Abstract
The assembly and egress of herpesviruses are complex processes that require the budding of viral nucleocapsids into the lumen of cytoplasmic compartments to form mature infectious virus. This envelopment stage shares many characteristics with the formation of luminal vesicles in multivesicular endosomes. Through expression of dominant-negative Vps4, an enzyme that is essential for the formation of luminal vesicles in multivesicular endosomes, we now show that Vps4 function is required for the cytoplasmic envelopment of herpes simplex virus type 1. This is the first example of a large enveloped DNA virus engaging the multivesicular endosome sorting machinery to enable infectious virus production.
The final stage in the assembly of enveloped viruses is the budding of viral cores across cellular membranes, in an extracellular direction, to allow the newly formed virions to be released from an infected cell. This release of enveloped viruses can be achieved either directly by budding across the plasma membrane into the extracellular space or alternatively by budding into the lumen of cellular organelles, with luminal virions released into the extracellular environment via an exocytic mechanism. This assembly process has been studied in detail for many enveloped RNA viruses, and a common theme has emerged in the acquisition of viral envelopes: the use of the cellular multivesicular endosome (MVE) budding machinery. Particularly through pioneering work with human immunodeficiency virus (HIV), it has been demonstrated that these enveloped RNA viruses can recruit endosomal sorting complexes required for transport (ESCRTs) via interaction with late-domain (L-domain) motifs contained within viral structural components (3, 6, 25). This step is crucial for these viruses to complete their budding through cellular membranes and has set a precedent for the utilization of ESCRT components by viral pathogens for assembly (3, 6, 25). However, these RNA viruses are relatively simple in terms of structure and composition, with only a limited number of viral proteins involved (e.g., Gag-Pol and Env in retroviruses). When considering the large enveloped DNA viruses, such as the Herpesviridae, which also need to bud across cellular membranes, the situation is significantly more complex, as these viruses are composed of in excess of 30 virally encoded proteins (24). We therefore posed the question of whether the use of the cellular ESCRT pathway for viral budding extends to such large complex viruses where so many proteins are assembled together.
Herpesviruses are ubiquitous pathogens of vertebrates, and infections by members of this family are associated with a wide variety of human and animal diseases (29). Herpesviruses are some of the most complex viruses known: they are composed of a double-stranded DNA genome encased in an icosahedral capsid, a layer of more than 15 different proteins known as the tegument, and a host-derived envelope containing 10 or more different viral glycoproteins (23). The assembly of herpesviruses begins in the nucleus of an infected cell, where newly synthesized genomes are packaged into preformed capsids. These nucleocapsids then undergo budding at the inner nuclear membrane to form an enveloped particle in the perinuclear space, followed by a fusion event at the outer nuclear membrane, releasing the nucleocapsids into the cytoplasm. Mature virus particles are then formed by the budding of the nucleocapsids and tegument into vesicles derived from the trans-Golgi network (TGN) or endosomes (33). Mature virions are released from cells by fusion of the vesicles containing mature virus with the plasma membrane, in a mechanism akin to exocytosis (reviewed in references 23 and 24). Evidence from a number of herpesviruses suggests that this assembly pathway is conserved among the three viral subfamilies Alpha-, Beta-, and Gammaherpesvirinae (13). Therefore, the mechanisms involved in herpesvirus assembly have been conserved for at least 400 million years of virus coevolution with host (21).
This complex assembly regimen for the production and release of new herpesviruses requires the budding of viral nucleocapsids and tegument proteins into the lumen of cytoplasmic organelles, and this remains a poorly understood process with no known requirement for cellular proteins. Several viral tegument proteins are thought to play critical roles in cytoplasmic envelopment, based mainly on evidence from the phenotypes of deletion mutants of herpes simplex virus type 1 (HSV-1) and pseudorabies virus (both alphaherpesviruses) and cytomegalovirus (a betaherpesvirus). These include homologues of the HSV-1 UL11 and UL36 genes (7, 11, 18, 30), the alphaherpesvirus UL48 and UL51 genes (10, 17, 26, 27), and the cytomegalovirus UL32 gene (1). In contrast, the role of viral envelope proteins in cytoplasmic envelopment appears to be subtle, with little effect of gene deletions unless multiple envelope proteins are absent (5, 8). Whether any of these viral proteins can function in a manner equivalent to that of the L-domain-containing proteins found in enveloped RNA viruses, to recruit ESCRT proteins, is unknown.
The cellular MVE budding machinery consists of the multiprotein complexes ESCRT-1, -2, and -3 together with several additional proteins (15), and this machinery is normally responsible for the sorting of proteins into MVE luminal vesicles that are destined for degradation in lysosomes (14, 31). As the budding of herpesvirus nucleocapsids into the lumen of TGN or endosome-derived vesicles shares similarities with the formation of luminal vesicles in MVEs, it seems possible that these viruses could have evolved mechanisms to use the ESCRT machinery in the formation of infectious viruses. Consistent with this hypothesis is the observation that assembled human cytomegalovirus virions have been observed in organelles showing MVE morphology (9). We now show that inhibition of activity of Vps4, an enzyme that is crucial for ESCRT function and MVE luminal vesicle formation (2), causes a severe defect in the production of infectious HSV-1. Analysis of the different stages of the viral life cycle demonstrates that functional Vps4 is required for the cytoplasmic envelopment of HSV-1.
