus 2 [GaHV-2]) and MDV-2 (GaHV-3). MDV-1 includes all the pathogenic strains and some vaccine strains derived from MDV, while MDV-2 consists of apathogenic strains originally isolated from apparently normal chickens (7). Extensive studies of MDV-2 strains have shown that they are nononcogenic (33, 38) and are widely used as vaccines against Marek's disease in combination with other strains.
Genome sequences of the MDV-1 strains Md5 and GA (19, 36) and the MDV-2 strain HPRS24 (17) show a similar organization composed of two unique sequences, one long (UL) and one short (US), flanked by inverted long and short internal repeats (IRL and IRS, respectively) and by long and short terminal repeats (TRL and TRS, respectively). The similarity of genome structures and sequences is also reflected in the close phylogenetic relationships of these viruses (22). While the Mardivirus lineage is thought to have diverged from the mammalian herpesviruses belonging to the genera Simplexvirus and Varicellovirus about 131 million years ago, MDV-1 and MDV-2 are estimated to have separated only about 26 million years ago (D. J. McGeoch, personal communication). The vast majority of the genes within the UL and US regions of these two viruses are homologous with a high degree of conservation (25). However, many open reading frames (ORFs), particularly those in the IRL/TRL region, are unique to MDV-1. As some of the genes within this region such as the meq and vIL-8 genes are known to be associated with oncogenicity (23, 26), the absence of these ORFs may account for the nononcogenic features of MDV-2.
In addition to the better-known gene regulatory pathways mediated through encoded proteins, noncoding RNAs, particularly microRNAs (miRNAs), are increasingly being recognized as important regulators of gene expression. Because of their tiny structures and nonimmunogenic features, several viruses have exploited this new regulatory mechanism for survival within cells (14, 35). Among the virus-encoded miRNAs, the vast majority have been identified in herpesviruses. The numbers of herpesvirus-encoded miRNAs listed in the latest Release 9.1 of the miRBase database (http://microrna.sanger.ac.uk) include 23 in Epstein-Barr virus (EBV), 16 in rhesus lymphocryptovirus (rLCV), 11 in human cytomegalovirus, 13 in Kaposi's sarcoma herpesvirus, 9 in murine herpesvirus 68, 1 in herpes simplex virus (HSV), and 8 in MDV-1 (13). It has been suggested that miRNA-mediated regulation of gene expression is particularly suited for the distinct biological features of herpesviruses, including nuclear replication and latency (30).
The eight MDV-1-specific miRNAs mapped to the MEQ and the latency-associated transcript (LAT) regions (8), and their expression in primary lymphomas as well as in transformed lymphoblastoid cell lines paralleled the pattern of the MEQ gene expression, suggesting a major role for these miRNAs in MDV latency and transformation. Since MDV-2 is nononcogenic but is antigenically and evolutionarily close to MDV-1, we wanted to find out whether MDV-2 encoded any miRNAs and, if so, whether there was any conservation of miRNA sequences between MDV-1 and MDV-2. For this purpose, we constructed a library using small RNA fractionated from an MSB-1 lymphoblastoid cell line known to be coinfected with the BC-1 strain of MDV-1 and the HPRS24 strain of MDV-2 (1, 15). In this study, we report the identification of novel MDV-2-specific miRNAs from the MSB-1 cell line. We also demonstrate that although there was conservation of the relative genomic positions of MDV-1 and MDV-2 miRNA clusters, there was no sequence conservation despite the close phylogenetic and antigenic relationship between the two viruses.
MATERIALS AND METHODS
Cells and virus.
Primary cultures of chicken embryo fibroblasts (CEF) prepared from 10-day-old specific-pathogen-free embryos obtained from flocks maintained at the Institute for Animal Health were used for the propagation of viruses. MDV-2 strains HPRS24 (7) and SB-1 (33) grown in CEF for 48 to 72 h were used for the preparation of RNA for Northern blotting analysis. MSB-1 (1) and 769T (generated from a testicular lymphoma of a bird infected with MDV-1 strain RB-1B derived from the pRB-1B [27] bacterial artificial chromosome clone) lymphoblastoid cell lines were grown at 37°C in 5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 10% tryptose phosphate, and 1% sodium pyruvate.
Construction of a small RNA library and sequencing of miRNAs.
