Abstract
Enhancers are defined as DNA elements that increase transcription when placed in any orientation relative to a promoter. The major immediate-early (MIE) enhancer region of murine cytomegalovirus is flanked by transcription units ie1/3 and ie2, which are transcribed in opposite directions. We have addressed the fundamental mechanistic question of whether the enhancer synchronizes transcription of the bidirectional gene pair (synchronizer model) or whether it operates as a genetic switch, enhancing transcription of either gene in a stochastic alternation (switch model). Clonal analysis of cytokine-triggered, transcription factor-mediated MIE gene expression from latent viral genomes provided evidence in support of the switch model.
Transcriptional enhancers are cis-acting genetic elements that can operate from a distance and irrespective of their orientation relative to the location of cognate or heterologous promoters (8, 14). Cytomegalovirus (CMV) major immediate-early (MIE) enhancers are essential for virus replication by enhancing MIE gene transcription to kick-start the viral transcriptional program in acute infection as well as in the initiation of virus reactivation from latency. MIE enhancers vary among different CMV species but all are composed of modules of transcription factor binding sites that form the structural basis for a dynamic interplay among a diverse array of viral and cellular transcription factors regulating enhancer activity (4, 6, 7, 9, 12, 15, 19, 22, 23, 29, 32, 33; for a recent review, see reference 34). The MIE locus of murine cytomegalovirus (mCMV) is special in that its enhancer region is flanked at one end by transcription unit ie1/3, driven by the core promoter P1/3, and at the other end by ie2, driven by P2 (Fig. 1) (12, 24, 35, 36). Transcription occurs in opposite directions, giving rise to spliced transcripts. Transcript IE1 (coding exons 2, 3, and 4) specifies the 76/89-kDa IE1 protein (25), which is involved in the early disruption of nuclear domain 10 (16; for a review, see reference 43). It acts as a transactivator of cellular genes involved in nucleotide metabolism (17, 28) and as a cotransactivator of viral early (E)-phase genes (35). Transcript IE3 (coding exons 2, 3, and 5) specifies the 88- to 90-kDa IE3 protein, which is the essential transactivator of viral E-phase genes (3, 35). Finally, transcript IE2 (coding exon 3) specifies the 43-kDa IE2 protein (36), whose function has yet to be identified (10, 30).
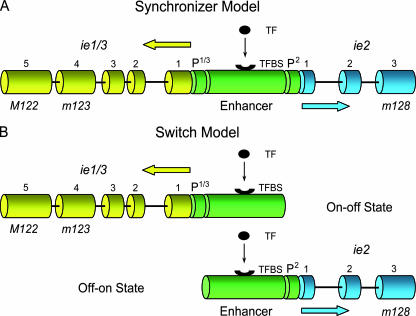
FIG. 1.
Models of MIE enhancer action in bidirectional gene pair transcription. (A) Synchronizer model. This model proposes that the enhancer region activates its flanking genes bidirectionally and simultaneously (the on-on state). As a consequence, transcriptions of ie1/3 and ie2 are expected to be correlated on the single genome level. (B) Switch model. This model proposes that the enhancer region activates its flanking genes bidirectionally but alternately (on-off and off-on states). As a consequence, transcriptions of ie1/3 and ie2 are expected to be anticorrelated on the single genome level, whereas alternate activation leads to correlated transcriptions on a multiple genome level. Bidirectional head-to-head architecture and exon-intron structure of the mCMV MIE locus (24, 35, 36) are illustrated. Open reading frames are designated according to the nomenclature proposed by Rawlinson et al. (37). Yellow and blue cylinders symbolize exons of ie1/3 and ie2, respectively. Yellow- and blue-colored arrows indicate the 5′-3′ direction of transcription on the respective DNA strand. P, promoter; TF, transcription factor(s); TFBS, transcription factor binding site(s).
Recently, reporter gene transfection assays have indicated that the mCMV MIE enhancer region consists of two bona fide enhancer elements in series (11). Notably, bidirectional gene pair organization, defined as two neighboring genes arranged head-to-head on opposite strands and regulated by a shared promoter/enhancer region, is a recently recognized common architectural feature in the human genome, often conserved among mouse orthologs and thought to provide a unique mechanism of regulation for a significant number of mammalian genes (1, 44). The existence of a bidirectional gene pair in the mCMV genome underlines the close host relatedness of this highly host-adapted viral pathogen.