MATERIALS AND METHODS
Cell lines, viruses, and antibodies.
Cos7 cells were grown in Glasgow minimal essential medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. HEK293 cells stably expressing the ecdysone receptor (EcR-293; Invitrogen), which allow ponasterone A (ponA)-induced transactivation of target genes under the control of ecdysone response elements, were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (complete Dulbecco's modified Eagle's medium) with the addition of 400 μg/ml Zeocin. HSV-1 lacking full-length UL36 expression (HSV-1ΔUL36) and a UL36-complementing cell line (HS30) were from P. Desai (Johns Hopkins University) (7). Antibodies to green fluorescent protein (GFP) were obtained from Abcam (Ab6556) and Clontech (JL8). Antibodies that recognize HSV-1 VP1/2 were generously provided by Peter O'Hare (Marie Curie Research Institute, Oxted, United Kingdom). Antiactin was obtained from Sigma (AC-40), and anti-VP16 (LP1) has been previously described (22).
Plasmids.
pEGFPC-Vps4WT and pEGFPC-Vps4EQ were from P Woodman (Manchester University) (4). pCR3.1-YFPVps4WT, pCR3.1-YFPVps4EQ, and pCR3.1-GFP were from J. Martin-Serrano (Kings College London). The enhanced GFP (EGFP) open reading frame was excised from pEGFPC2 (Clontech) with NheI and XbaI and ligated into pIND(SP1) (Invitrogen), which had been cut with the same enzymes, to make pIND-GFP. The Vps4WT and Vps4EQ coding sequences were excised from pEGFP-Vps4WT and pEGFP-Vps4EQ, respectively, with BsrGI and BamHI and ligated into pIND-GFP, which had been cut with the same enzymes, to make pIND-GFPVpsWT and pIND-GFPVps4EQ, respectively. The 5′ end of HSV-1 UL36 was excised from pSG124 (pBR325 containing EcoRI fragment A of the HSV-1 strain KOS genome) (12) with HindIII and EcoRI and ligated into pcDNA3 to generate pcDNA3-UL36-5′end. The 3′ end of UL36 was excised from pSG3 (pBR325 containing EcoRI fragment L of the HSV-1 strain KOS genome) (12) with EcoRI, ligated into pcDNA3-UL36-5′end cut with EcoRI, and screened for correct orientation. The resulting pcDNA3-UL36 plasmid contained the entire HSV-1 UL36 open reading frame with 170 bp of 5′ genomic sequence and 1,352 bp of 3′ genomic sequence.
Complementation assay.
Cos7 cells were seeded into six-well dishes at 2 × 105 cells per well and transfected with 2 μg pCR3.1-GFP alone or 1 μg pcDNA3-UL36 together with 1 μg of pCR3.1-GFP, pCR3.1-YFPVps4WT, or pCR3.1-YFPVps4EQ. After 24 h, cells were infected with HSV-1ΔUL36 at 10 PFU/cell, and unabsorbed virus was inactivated by treatment with acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0). At 16 h postinfection (hpi) samples were harvested by scraping. Three samples of each condition were sonicated for 20 seconds at 40% amplitude followed by freeze-thawing to liberate cell-associated virus. Infectious viral titers were determined by plaque assays on HS30 cells (7). The cells from a fourth sample of each condition were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris, pH 7.9, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) supplemented with protease inhibitor cocktail (Roche) and incubated on ice for 20 min. Lysates were clarified by centrifugation at 13,000 rpm for 20 min at 4°C. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to nitrocellulose and the expression levels of VP1/2- and GFP-tagged proteins were determined using anti-VP1/2 or anti-GFP (JL8) and chemiluminescence detection.
Ecdysone-responsive stable cell lines.
EcR-293 cells were transfected with pIND-GFPVps4WT or pIND-GFPVps4EQ and placed under selection in complete Dulbecco's modified Eagle's medium supplemented with 400 μg/ml Zeocin and 800 μg/ml G418. Single clones were isolated by limiting dilution, and samples of each clone were screened by fluorescence microscopy and Western blotting after addition of 1 μM ponA. Clones demonstrating GFP fluorescence in all cells were maintained. One clone of each cell line showing comparable levels of inducible expression in the presence of ponA by Western blotting was chosen for further experiments. 293-Vps4WT and 293-Vps4EQ cells were treated with or without 1 μM ponA for various times, and cellular proteins were extracted as described above. Following SDS-PAGE, proteins were transferred to nitrocellulose and the expression of Vps4 proteins was determined with anti-GFP (Ab6556) and chemiluminescence detection. Cells grown on glass coverslips were fixed 16 h after ponA addition and analyzed with an Olympus IX70 fluorescence microscope. Images were captured using a Reitga 2000R charge-coupled device camera and ImageQ software and processed using Adobe Photoshop.
HSV-1 growth assays.