The cloning of small RNAs from MSB-1 cells was conducted by following the protocols described previously (29). Briefly, total RNA was extracted from MSB-1 cells using TRIzol reagent (Invitrogen, Paisley, United Kingdom), and 500 μg of total RNA was spiked with 0.5 nM of radiolabeled ([γ-32P]ATP [Amersham]) 19- and 24-nucleotide oligoribonucleotides containing the PmeI restriction site (5′-CGUACGCGGGUUUAAACGA-3′ and 5′-CGUACGCGGAAUAGUUUAAACUGU-3′) and then separated on a 15% denaturing polyacrylamide gel. A gel slice containing RNAs of 19- to 24-nucleotide size was excised and eluted overnight in 0.3 M NaCl at 4°C. RNA was recovered after ethanol precipitation and ligated sequentially to a 5′-end-adenylated 3′ adapter oligonucleotide (5′-rAppTTTAACCGCGAATTCCAG/3ddC/-3′, where r is ribonucleotide [Integrated DNA Technologies]) and a 5′ adapter chimeric DNA/RNA oligonucleotide (5′-ACGGAATTCCTCACTrArArA-3′). Reverse transcription (RT)-PCR was performed with a 3′ primer (5′-GACTAGCTGGAATTCGCGGTTAAA-3′) and a 5′ primer (5′-CAGCCAACGGAATTCCTCACTAAA-3′). The purified PCR products were digested with PmeI to eliminate size marker sequences. In order to introduce BanI restriction sites, a second PCR was performed using the primer pair 5′-CAGCCAACAGGCACCGAATTCCTCACTAAA-3′ and 5′-GACTAGCTTGGTGCCGAATTCGCGGTTAAA-3′, followed by concatamerization after BanI digestion and T4 DNA ligation. Subsequently, the 3′ ends of the concatemers were filled in by incubation for 15 min at 72°C with Taq polymerase in a standard PCR mixture and used directly for ligation into pGEM-T Easy vector (Promega, Southampton, United Kingdom). Selected clones were sequenced and analyzed.
Northern blotting analysis of miRNAs.
Total RNA was extracted from different cells using TRIzol reagent (Invitrogen). Samples of 20 μg total RNA were resolved using a 15% polyacrylamide-1× Tris-borate-EDTA-8 M urea gel and blotted to a GeneScreen Plus membrane (Perkin-Elmer). DNA oligonucleotides with sequences exactly complementary to candidate miRNAs were end labeled with [γ-32P]ATP (Amersham, Bucks, United Kingdom) and T4 polynucleotide kinase (New England Biolabs, Herts, United Kingdom) to generate high-specific-activity probes. Hybridization, washing, and autoradiography were carried out as previously described (29).
Immunofluorescence and Western blotting.
CEF were cocultivated with MSB-1 cells for 6 to 7 days, and MDV-specific plaques were stained with MDV-1-specific monoclonal antibody (MAb) BD1 (20) or MDV-2-specific MAb Y5 (18). The specific cell populations detected by the two antibodies were visualized with anti-mouse immunoglobulin G (IgG) conjugates of Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes), using a DM IRB model microscope (Leica Microsystems, Milton Keynes, United Kingdom). For Western blotting, 106 MSB-1 cells, uninfected CEF, or CEF infected with MSB-1 MDV isolates or SB-1 were lysed in protein gel sample buffer (8 M urea, 2% sodium dodecyl sulfate, 10 mM Tris-HCl [pH 6.8], 0.05% bromophenol blue) and separated on a NuPAGE 10% bis-Tris gel (Invitrogen) and transferred onto nitrocellulose membranes using an iBlot gel transfer system (Invitrogen). Western blotting (WB) was performed with MAb Y5, followed by an anti-mouse IgG peroxidase conjugate (Sigma-Aldrich, Dorset, United Kingdom). The membrane was finally developed with an ECL Western blotting analysis system (Amersham).
miRNA sequence accession numbers.
MDV-2 miRNAs miRNA-1 to miRNA-17 described here are annotated as mdv2-miR-M14 to mdv2-miR-M30, respectively, in the miRBase Sequence Database (http://microrna.sanger.ac.uk/sequences).
RESULTS
MSB-1 lymphoblastoid cell line is coinfected with MDV-1 and MDV-2.
Cocultivation of MSB-1 cells on CEF produced MDV-specific plaques 6 to 7 days after incubation. The specific identity of viral plaques on CEF was examined using an immunofluorescence (IF) test with MDV-1- or MDV-2-specific antibodies. Simultaneous detection of distinct IF-positive plaques stained with MDV-1-specific MAb BD1 and MDV-2-specific MAb Y5 confirmed the presence of both viruses in MSB-1 cells (not shown). On the basis of the number of plaques stained with each of the MAbs, the level of MDV-2 appeared to be at least 10-fold higher than MDV-1, perhaps reflecting the faster replication of MDV-2 in CEF. The presence of MDV-2 in MSB-1 cells was also confirmed by WB using MDV-2-specific MAb Y5, which detected specific bands on MSB-1 cells as well as in CEF infected with MSB-1-derived viruses (not shown). CEF infected with MDV-2 strain SB-1 was used as a control for the specificity of the MAb Y5. On the basis of these findings, which supported the presence of replicating MDV-2, we chose to use MSB-1 cells for constructing a small RNA library for cloning the MDV-2-specific miRNAs.