As shown by Trinklein et al. (44) for mammalian genes, bidirectional organization is associated mostly with coexpression of the gene pair, whereas in a minority of examples, transcription of one gene of the pair is induced while transcription of the other is inhibited. The bidirectional MIE locus architecture of mCMV prompted us to ask whether the enhancer region-flanking transcription units are transcribed synchronously upon activation of the enhancer region (on-on and off-off states of the gene pair), referred to here as the “synchronizer model” of enhancer action (Fig. 1A) or whether the enhancer operates as a genetic switch that decides between mutually exclusive but alternate states (on-off and off-on states of the gene pair), referred to here as the “switch model” of enhancer action (Fig. 1B).
The two models can be distinguished experimentally only on a single genome level, because statistical ensembles of high numbers of genomes or bidirectional reporter gene pair constructs, as is always the case in acute infection or upon transfection in cell culture models, would unavoidably mimic an on-on synchronicity due to the superposition of many individual on-off and off-on states. As discussed by Fiering et al. (14), this problem is inherent in all bulk read-out assays.
We have previously shown for the BALB/c mouse model of mCMV latency in the lungs that in situ MIE locus activity occurs randomly, is independently distributed, with a very low point prevalence of only 10 to 20 transcriptional events per 106 latent viral genomes (18, 26; reviewed in reference 41), and provides the molecular basis for immune surveillance of latency by IE1 epitope-specific CD8 T cells (39). This stochastic MIE locus activity can be described by Poisson distribution statistics (18) and is reminiscent of the variegated expression, also known as mosaic expression, of transgenes thought to involve chromatin remodeling (8, 14). Likewise, silencing and desilencing by local closing and opening of viral chromatin structure is thought to be the mechanistic basis for CMVs’ MIE locus activity (31, 38; for reviews, see references 5, 41, and 42). As the rare and stochastic transcription events most likely represent MIE locus activities from single viral genomes, the mouse model of pulmonary latency is unique in that it fulfills the conditions for testing the two proposed models of mCMV MIE enhancer action.
In our previous experiments, the question of whether the MIE enhancer was involved at all in the observed sporadic MIE locus activity remained an open one. Alternatively, it might have reflected a stochastic, basal activity of the independent core promoters P1/3 and P2 that did not involve the enhancer (18, 26). Thus, in order to decide between the two models, the enhancer must be activated experimentally. Tumor necrosis factor alpha (TNF-α) activates the mCMV enhancer through transcription factors NF-κB and AP-1 (20, 21), resulting in an enhanced frequency of MIE gene transcription in latently infected lungs (40; reviewed in reference 41). This established in vivo approach of TNF-induced enhancer activity was used here for a quantitative analysis of MIE gene transcription by real-time reverse transcriptase (RT)-PCRs specific for spliced IE1 and IE2 transcripts.
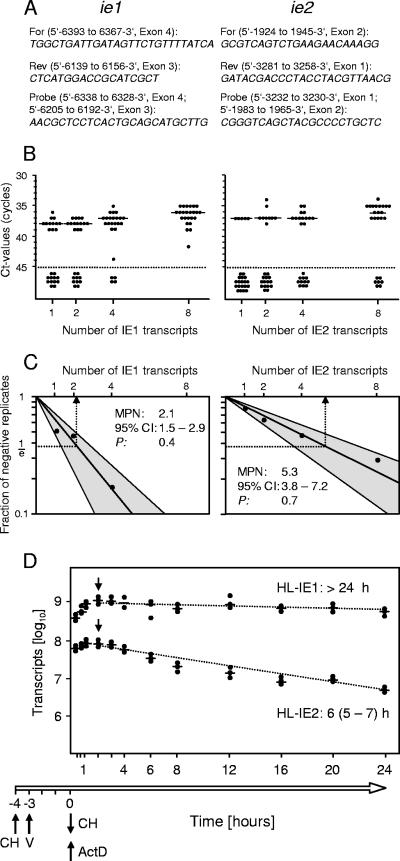
The sensitivity of the RT-PCR, which is defined mainly by the RT step rather than by cDNA amplification efficacy, was determined by a limiting dilution assay (27, 39) using graded numbers of synthetic transcripts (Fig. 2A to C). Although IE1 RT-PCR proved to be slightly more sensitive, cycle threshold (CT) values clearly discriminated between positive and negative samples for both types of RT-PCR. Transcript stability is another factor pertinent to transcript detectability. As shown in Fig. 2D for natural transcripts present in acutely infected fibroblasts, IE1 transcripts are extraordinarily stable, and IE2 transcripts are still fairly long-lived. Thus, a high-frequency oscillation between on-off and off-on states of the bidirectional gene pair should give an on-on result indistinguishable from the synchronizer model.