293-Vps4WT and 293-Vps4EQ cells were treated with or without 1 μM ponA for 16 h and infected with HSV-1 (SC16) at 10 PFU/cell, and unabsorbed virus was inactivated with acid wash. At various times, samples were harvested as described above. Infectious viral titers were determined by plaque assays on Vero cells. The expression levels of GFP-Vps4 proteins, VP16, and actin in protein extracts from samples at various times postinfection were analyzed by Western blotting with anti-GFP (JL8), anti-VP16 (LP1), or antiactin (AC-40) as described above.
Quantitative real-time PCR.
293-Vps4WT and 293-Vps4EQ cells were treated with or without 1 μM ponA for 16 h and infected with HSV-1 (SC16) at 10 PFU/cell, and unabsorbed virus was removed by acid wash. Cells were harvested at 16 hpi and treated with 10 mM Tris, pH 8.0, 50 mM EDTA, 0.5% SDS, 0.2 mg/ml proteinase K for 18 h at 37°C. Samples were sonicated three times for 30 seconds each at 40% amplitude, and DNA was extracted with a QIAquick PCR purification kit (QIAGEN). Real-time PCR was performed using a Rotorgene (Corbett Research) in triplicate for each sample. Primers and high-pressure liquid chromatography-purified probes were designed by Tib-MolBiol (Berlin). A primer set for the detection of HSV-1 genomes was based on the ICP0 gene promoter region with the following HSV-1 sequence coordinates: forward primer, 2072 to 2090; reverse primer, 2209 to 2139; and TaqMan probe, 2115 to 2134, where the ICP0 transcription start site is 2115. A primer set for the detection of cellular genomes was based on the human glyceraldehyde-3-phosphate dehydrogenase gene promoter region with the following gene coordinates: forward primer, 1725 to 1744; reverse primer, 1913 to 1894; and TaqMan probe, 1913 to 1894, where numbers are relative to a transcription start site of 1874 based on comparison of genomic and cDNA sequences (accession no. AY340484). Results from the real-time PCRs were quantified as copy number per sample using the Rotorgene software from standard curves that were generated using known amounts of plasmids containing each of the relevant DNA regions. The mean of each triplicate data set for ICP0 was divided by the mean of the triplicate glyceraldehyde-3-phosphate dehydrogenase data set from the same DNA sample to obtain estimates of HSV-1 genome copies present per cell.
[35S]methionine labeling.
293-Vps4WT and 293-Vps4EQ cells were treated with 1 μM ponA for 16 h and infected with HSV-1 (SC16) at 10 PFU/cell, and unabsorbed virus was inactivated by acid wash. At 7 hpi, cells were washed and incubated in methionine-free medium for 1 h followed by [35S]methionine labeling for 16 h. Cells were harvested and lysed as described above, and protein extracts were analyzed by SDS-PAGE and autoradiography.
Electron microscopy (EM).
Infected-cell samples were harvested and fixed in either 2.5% glutaraldehyde and 2% paraformaldehyde in 100 mM sodium cacodylate or 4% glutaraldehyde in 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 7.4] containing 2 mM CaCl2 for 2 h at room temperature. Cells were washed in 50 mM Tris (pH 7.4) containing 2 mM CaCl2, treated with 1% osmium tetroxide for 30 min, and rinsed in distilled water. Samples were then stained with 2% uranyl acetate for 30 min, rinsed with distilled water, dehydrated in methanol, infiltrated with Spurr's epoxy resin over 5 days, and cured at 60°C. Fifty-nanometer sections were cut using a Leica Ultracut S microtome and mounted on uncoated copper grids. Samples were counterstained with uranyl acetate and lead citrate and viewed with a Philips CM100 transmission electron microscope, and images were recorded on Kodak 4489 cut film. Negatives were scanned at 1,200 dots per inch and processed using Adobe Photoshop.
RESULTS
One of the essential proteins for cargo sorting and MVE luminal vesicle formation is Vps4, a member of the AAA ATPase family. Vps4 is required to dissociate the ESCRT proteins from MVE membranes and allow their recycling for further rounds of vesicle formation (2). Expression of ATPase-defective (dominant-negative) mutants of either homologue (Vps4A or Vps4B) where the active site glutamate has been mutated to glutamine leads to defective MVE sorting and formation of large membrane structures known as class E compartments (4, 36). The term class E compartment was originally used to describe the multilamellar perivacuolar compartment found in yeast cells expressing ATPase-defective Vps4 (2). We decided to analyze the effect of inhibiting Vps4 activity on the envelopment of HSV-1.
Transient expression of nonfunctional Vps4 reduces the yield of infectious HSV-1.
As an initial approach to determine whether Vps4 plays a role in the assembly of HSV-1 virions, cells transfected with plasmids expressing wild-type or dominant-negative Vps4 were infected with HSV-1 and the progeny virus was assayed. To avoid problems associated with variable transfection efficiencies and replication of HSV-1 in untransfected cells, a complementation assay was used. HSV-1 lacking the essential gene UL36 (HSV-1ΔUL36) does not produce infectious virus unless the defect is complemented through provision of UL36 expression in trans (7). If the provision of UL36 expression is through transient transfection, then transfected cells will be the only source of infectious progeny. Therefore, cotransfection of UL36 and Vps4 expression plasmids, followed by infection with HSV-1ΔUL36, will mean that the vast majority of progeny virus will have to be produced in the presence of plasmid-expressed Vps4, and so the effect of dominant-negative Vps4 can be assessed more reliably.