Identification of MDV-2-specific miRNAs from the MSB-1 cell line.
For the construction of the small RNA library from MSB-1 cells, a size-fractionated 19- to 24-nucleotide small RNA population was used. After adapter ligation, amplification, and restriction digestion with BanI, the amplicons were cloned as cDNA concatemers into pGEM-T Easy vector. Sequencing of ∼1,200 clones using vector-specific primers identified a total of 5,099 high-quality reads containing small RNA sequences with both the 5′ and 3′ adapters. BLAST (2) homology searches of the sequences were performed against the published HPRS24 strain sequence (GenBank accession number AB049735). A total of 518 sequences (about 10% of the total), representing 17 candidate miRNAs, perfectly matched the HPRS24 sequence. In addition, the MSB-1 library also revealed all the recently described (8) MDV-1 miRNAs (not shown).
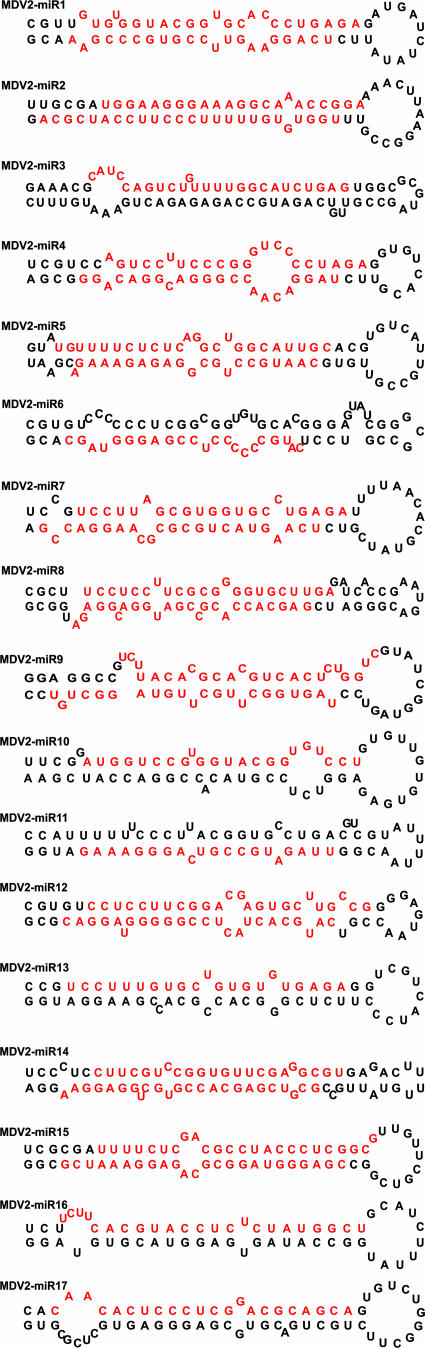
During the biogenesis of miRNAs, only one of the strands of the ∼20-bp miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) and guides gene regulation. Even though the non-miRNA passenger strand of the duplex is rapidly degraded (6), these strands could still be captured during cloning. Moreover, for some miRNAs, both strands can be incorporated into the RISC with comparable frequency. In the HPRS24 library, two mature forms, representing both strands of the duplex, were found for 10 out of the 17 candidate miRNAs, increasing the total number of cloned mature MDV-2-encoded miRNAs to 27. In most of these cases, the relative frequency of the passenger strand was much lower than that of the miRNA strand (Table 1). However, in some cases, both mature forms were incorporated with similar frequencies, suggesting the possibility that both strands may be functional and are incorporated into the RISC. Designations of the miRNAs were made sequentially according to their locations in the HPRS24 genome, and the suffix 5p or 3p was added to indicate the 5′ or 3′ arm, respectively, of the stem-loop precursor from which it was derived (Table 1). Some of the mature miRNAs showed variations in length (Table 1, variations are shown in parentheses). For further validation of HPRS24-encoded candidate miRNAs, each of the sequences including 60 to 80 bases of the surrounding sequence in the viral genome was subjected to MFOLD calculation (39), and secondary structures were drawn according to RNADRAW software (21). As shown in Fig. 1, the miRNA precursors showed an average length of ∼70 nucleotides and were able to form a hairpin structure. Moreover, 8 of the 17 candidates (Table 1) reside in pre-miRNAs predicted from this virus by using a computational method that was previously used for identification of miRNAs in other herpesviruses (28). These findings support the conclusion that the cloned sequences are MDV-2-encoded miRNAs.