FIG. 2.
Detection limits of IE1- and IE2-specific real-time quantitative RT-PCRs. (A) Primers and probes. For, forward primer; Rev, reverse primer. Map positions are given according to GenBank accession no. L06816. RT-PCRs were performed on an ABI Prism 7500 (Applied Biosystems), with reaction conditions as described previously (39), except that the primer concentration was 0.6 μM and the 5-carboxy-X-rhodamine concentration was 0.132 μM. (B) Limiting dilution assay. Graded numbers of synthetic polyadenylated IE1 and IE2 transcripts (18) in 24 replicates were amplified by the respective real-time RT-PCRs. Dots represent the numbers of cDNA amplification CT values required for detection, with the median values for positive replicates marked by horizontal bars. The dotted lines indicate the cutoff CT value, defining a sample as negative if no signal above water control was obtained after 45 amplification cycles. (C) Poisson distribution analysis (27) based on the experimentally determined fractions of negative replicates (see panel B). The log-linear plots show the Poisson distribution graphs calculated with the maximum-likelihood method (13). Ninety-five percent confidence interval (CI) regions are shown shaded. The most probable number (MPN value) for the detection limit, representing the reciprocal of the Poisson distribution parameter λ, is revealed as the abscissa coordinate (dashed arrow) of the point of intersection between 1/e and the respective calculated regression line. CI, 95% confidence interval of MPN; P, probability value indicating the goodness of fit, which needs to be >0.05 for accepting the null hypothesis. (D) Transcript stability was measured as described previously in greater detail (39). The schedule for inhibitor treatment of infected mouse embryo fibroblasts is indicated: CH, cycloheximide for inhibition of protein synthesis restricting transcription to IE genes; V, infection with mCMV at a multiplicity of infection of 4 (centrifugal infection with 0.2 PFU/cell); ActD, actinomycin D to prevent further transcription. At the indicated time points after replacement of CH by ActD, total RNA was isolated from triplicate cultures, and IE1 and IE2 transcripts were quantitated by the respective real-time RT-PCRs. Dots represent data from triplicate cultures with the median value marked. Half-lives (HL; 95% confidence intervals of half-lives) HL-IE1 and HL-IE2 of the respective transcripts were determined from the negative slopes (95% confidence intervals of slopes) of the log-linear regression lines. Because of a gap period in the ActD effect, regression analysis included only data from day 2 onward (arrows). Calculations were performed using Mathematica Statistics linear-regression software, version 5.1 (Wolfram Research, Inc., Champaign, IL).
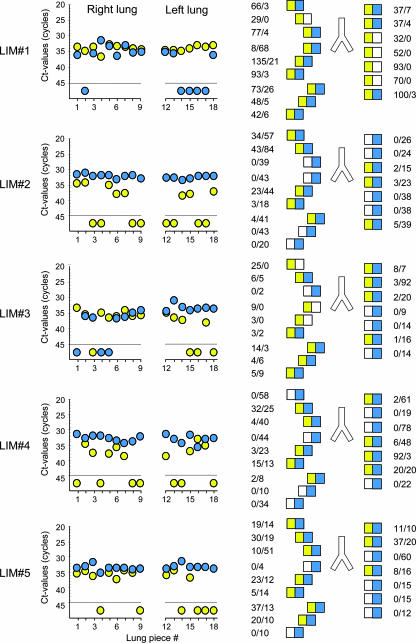
For the analysis of variegated MIE gene expression in vivo (18, 26), latently infected lungs were subdivided into pieces, and IE1 as well as IE2 transcripts were quantitated for each individual tissue piece (Fig. 3). Notably, similar to the findings shown above (Fig. 2B) for synthetic transcripts under limiting dilution conditions, positive and negative samples were clearly discriminated by a distance of at least 5 CT values, with a CT value range of <40 to >45, respectively (Fig. 3, left panel), leading to binary on-or-off data summarized in lung pictograms (Fig. 3, right panel). The existence of a considerable number of pieces selectively containing either IE1 or IE2 transcripts clearly rejects the synchronizer model and also excludes the possibility of a high-frequency oscillation between on-off and off-on states of the same genome. A bias that does not favor IE2 due to slightly lower sensitivity of the respective RT-PCR and somewhat shorter half-life of the IE2 RNA cannot explain the data, since IE1 transcripts were less frequently detected despite their longer half-life and the higher sensitivity of the respective RT-PCR.