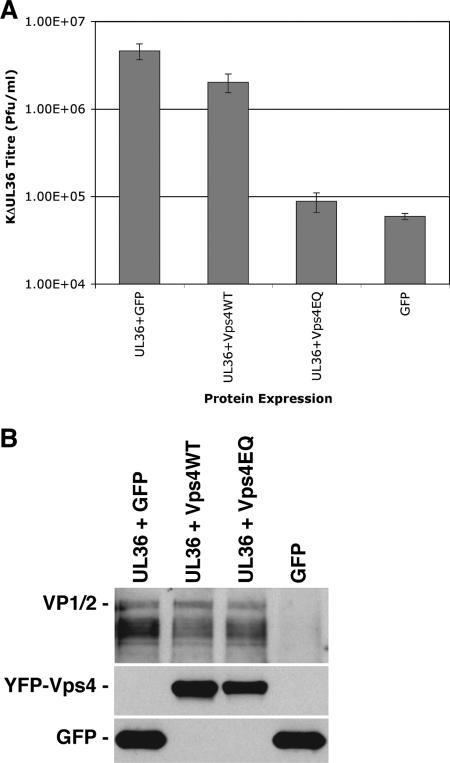
Cells were transfected with plasmids expressing UL36 together with wild-type or dominant-negative Vps4 (Vps4WT or Vps4EQ, respectively) or control plasmids and subsequently infected with HSV-1ΔUL36. The virus yields were assayed on a UL36-complementing cell line to allow plaque formation (7). The UL36 complementation assay system demonstrated approximately an 80-fold difference in the production of infectious HSV-1 progeny between cells expressing GFP together with UL36 in trans and cells expressing GFP alone (with no UL36) (Fig. 1A). Expression of yellow fluorescent protein (YFP)-Vps4EQ with UL36 resulted in a reduction in progeny HSV-1 titer by ∼50-fold compared to expression of GFP with UL36. Expression of UL36 with YFP-Vps4WT showed a small reduction in HSV-1 titer (∼2-fold). GFP, YFP-Vps4WT, and YFP-Vps4EQ were all expressed to similar levels in these assays, and the expression of UL36 from transfected plasmids was unaffected by the coexpression of Vps4WT or Vps4EQ (Fig. 1B). These results are similar to those reported using HIV: namely, overexpression of wild-type Vps4 gave a minor dominant-negative effect, resulting in a slightly smaller yield of HIV, whereas a major dominant-negative effect resulting in a large reduction in HIV yield was observed in the presence of Vps4EQ (34). Similar complementation experiments were performed using a glycoprotein-H-null HSV-1 and transfection of cells with glycoprotein-H-expressing plasmids as previously described (35). These experiments generated very similar results, with coexpression of YFP-Vps4EQ leading to a significant decrease in infectious HSV-1 (∼10-fold). These data suggest that the inhibition of HSV-1 growth in the presence of Vps4EQ is independent of which complementation system is used. Therefore, functional Vps4 appears to be required for the production of infectious HSV-1 particles.
FIG. 1.
The effect of transient Vps4EQ expression on the production of infectious HSV-1. Cells transfected with a UL36 expression plasmid in the presence of GFP, YFP-Vps4WT, or YFP-Vps4EQ expression plasmids or cells transfected with a GFP expression plasmid alone were infected with HSV-1ΔUL36. (A) Progeny viral titers were determined by plaque assay on a UL36-complementing cell line. Bars represent mean PFU/ml, and error bars represent 1 standard deviation from the mean of triplicate samples. (B) Protein extracts were analyzed by Western blotting with VP1/2- and GFP-specific antibodies.
Stable cell lines expressing dominant-negative Vps4 demonstrate a decreased growth of HSV-1.