TABLE 1.
Sequences and genomic positions of MDV-2 miRNAs
| MDV-2 miRNAa | Sequence (5′ to 3′)c | Length (nt) | Hits | nt positions |
|---|---|---|---|---|
| miR-1-5p | GUGUGGUACGGUGCACCCUGAGA | 23 | 1 | 126649-126671 |
| miR-1-3p | UCAGGAAGUUCCGUGCCCGAA | 21 | 1 | 126686-126706 |
| miR-2-5pb | UGGAAGGGAAAGGCAAACCGGA | 22 | 1 | 126881-126902 |
| miR-2-3pb | (UGG)UGUGUUUUUCCCUUCCAUC(GCA) | 20-23 | 30 | 126918-126940 |
| miR-3 | (CAU)CCAGUCUGUUUUGGCAUCUG(A) | 21-23 | 93 | 128012-128035 |
| miR-4-5p | AGUCCUUCCCGGGUCCCCUAGA | 22 | 3 | 128401-128422 |
| miR-4-3p | (U)AGGACAACCGGGACGGACAG(G) | 21-22 | 36 | 128435-128456 |
| miR-5-5p | (UG)UUUUCUCUCAGGCUGGCAUUGC | 22-24 | 6 | 128558-128581 |
| miR-5-3p | CAAUGCCUGCGGAGAGAAAGA | 21 | 4 | 128601-128621 |
| miR-6 | CAUGCCCCCCUCCGAGGGUAG(C) | 21-22 | 9 | 128757-128778 |
| miR-7-5p | UCCUUAGCGUGGUGCCUGAGA | 21 | 1 | 128868-128888 |
| miR-7-3p | UCAAGUACUGCGCGCAAGG(ACCG) | 19-23 | 11 | 128906-128928 |
| miR-8-5p | UCCUCCUUCGCGGGGUGCUUG(A) | 21-22 | 4 | 129002-129023 |
| miR-8-3p | GAGCACCACGCCGAUGGACGGA(GA) | 22-24 | 10 | 129044-129067 |
| miR-9-5p | UCUUACACGCACGUCACUCUGG(UC) | 22-24 | 15 | 129281-129304 |
| miR-9-3p | UAGUGGCUUGCUUGUAGGCUGU | 22 | 3 | 129318-129339 |
| miR-10b | AUGGUCCGUGGUACGGUGUCCU | 22 | 1 | 129431-129452 |
| miR-11b | UUAGAUGCCGUCAGGGAAAGA(U) | 21-22 | 3 | 130058-130079 |
| miR-12-5pb | CCUCCUUCGGACGAGUGCUUGCCG | 24 | 1 | 130170-130193 |
| miR-12-3pb | (CA)UGCACUACUCCGGGGGUAGGAC | 22-24 | 4 | 130206-130229 |
| miR-13 | (UCC)UUUGUGCUGUGUGUGAGA | 18-21 | 15 | 130304-130324 |
| miR-14-5p | CUUCGUCCGGUGUUCGAGGC(GU) | 20-22 | 15 | 130441-130462 |
| miR-14-3p | (G)CGUCGAGCACCGUGCUGGAG(GAA) | 20-23 | 17 | 130480-130502 |
| miR-15-5pb | (UU)UUCUCGACGCCUACCCUCG(GCG) | 20-23 | 100 | 130659-130681 |
| miR-15-3pb | CGAGGGUAGGCGCAGAGGAAAUCG | 24 | 1 | 130695-130718 |
| miR-16 | (U)CUUCACGUACCUCUCUAUGG(CU) | 21-22 | 118 | 130803-130824 |
| miR-17 | CAACACUCCCUCGGACGCAGC(A) | 21-22 | 15 | 136516-136537 |
The miRNAs (miR) derived from a single primary miRNA stem-loop precursor are indicated by a 5p (5′ arm) or a 3p (3′ arm) suffix.
miRNAs predicted using the computational method described previously (28).
Sequence variations surrounding the recovered HPRS24 strain miRNAs are indicated by variable nucleotides (nt) in parentheses.
FIG. 1.
Predicted secondary structures of MDV-2 pre-miRNAs These structure predictions were derived using the MFOLD algorithm (39). The mature miRNAs are indicated in red.
MDV-2-specific miRNAs are clustered in two regions of the genome.