FIG. 3.
Contextual analysis of MIE locus transcription patterns in latently infected lungs. The BALB/c mouse model of syngeneic bone marrow transplantation and infection with the mCMV wild-type Smith (ATCC VR-194) strain was employed to establish viral latency in the lungs of transplantation recipients (41). Transcription analysis for five latently infected mice (designated LIM#1 through LIM#5) was performed at 12 months after transplantation and at 24 h after the in vivo activation of MIE gene expression by 1 μg of recombinant murine TNF-α administered intravenously (40). For the analysis of variegated MIE gene expression, lungs were cut into pieces, specifically into nine pieces derived from the superior, middle, and inferior lobes of the right lung and seven pieces derived from the left lung. Two pieces (pieces 10 and 11) of the postcaval lobe were used to control for the presence and load of latent viral DNA (not shown). Transcripts IE1 and IE2 were quantified for each of the total number of 80 tissue pieces by the respective real-time RT-PCRs from 10% aliquots of the yields of total RNA purified with a QIAGEN RNeasy Plus kit. (Left panel) raw data given as CT values for the 16 lung pieces (pieces 1 to 9 and 12 to 18) tested per mouse. The dashed line marks the cutoff CT value separating negative from positive samples. Data for IE1 and IE2 are shown as yellow- and blue-filled circles, respectively. (Right panel) corresponding lung pictograms in anatomical view. Results are expressed in numbers of IE1/IE2 transcripts per test aliquot. In accordance with the MIE locus architecture illustrated in Fig. 1, yellow and blue boxes symbolize the presence of IE1 and IE2 transcripts, respectively. Open boxes symbolize the absence of transcripts.
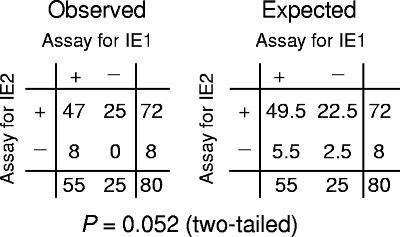
Finally, statistical correlation analysis (Fig. 4) showed that the prevalence of double-positive pieces is in accordance with the prevalence expected for a random distribution of viral genomes expressing IE1 or IE2 in a mutually exclusive manner compatible with the switch model. As we consider it unlikely that the viral genome exists in two physically distinct forms from which either IE1/3 or IE2 can be transcribed, we propose, in agreement with the stochastic nature of enhancer action discussed by Fiering and colleagues (14), that a single signaling event to the enhancer stochastically activates either of the two genes of the bidirectional gene pair with alternation possible for the physically same genome upon a second signaling event to the enhancer.
FIG. 4.
Correlation analysis. The binary transcription-on and transcription-off data (dichotomous variables) derived from Fig. 3 were arranged in a two-by-two contingency table (Observed) and compared with those in the table expected for independent distribution (null hypothesis) of IE1 and IE2 transcription (Expected). Fisher's exact test was used to calculate the two-tailed P value (method of the sum of small P values) following the recommendations provided by Simple Interactive Statistical Analysis (2, 45). The variables are considered to be positively correlated only if the P value is <0.01 and under the condition that the number of double positives observed is greater than the number of double positives expected, both of which were not fulfilled. The hypothesis of independent distribution cannot be rejected if P is >0.05, which was the case.
In conclusion, the data are consistent with the switch model of enhancer action and have definitely excluded the synchronizer model. These findings were not predicted and provide us with a novel detail for a better understanding of the way enhancers work. It is particularly intriguing that mCMV shares with mammalian genomes the architectural feature of bidirectional gene organization and, even more, that this occurs at a gene locus of outstanding regulatory importance. It is a challenge to identify a function for IE2 and to understand why its expression is paired with IE1 and IE3 in bidirectional gene organization, an architecture used in mammalian genomes most frequently for genes involved in DNA repair.