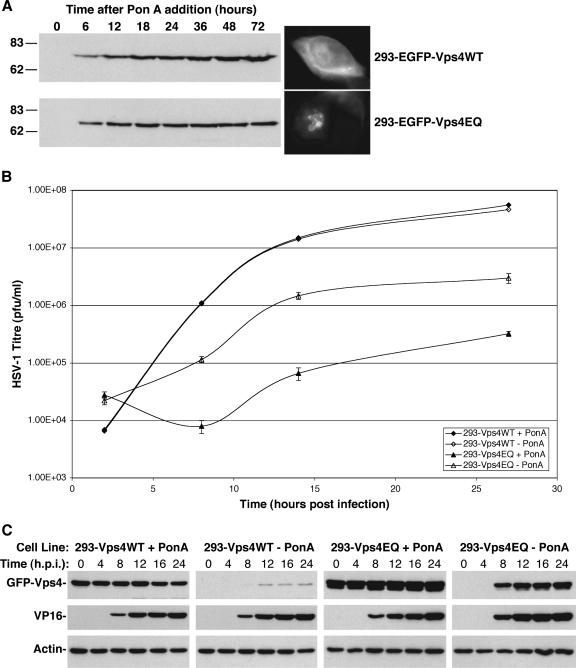
To further investigate the role of Vps4 in HSV-1 assembly, stable cell lines expressing Vps4WT or Vps4EQ were constructed to overcome the limitations of transient transfection. Although there were no toxicity issues during the time frame of the experiments in this publication, we have observed that long-term expression of dominant-negative Vps4EQ, but not Vps4WT, is eventually toxic to cells (unpublished observations). Therefore, an inducible system was deemed necessary for the production of such stable cell lines. HEK293 cells harboring the ecdysone receptor were transfected with expression plasmids encoding GFP-Vps4WT or GFP-Vps4EQ, under control of the ecdysone response element, and a constitutively expressed selectable gene. GFP-tagged Vps4 proteins were used for ease of selection and detection. Stable clones were selected and isolated by limiting dilution. One clone of each cell line that showed comparable levels of protein expression after addition of ponA was used for all subsequent experiments, although any effects on viral replication were confirmed with independent clones (unpublished data). No Vps4 protein expression was observed in the cells in the absence of ponA, and expression increased steadily over time after induction (Fig. 2A). Fluorescence microscopy showed GFP-Vps4WT to be localized in a diffuse pattern throughout the cytoplasm, whereas GFP-Vps4EQ was found associated with large swollen membrane structures (Fig. 2A). These observations are similar to previous reports, which have demonstrated that expression of GFP-Vps4EQ sequesters wild-type Vps4 and causes the formation of large vacuoles in mammalian cells (4, 36).
FIG. 2.
Single-step growth curves of HSV-1 in Vps4WT- and Vps4EQ-expressing cell lines. Clonal 293 cell lines expressing GFP-Vps4WT or GFP-Vps4EQ under the control of the ecdysone response element were isolated. (A) Cells were treated with 1 μM ponA for various times, and protein extracts were analyzed by Western blotting with a GFP-specific antibody. Fluorescence microscope images were collected at 16 h after ponA addition. Numbers at left are molecular masses in kilodaltons. (B) Cells were treated with or without ponA for 16 h and infected with HSV-1, and progeny virus was harvested at various times. Infectious viral titers were determined by plaque assay. Data represent mean PFU/ml, and error bars represent 1 standard deviation from the mean of triplicate samples. (C) Protein extracts were harvested from infected cells at various times postinfection and analyzed by Western blotting with GFP-, VP16-, and actin-specific antibodies.
One-step growth analysis of HSV-1 was performed in the Vps4 stable cell lines. Untreated cells or cells treated with ponA were infected with HSV-1. At various times postinfection, samples were harvested and the titer of infectious HSV-1 progeny was determined. HSV-1 replicated efficiently in 293-Vps4WT cells irrespective of ponA-induced protein expression (approximately 3.5-log growth; Fig. 2B). However, 293-Vps4EQ cells showed substantially reduced HSV-1 replication in the presence of ponA (approximately a 2-log reduction), suggesting that Vps4EQ expression inhibits the replication of HSV-1 (Fig. 2B). Surprisingly 293-Vps4EQ cells also demonstrated reduced HSV-1 replication in the absence of ponA (approximately 1-log reduction). However, when protein samples were analyzed by Western blotting, it was apparent that infection of these cell lines with HSV-1 caused transactivation of the ecdysone-responsive Vps4 genes (Fig. 2C). The expression of Vps4 proteins in the absence of ponA was observed at 8 to 12 hpi. Surprisingly, Vps4EQ expression was induced to a greater level by HSV-1 infection than that of Vps4WT, although the reason for this is unclear. Nevertheless, “uninduced” 293-Vps4EQ cells were in fact expressing high levels of Vps4EQ within 8 h of HSV-1 infection, explaining the reduction of HSV-1 growth observed under these conditions. Irrespective of the induction of Vps4 expression by HSV-1 infection, these data demonstrate that the expression of dominant-negative Vps4EQ causes a ≥100-fold reduction in the production of infectious HSV-1.
The effect of dominant-negative Vps4 on viral genome and protein synthesis.
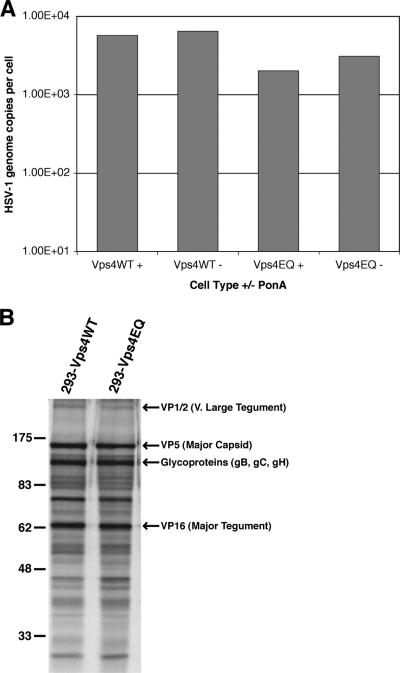
Although Vps4EQ inhibits ESCRT-mediated membrane budding, the inhibition of HSV-1 replication could potentially be due to secondary effects on other aspects of the virus life cycle, rather than a specific effect on cytoplasmic envelopment of nucleocapsids. As a first step to identify the stage of HSV-1 infection that is inhibited in these cells, the viral genome load was analyzed. Cells were treated with or without ponA and then infected with HSV-1. DNA was isolated from each sample at 16 hpi, and the HSV-1 and cellular genome copy numbers were determined by real-time PCR. Induction of Vps4EQ appeared to have a minor effect on viral genome synthesis (≤3-fold reduction; Fig. 3A), but this is unlikely to account for the large reduction in infectious virus yield.