We then examined the positions of the miRNAs in the 164,270-bp HPRS24 genome. Out of the 17 candidate MDV-2 miRNAs, 16 were clustered together in the same orientation in a 4.2-kb region (position 126649 to 130824 in the TRL/IRL region). Within this miRNA cluster, there were four putative ORFs: R-LORF2, R-LORF3, R-LORF4, and R-LORF5. Eight miRNAs in this cluster were located in the coding regions of these ORFs (Fig. 2). These included miRNA-2 (R-LORF5); miRNA-4, miRNA-5, and miRNA-6 (R-LORF4); and miRNA-10 (R-LORF3) and miRNA-13, miRNA-14, and miRNA-15 (R-LORF2). The orientations of miRNA-2 and miRNA-4, -5, and -6 are the same as that for R-LORF5 and R-LORF4, respectively. On the other hand, miRNA-10 and miRNA-13, -14, and -15 are located in an orientation opposite to that of R-LORF3 and R-LORF2, respectively. The only MDV-2 miRNA outside this cluster, miRNA-17, was located in the IRS/TRS region (position 136516 to 136537) in the opposite orientation to the coding region of ICP4 between amino acids STPSHVSRSPDTAASEG (position 1629 to 1645). The ICP4 secondary structure prediction (PSI BLAST) showed that this miRNA is located in a loop region between two alpha helices that showed no conservation with MDV-1, HVT, or HSV-1.
FIG. 2.
Genomic location of MDV-1 and MDV-2 miRNAs. (A) Schematic diagram showing the loci in the MDV-2 genome to which the miRNAs (miR) identified in this report map. The TRL and IRL flanking the unique long region as well as the IRS and TRS flanking the unique short region are shown. Genomic positions and the orientation of the ORFs contained in the miRNA loci are indicated. Small arrowheads indicate the locations of the MDV-2 miRNAs. (B) Schematic diagram showing the relative position of the miRNA-containing loci of MDV-1 genome reported recently (8).
The clustering of the 16 miRNAs in the same orientation in the 4.2-kb region of the TRL/IRL suggests that they may be processed from a single long pre-miRNA transcribed from a unique promoter. One would thus expect similar levels of expression of these miRNAs. However, the numbers of copies of each miRNA, taken as an indication of the abundance of the miRNAs in the library, showed marked differences. miRNA-3, miRNA-15-5p, and miRNA-16 were the most frequently identified, with approximately 100 copies each. On the other hand, miRNAs such as miRNA-1, miRNA-2-5p, miRNA-7-5p, miRNA-10, miRNA-12-5p, and miRNA-15-3p were represented only once (Table 1). This difference in the abundance of the miRNAs is most likely due to differences in the Drosha processing and to miRNA stability.
Demonstration of miRNA expression by Northern blotting.
For further confirmation of expression of MDV-2 miRNAs in MSB-1 and MDV-infected CEF, Northern blotting hybridization with individual miRNA probes was carried out. For those miRNAs where both strands were cloned, probes only from the 5p strand was used in Northern blotting. As a negative control for the dual-virus-infected MSB-1 cells, a lymphoblastoid cell line 769T infected only with MDV-1 (RB-1B strain) was used. Uninfected CEF as well as CEF lytically infected with MDV-2 strain HPRS24 were also included in the analysis. Northern blotting detected 15 out of the 17 MDV-2 miRNAs in MSB-1 cells as well as in HPRS24-infected CEF. No miRNAs were detected with RNA extracted from the uninfected CEF or the MDV-1-infected 769T cell line (Fig. 3). There were differences in the levels of expression (based on the intensity of the signals) of miRNAs between the two cell types, with MSB-1 cell line expressing most of the miRNAs at higher levels. Both pre-miRNA and mature miRNAs could be detected in both cell types with Northern blotting. In some cases, such as miRNA-5, miRNA-12-5p, and miRNA-14-5p, the amount of pre-miRNAs was equal to or more abundant than the mature miRNAs, suggesting inefficient processing. This was also evident with miRNA-7, where such differences in the efficiency of processing were evident between the two cell types. In the case of miRNA-15-5p, two bands possibly representing two mature miRNA species were observed with both MSB-1- and HPRS24-infected CEF. In order to confirm that the MDV-2 miRNAs reported here are not unique to just the HPRS24 strain of MDV-2, we also carried out Northern blotting analysis of CEF infected with another MDV-2 strain, SB-1 (33). These studies confirmed that the SB-1 strain also expressed these miRNAs (not shown).
FIG. 3.
Northern blotting analysis for determining the expression of HPRS24 miRNAs. Twenty micrograms of total RNA from MSB-1 (lane 1), pRB-1B virus-transformed 769T cell line (lane 2), HPRS24-infected CEF (lane 3), and uninfected CEF (lane 4) was separated on a 15% denaturing polyacrylamide gel, blotted, and probed with an end-labeled antisense oligonucleotide to the indicated miRNA. Size markers to indicate the positions of the pre-miRNA and the mature miRNA are shown. The cellular U6 snRNA served as the loading control (a representative blot of this set is shown).