FIG. 3.
HSV-1 genome and protein synthesis. 293-Vps4WT and 293-Vps4EQ cells were treated with 1 μM ponA or not and infected with HSV-1. (A) DNA samples were harvested at 16 hpi, and the copy numbers of HSV-1 and cellular genomes were determined by real-time PCR. Data are shown as mean numbers of HSV-1 genome copies per cell. (B) Infected cells were labeled with [35S]methionine, and protein samples were harvested at 24 hpi. Radiolabeled proteins were detected by autoradiography following SDS-PAGE. The positions of various viral proteins, as expected from previous publications, are shown. Numbers at left are molecular masses in kilodaltons.
To determine whether viral protein synthesis was affected by Vps4EQ expression, cells were treated with ponA, infected with HSV-1, and then metabolically labeled with [35S]methionine from 8 to 24 hpi to monitor the total accumulation of viral proteins. HSV-1 infection results in a potent shutdown of host cell protein synthesis, and so virtually all de novo protein expression during HSV-1 infection is of viral origin (32). No significant difference was observed in the pattern or levels of protein synthesis during HSV-1 infection of Vps4WT- or Vps4EQ-expressing cell lines, and all major viral gene products could be identified at equivalent levels (Fig. 3B). Furthermore, no significant difference was observed in the rate of synthesis of VP16 in either Vps4WT- or Vps4EQ-expressing cell lines in the presence or absence of ponA (Fig. 2C).
Taken together, these data demonstrate that expression of Vps4EQ has little or no effect on viral genome synthesis, the overall accumulation of viral proteins, or the rate of expression of a major viral protein, VP16. Therefore, Vps4EQ expression does not appear to inhibit viral entry or the production of the viral structural components.
Dominant-negative Vps4 inhibits the production of enveloped virus particles.
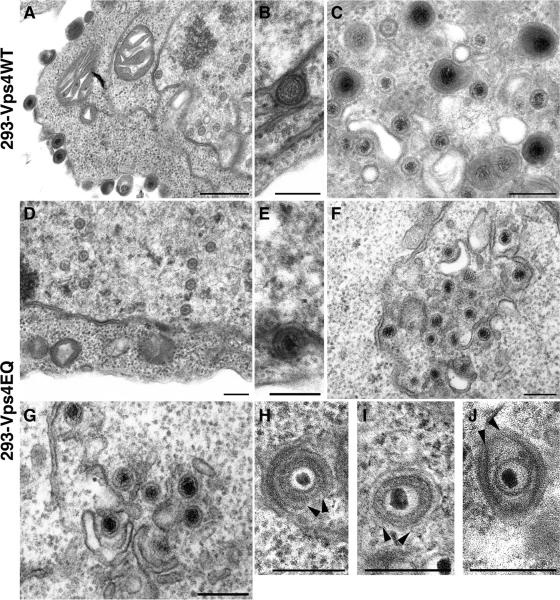
To determine the effect of Vps4EQ expression on HSV-1 assembly at the ultrastructural level, cells expressing Vps4WT or Vps4EQ were infected with HSV-1 and analyzed by EM. In Vps4WT-expressing cells, all stages of HSV-1 assembly could be visualized: nucleocapsids in the nucleus and extracellular virions (Fig. 4A), perinuclear enveloped particles (Fig. 4B), and fully enveloped virions contained within vesicles and some unenveloped capsids in the cytoplasm (Fig. 4C). In Vps4EQ-expressing cells nucleocapsids in the nucleus and perinuclear enveloped particles could be identified at a frequency similar to that in Vps4WT cells (Fig. 4D and E). However, no extracellular virus particles were observed, and only unenveloped capsids were present within the cytoplasm with an almost complete absence of any fully enveloped virions (Fig. 4F to J). Interestingly, cytoplasmic capsids within Vps4EQ cells were usually associated with membranes, with some virions almost completely enveloped but not fully surrounded by membrane (Fig. 4H to J; arrowheads indicate the membrane edges). These data demonstrate two points: firstly, inhibition of Vps4 function appears to have no effect on capsid assembly or nucleocapsid egress from the nucleus, and secondly, expression of Vps4EQ inhibits HSV-1 final envelopment in the cytoplasm, and this inhibition of cytoplasmic envelopment can occur at late stages of assembly.
FIG. 4.
EM analysis of infected cells. 293-Vps4WT and 293-Vps4EQ cells were induced with ponA and infected with HSV-1. At 15 hpi, cells were fixed, processed, and analyzed by transmission EM. Bars, 500 nm (A) or 200 nm (B to J).
As individual electron microscope images can be subjective and represent only a snapshot of events in a very small section of any given cell, these data were quantified by counting large numbers of particles at various stages of viral assembly from several cells. Thirty cells from each cell type were viewed, and all the viral particles were scored as either capsids located in the nucleus, unenveloped capsids in the cytoplasm, fully enveloped virions in the cytoplasm (contained in membrane vesicles), and extracellular enveloped virus. These data show very little difference in the numbers of capsids in the nucleus or unenveloped capsids in the cytoplasm between cells expressing Vps4WT and those expressing Vps4EQ (Table 1). However, a large difference in the number of fully enveloped virus particles was observed: enveloped particles could be readily identified in the cytoplasm and extracellular space of Vps4WT cells, but virtually no enveloped particles were observed in Vps4EQ cells (Table 1). Even the very low number of enveloped cytoplasmic virions that were scored in the cytoplasm of Vps4EQ cells may have been capsids that were almost fully enveloped (such as those shown in Fig. 4H to J) but sectioned in a different plane. The majority (≤80%) of unenveloped cytoplasmic capsids observed in both cell lines were physically close to membranes, suggesting that they were partially assembled. Interestingly, of these membrane-associated capsids, very few appeared almost fully enveloped (as in Fig. 4H to J) in Vps4WT cells, whereas many membrane-associated capsids (∼12%) in the cytoplasm of Vps4EQ cells demonstrated this type of morphology. Taken together, these EM data suggest that the assembly of infectious HSV-1 requires Vps4 activity for the successful budding of virions at cytoplasmic membranes.
TABLE 1.
Quantification of HSV-1 particles in Vps4WT and Vps4EQ cells
| Cell line | No. of nuclear capsids | No. of unenveloped cytoplasmic capsids | No. of enveloped cytoplasmic virions | No. of extracellular virus particles |
|---|---|---|---|---|
| Vps4WT | 812 | 385 | 113 | 48 |
| Vps4EQ | 1,003 | 379 | 5 | 0 |
DISCUSSION
It is now well accepted that many families of enveloped RNA viruses (including the Retroviridae, Filoviridae, Rhabdoviridae, Arenaviridae, and Paramyxoviridae) require ESCRT proteins for viral budding at the plasma membrane or into the lumen of cytoplasmic compartments (3, 6, 25, 28). Such common usage of the ESCRT proteins by completely unrelated virus families demonstrates the importance of this cellular membrane budding machinery for enveloped viral pathogens and how convergent evolution has occurred in these virus families to engage this cellular machinery for the common goal of budding newly synthesized infectious virus out of the cytoplasm of cells. The first demonstration of an involvement of ESCRT proteins in assembly of small DNA viruses has recently been shown for hepatitis B virus (16). Our data now demonstrate that the replication of a large, complex DNA virus, HSV-1, is inhibited by the expression of dominant-negative Vps4, an ATPase that is essential for the function of ESCRT proteins. These results may be best explained by a direct involvement of Vps4 in HSV-1 budding. To our knowledge, these data are the first observation that ESCRT protein function may be required for the membrane budding of a large, complex, enveloped DNA virus.
In experiments demonstrating a reduction in infectious virus yields it is always important to determine whether the observed reduction is due to the failure to assemble virus particles or due to the assembly of particles that have reduced infectivity. Our data show that the inhibition of Vps4 function does indeed inhibit viral assembly: EM quantification demonstrates an almost total lack of enveloped virus when Vps4 activity is inhibited. A further question that is difficult to answer unequivocally is whether defects in viral assembly that are caused by inhibiting membrane traffic pathways (such as ESCRT protein inhibition) are due to a direct inhibition of the membrane budding mechanism or arise from secondary effects of mislocalizing a subset of viral components to the incorrect compartment(s) so that they are not available to participate in assembly. Our EM observations indicate that envelopment of HSV-1 can initiate in Vps4EQ cells and can proceed almost to completion. These “partially enveloped” virions are otherwise morphologically similar to those observed in normal cells. It appears, therefore, that in Vps4EQ cells all the viral proteins that are necessary for the interaction of capsids and tegument with membranes to drive envelopment to late stages must be present on these structures, but envelopment cannot be completed. The obvious interpretation is that functional Vps4 is directly involved in completion of the envelopment (i.e., budding) process of HSV-1. It is also possible that inhibition of certain membrane traffic pathways could result in a block of secretion, thereby interfering with the release of HSV-1 from cells. However, it seems unlikely that expression of dominant-negative Vps4 is reducing HSV-1 titer simply by inhibiting the release of enveloped virions via secretion; otherwise, similar or even greater numbers of enveloped virions contained within cytoplasmic vesicles would be expected, whereas we observe a dramatically reduced number of enveloped virions in the cytoplasm (>20-fold). Furthermore, previous reports have demonstrated that the expression of dominant-negative Vps4 does not affect the localization of secretory pathway proteins, including markers of the endoplasmic reticulum, Golgi compartment, and TGN (4, 36).