DISCUSSION
As nonimmunogenic, small regulatory molecules that require very little coding space in the genomes, miRNAs have been used as attractive machinery for modulating gene expression by several viruses. Members of the family Herpesviridae, whose life cycle alternates between the stages of nuclear replication and latency, appear to particularly exploit this new tier of regulation. At least 81 miRNAs have been identified (http://microrna.sanger.ac.uk) in this family of viruses. The majority of the herpesvirus-encoded miRNAs do not share any sequence homologies with each other or with the host miRNAs, suggesting their independent evolution (24, 28). However, the herpesviruses used in such comparisons belong to different genera, and the absence of homology among miRNAs might be a reflection of the herpesvirus diversity itself. On the other hand, when the closely related lymphocryptoviruses rLCV and EBV were compared, there was sequence conservation in some of their miRNAs (9). As rLCV and EBV are separated by at least 13 million years of evolution, it has been suggested that conservation of miRNAs in these viruses may be attributed to selection imposed by their putative target sequences. In order to investigate whether such selection might also operate in other closely related herpesviruses, we examined the miRNAs encoded by members of the Mardivirus genus, MDV-1 and MDV-2, which are separated by at least 26 million years of evolution. MDV-1 is a highly oncogenic alphaherpesvirus that infects chickens and has recently been shown to encode at least eight miRNAs that map to the MEQ and the LAT regions of the viral genome (8). MDV-2 is a nononcogenic virus widely used as a live vaccine against Marek's disease. Analysis of the miRNAs encoded by these viruses could provide insights into the relationship between miRNAs and oncogenesis.
The MDV-transformed lymphoblastoid cell line MSB-1 (1) has been previously reported to be infected with both MDV-1 strain BC-1 and MDV-2 strain HPRS24 (15), and we have confirmed this using IF and WB with specific MAbs. Analysis of the small RNA library from the MSB-1 cell line showed that nearly 10% (518 of 5,099) of the cloned sequences were attributed to 17 candidate HPRS24-encoded miRNAs (Table 1). These candidate sequences conformed to most of the miRNA validation criteria (4) such as (i) their cloning from a cDNA library, (ii) the detection of either strand of the putative double-stranded miRNA precursor, and (iii) their identification by Northern blotting. Nearly all of the predicted miRNAs could also be folded into hairpin structures (Fig. 1). However, some of the miRNAs, such as MDV-2 miRNA-6, which were cloned at least nine times in the library, showed slightly uncharacteristic central bulging and a small terminal bulge using an MFOLD structure prediction. The identification of these novel MDV-2-encoded miRNAs adds to the growing list of herpesvirus-encoded miRNAs.
MDV-2 has a typical alphaherpesvirus genome structure with a colinear arrangement of genes, as in MDV-1 (25). Although both viruses have numerous genes showing conservation in primary sequence and in genomic positions across the genome, several genes located in the IRL/TRL region are unique to each virus. For example, the genes such as meq and vIL-8 that are located in this region and are associated with the oncogenicity of MDV-1 are absent in MDV-2. On the other hand, MDV-2 has several ORFs within this region that are absent in MDV-1. Previous studies have indicated that the IRL/TRL region in MDV-1 is transcriptionally active in latently infected tumors and transformed cells (31, 34). Furthermore, five out of the eight miRNAs identified recently in MDV-1 within this region are also expressed at very high levels in transformed cell lines and tumor cells (8), confirming that this region is transcriptionally active even during stages of latency and transformation. It is remarkable that all but one of the 17 MDV-2-encoded miRNAs reported here are clustered in a 4.2-kb fragment of the IRL/TRL region spanning the putative ORFs R-LORF2, R-LORF3, R-LORF4, and R-LORF5 (Fig. 2). Although there have not been any experimental data to confirm the existence, the transcriptional profile, or the functions of these ORFs, high levels of expression of MDV-2 miRNAs from this region, as demonstrated by direct cloning and Northern blotting, would indicate that this region is transcriptionally active during latency. Compared to the location of the majority of the herpesvirus-encoded miRNAs in the noncoding region of different genes, the identification of 8 out of 17 MDV-2 miRNAs in the coding regions of the ORFs was quite striking. R-LORF2 and R-LORF3 have opposite orientations to R-LORF4 and R-LORF5. The miRNA cluster shares the same orientation as R-LORF5 and R LORF4, with MDV-2-miR-1 located upstream of the R-LORF5 promoter region.