Enveloped RNA viruses recruit the ESCRT machinery to sites of viral budding through the interaction of viral L-domain motifs with ESCRT proteins. At least three classes of L-domain motifs have been identified: P-T/S-A-P motifs, Y-P-x-L motifs, and P-P-x-Y motifs (3, 6, 25). The majority of these RNA viruses express proteins that contain one or two L-domain motifs that are necessary and sufficient for the recruitment of ESCRT proteins. The situation with herpesviruses is likely to be more complex. All three classes of L-domain motifs are present in several HSV-1 structural proteins including major capsid protein (VP5), the tegument protein VP16, the very large tegument protein VP1/2, the cytoplasmic domain of glycoprotein E, and at least three additional tegument proteins. This frequency of L-domain motif occurrence in HSV-1 virion components may signify the importance of engaging ESCRT proteins for the assembly of infectious virions and may also reflect the functional redundancy of different proteins in herpesvirus assembly (24). Such redundancy of function may allow for the loss of any one or more L-domain motifs without inhibiting virus production but complicates the analysis of the role of such motifs by mutagenesis. Interestingly, of all the HSV-1 tegument and envelope proteins containing potential L-domain motifs, VP1/2 is the only one conserved throughout all alpha-, beta-, and gammaherpesviruses, and mutants lacking VP1/2 are defective in cytoplasmic envelopment (7, 11). The VP1/2 protein of HSV-1 contains all three classes of L-domain motifs that could potentially interact with ESCRT proteins, and at least one L-domain motif appears to be present in all VP1/2 homologues analyzed, an observation of some significance given that the respective genes diverged in evolution approximately 400 million years ago (21). It is therefore tempting to speculate that VP1/2 may function in part by recruiting ESCRT components to sites of herpesvirus envelopment.
In summary, it appears that the ESCRT pathway provides the budding machinery for a wide variety of enveloped viruses, which we suggest include HSV-1, where the interactions of a large number of virus-specific proteins are involved in assembly and envelopment. In some cases, budding of viruses has been observed at MVEs, the normal sites of action of the ESCRT machinery (9, 19), but in other cases it is clear that viruses can recruit ESCRT components to alternative compartments, such as the plasma membrane (20). In the case of herpes simplex virus it is apparent that the budding compartment lacks the morphological characteristics of MVEs, but the precise nature of the compartment remains unknown.
Acknowledgments
We thank P. O'Hare (Marie Curie Research Institute) for the generous gift of anti-VP1/2, J. Martin-Serrano (Kings College London) and P. Woodman (Manchester University) for expression plasmids, P. Desai (Johns Hopkins University) for HSV-1ΔUL36 and HS30 cells, J. Powell (Multi-Imaging Centre, Cambridge University) for help with EM sample preparation, and H. Browne for helpful discussion and critical reading of the manuscript.
This work was funded by the Royal Society (University Research Fellowship to C.M.C.) and the Wellcome Trust (program grant no. 68075).
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 7.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldin, A. L., R. M. Sandri-Goldin, M. Levine, and J. C. Glorioso. 1981. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J. Virol. 38:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317-323. [DOI] [PubMed] [Google Scholar]
- 15.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kian Chua, P., M. H. Lin, and C. Shih. 2006. Potent inhibition of human hepatitis B virus replication by a host factor Vps4. Virology 354:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, R. Klopfleisch, W. Fuchs, M. Kopp, M. Lenk, and T. C. Mettenleiter. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer, B., A. Pelchen-Matthews, M. Deneka, E. Garcia, V. Piguet, and M. Marsh. 2005. HIV interaction with endosomes in macrophages and dendritic cells. Blood Cells Mol. Dis. 35:136-142. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 22.McLean, C., A. Buckmaster, D. Hancock, A. Buchan, A. Fuller, and A. Minson. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297-305. [DOI] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 25.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 26.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozawa, N., Y. Kawaguchi, M. Tanaka, A. Kato, A. Kato, H. Kimura, and Y. Nishiyama. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 79:6947-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelchen-Matthews, A., G. Raposo, and M. Marsh. 2004. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 12:310-316. [DOI] [PubMed] [Google Scholar]
- 29.Roizman, B., and E. Pellet. 2001. The family herpesviridae: a brief introduction, p. 2381-2399. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, PA.
- 30.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slagsvold, T., K. Pattni, L. Malerod, and H. Stenmark. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 16:317-326. [DOI] [PubMed] [Google Scholar]
- 32.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turcotte, S., J. Letellier, and R. Lippé. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, D. W., N. Davis-Poynter, and A. C. Minson. 1994. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J. Virol. 68:6985-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimori, T., F. Yamagata, A. Yamamoto, N. Mizushima, Y. Kabeya, A. Nara, I. Miwako, M. Ohashi, M. Ohsumi, and Y. Ohsumi. 2000. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11:747-763. [DOI] [PMC free article] [PubMed] [Google Scholar]