The single other miRNA identified outside the cluster, MDV-2 miRNA-17, is located in the IRS/TRS region. Interestingly, MDV-1 also has three miRNAs located in an equivalent position (8). These three MDV-1 miRNAs map to a large intron in the 5′ end of the latency-associated MSR transcript, antisense to the ICP4 gene (10). The MDV-2 miRNA-17 is also in an antisense orientation to ICP4 but overlaps with the coding region, in a region corresponding to amino acids 1,629 to 1,645 that are predicted to form a loop structure between two alpha helices not conserved between MDV-1 and MDV-2. No data for either the existence or for the transcriptional regulation of LAT/ICP4 in MDV-2 have been examined, and until such data are available, the role of this single MDV-2 miRNA in the regulation of MDV-2 latency remains speculative.
Despite the demonstration of relative conservation of miRNA loci in these two closely related viruses, there was no evidence of conservation of primary miRNA sequences of MDV-1 and MDV-2, suggesting their independent evolution. As the majority of the herpesvirus-encoded miRNAs did not show sequence conservation beyond very closely related viruses (9, 24, 28), the lack of sequence homology between MDV-1 and MDV-2 miRNAs was not entirely surprising. Even between closely related EBV and rLCV, the only example where such conservation of miRNA sequences has been demonstrated, only 7 out of 16 miRNAs showed sequence conservation, indicative of a selection probably imposed by the target genes (9). Although MDV-1 and MDV-2 are closely related and separated by only approximately 26 million years, the absence of sequence conservation in any of the miRNA sequences may reflect rapid evolution toward new targets, possibly related to the differences in pathogenicity between the two viruses. Specifically, while MDV-1 is oncogenic, MDV-2 shows other distinctive features such as the induction of cytolytic lesions in immunosuppressed birds (32), protective synergism with serotype 3 vaccines (37), enhancement of lymphoid leukosis (5), and reticuloendotheliosis (3), as well as persistence in transformed B cells (11, 12). Future studies will elucidate whether the virus-encoded miRNAs participate in these processes. Prediction of the putative target genes of virus-encoded miRNAs is particularly difficult, and despite the availability of numerous computational programs for target prediction, targets of only a very few have been validated. Nevertheless, as MDV-2 miRNA-10, miRNA-13, miRNA-14, and miRNA-15 are transcribed antisense to R-LORF2 and R-LORF3 transcripts, these miRNAs could potentially target these transcripts for cleavage as small interfering RNA because of the perfect complementarity of the mRNA sequences. However, as the functions of these novel ORFs are largely unknown at this point, it is difficult to evaluate the impact that the miRNAs could have on the biology of these viruses. Our recent success with the construction of the complete genome of the SB-1 strain of MDV-2 as an infectious bacterial artificial chromosome clone (unpublished data) will allow us to carry out functional analysis of the role of these miRNAs in the biology of MDV-2 in natural infection models.
Acknowledgments
We thank Mick Watson for assisting in sequence analysis, Mick Gill for digital imaging and graphics, and D. J. McGeoch for sharing the data for phylogenetic relationships of herpesviruses.
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17:105-116. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aly, M. M., R. L. Witter, and A. M. Fadly. 1996. Enhancement of reticuloendotheliosis virus-induced bursal lymphomas by serotype 2 Marek's disease virus. Avian Pathol. 25:81-94. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V., B. Bartel, D. P. Bartel, C. B. Burge, J. C. Carrington, X. Chen, G. Dreyfuss, S. R. Eddy, S. Griffiths-Jones, M. Marshall, M. Matzke, G. Ruvkun, and T. Tuschl. 2003. A uniform system for microRNA annotation. RNA 9:277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon, L. D., R. L. Witter, and A. M. Fadly. 1989. Augmentation of retrovirus-induced lymphoid leukosis by Marek's disease herpesviruses in White Leghorn chickens. J. Virol. 63:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 7.Biggs, P. M., and B. S. Milne. 1972. Biological properties of a number of Marek's disease virus isolates., p. 88-94. In P. M. Biggs, G. de The, and L. N. Payne (ed.), Oncogenesis and herpesviruses. IARC, Lyon, France.
- 8.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 80:8778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantello, J. L., M. S. Parcells, A. S. Anderson, and R. W. Morgan. 1997. Marek's disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J. Virol. 71:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fynan, E., T. M. Block, J. DuHadaway, W. Olson, and D. L. Ewert. 1992. Persistence of Marek's disease virus in a subpopulation of B cells that is transformed by avian leukosis virus, but not in normal bursal B cells. J. Virol. 66:5860-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fynan, E. F., D. L. Ewert, and T. M. Block. 1993. Latency and reactivation of Marek's disease virus in B lymphocytes transformed by avian leukosis virus. J. Gen. Virol. 74:2163-2170. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, A., J. J. Gartner, P. Sethupathy, A. G. Hatzigeorgiou, and N. W. Fraser. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 442:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Hirai, K., M. Yamada, Y. Arao, S. Kato, and S. Nii. 1990. Replicating Marek's disease virus (MDV) serotype 2 DNA with inserted MDV serotype 1 DNA sequences in a Marek's disease lymphoblastoid cell line MSB1-41C. Arch. Virol. 114:153-165. [DOI] [PubMed] [Google Scholar]
- 16.International Committee on Taxonomy of Viruses. 2006. 00.031.1.03 Mardivirus. In C. Búchen-Osmond (ed.), ICTVdB: the Universal Virus Database, version 4. Columbia University, New York, NY.
- 17.Izumiya, Y., H. K. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255:191-221. [DOI] [PubMed] [Google Scholar]
- 18.Lee, L. F., X. Liu, and R. L. Witter. 1983. Monoclonal antibodies with specificity for three different serotypes of Marek's disease viruses in chickens. J. Immunol. 130:1003-1006. [PubMed] [Google Scholar]
- 19.Lee, L. F., P. Wu, D. Sui, D. Ren, J. Kamil, H. J. Kung, and R. L. Witter. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc. Natl. Acad. Sci. USA 97:6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, D., P. F. Green, M. A. Skinner, C. Jiang, and N. Ross. 1994. Use of recombinant pp38 antigen of Marek's disease virus to identify serotype 1-specific antibodies in chicken sera by Western blotting. J. Virol. Methods 50:185-195. [DOI] [PubMed] [Google Scholar]
- 21.Matzura, O., and A. Wennborg. 1996. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci. 12:247-249. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 23.Nair, V., and H. J. Kung. 2004. Marek's disease virus oncogenicity: molecular mechanisms, p. 32-48. In F. Davison and V. Nair (ed.), Marek's disease, an evolving problem. Elsevier Academic Press, Oxford, United Kingdom.
- 24.Nair, V., and M. Zavolan. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14:169-175. [DOI] [PubMed] [Google Scholar]
- 25.Osterrieder, K., and J. F. Vautherot. 2004. The genome content of Marek's disease-like viruses, p. 17-31. In F. Davison and V. Nair (ed.), Marek's disease, an evolving problem. Elsevier Academic Press, Oxford, United KIngdom.
- 26.Osterrieder, N., J. P. Kamil, D. Schumacher, B. K. Tischer, and S. Trapp. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4:283-294. [DOI] [PubMed] [Google Scholar]
- 27.Petherbridge, L., A. C. Brown, S. J. Baigent, K. Howes, M. A. Sacco, N. Osterrieder, and V. K. Nair. 2004. Oncogenicity of virulent Marek's disease virus cloned as bacterial artificial chromosomes. J. Virol. 78:13376-13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 29.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 30.Sarnow, P., C. L. Jopling, K. L. Norman, S. Schutz, and K. A. Wehner. 2006. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat. Rev. Microbiol. 4:651-659. [DOI] [PubMed] [Google Scholar]
- 31.Schat, K. A., A. Buckmaster, and L. J. Ross. 1989. Partial transcription map of Marek's disease herpesvirus in lytically infected cells and lymphoblastoid cell lines. Int. J. Cancer 44:101-109. [DOI] [PubMed] [Google Scholar]
- 32.Schat, K. A., and B. W. Calnek. 1978. Characterization of an apparently nononcogenic Marek's disease virus. J. Natl. Cancer Inst. 60:1075-1082. [DOI] [PubMed] [Google Scholar]
- 33.Schat, K. A., and B. W. Calnek. 1978. Protection against Marek's disease-derived tumor transplants by the nononcogenic SB-1 strain of Marek's disease virus. Infect. Immun. 22:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugaya, K., G. Bradley, M. Nonoyama, and A. Tanaka. 1990. Latent transcripts of Marek's disease virus are clustered in the short and long repeat regions. J. Virol. 64:5773-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 36.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witter, R. L., and L. F. Lee. 1984. Polyvalent Marek's disease vaccines: safety, efficacy and protective synergism in chickens with maternal antibodies. Avian Pathol. 13:75-92. [DOI] [PubMed] [Google Scholar]
- 38.Witter, R. L., L. F. Lee, and J. M. Sharma. 1990. Biological diversity among serotype 2 Marek's disease viruses. Avian Dis. 34:944-957. [PubMed] [Google Scholar]
- 39.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]