Acknowledgments
This work was supported by German Research Council collaborative research grant 490, individual project E2, Immunological Control of Latent CMV Infection.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109:807-809. [DOI] [PubMed] [Google Scholar]
- 2.Agresti, A. 1992. A survey of exact inference for contingency tables. Stat. Sci. 7:131-177. [Google Scholar]
- 3.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 6.Baskar, J. F., P. P. Smith, G. S. Ciment, S. Hoffmann, C. Tucker, D. J. Tenney, A. M. Colberg-Poley, J. A. Nelson, and P. Ghazal. 1996. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J. Virol. 70:3215-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baskar, J. F., P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 70:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:60-63. [DOI] [PubMed] [Google Scholar]
- 9.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Häsler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate-early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 10.Cardin, R. D., G. B. Abenes, C. A. Stoddard, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236-241. [DOI] [PubMed] [Google Scholar]
- 11.Chatellard, P., R. Pankiewicz, E. Meier, L. Durrer, C. Sauvage, and M. O. Imhof. 2007. The IE2 promoter/enhancer region from mouse CMV provides high levels of therapeutic protein expression in mammalian cells. Biotechnol. Bioeng. 96:106-117. [DOI] [PubMed] [Google Scholar]
- 12.Dorsch-Häsler, K., G. M. Keil, F. Weber, M. Jasin, W. Schaffner, and U. H. Koszinowski. 1985. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. USA 82:8325-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazekas de St. Groth, S. 1982. The evaluation of limiting dilution assays. J. Immunol. Methods 49:R11-R23. [DOI] [PubMed] [Google Scholar]
- 14.Fiering, S., E. Whitelaw, and D. I. K. Martin. 2000. To be or not to be active: the stochastic nature of enhancer action. BioEssays 22:381-387. [DOI] [PubMed] [Google Scholar]
- 15.Ghazal, P., M. Messerle, K. Osborn, and A. Angulo. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazal, P., A. E. Visser, M. Gustems, R. García, E. M. Borst, K. Sullivan, M. Messerle, and A. Angulo. 2005. Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J. Virol. 79:7182-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gribaudo, G., L. Riera, D. Lembo, M. De Andrea, M. Gariglio, T. L. Rudge, L. F. Johnson, and S. Landolfo. 2000. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J. Virol. 74:4979-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grzimek, N. K. A., D. Dreis, S. Schmalz, and M. J. Reddehase. 2001. Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs. J. Virol. 75:2692-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grzimek, N. K. A., J. Podlech, H.-P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hummel, M., and M. I. Abecassis. 2002. A model for reactivation of CMV from latency. J. Clin. Virol. 25:S123-S136. [DOI] [PubMed] [Google Scholar]
- 21.Hummel, M., Z. Zhang, S. Yan, I. Deplaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isomura, H., and M. F. Stinski. 2003. Effect of substitution of the human cytomegalovirus enhancer or promoter on replication in human fibroblasts. J. Virol. 77:3602-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keil, G. M., A. Ebeling-Keil, and U. H. Koszinowski. 1987. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J. Virol. 61:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keil, G. M., M. R. Fibi, and U. H. Koszinowski. 1985. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J. Virol. 54:422-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurz, S. K., M. Rapp, H.-P. Steffens, N. K. A. Grzimek, S. Schmalz, and M. J. Reddehase. 1999. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefkovits, I., and H. Waldman. 1979. Limiting dilution analysis of cells in the immune system, p. 38-82. Cambridge University Press, Cambridge, England.
- 28.Lembo, D., G. Gribaudo, A. Hofer, L. Riera, M. Cornaglia, A. Mondo, A. Angeretti, M. Gariglio, L. Thelander, and S. Landolfo. 2000. Expression of an altered ribonucleotide reductase activity associated with the replication of murine cytomegalovirus in quiescent fibroblasts. J. Virol. 74:11557-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Löser, P., G. S. Jennings, M. Strauss, and V. Sandig. 1998. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NF-κB. J. Virol. 72:180-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning, W. C., and E. S. Mocarski. 1988. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent α gene (ie2) is dispensable for growth. Virology 167:477-484. [PubMed] [Google Scholar]
- 31.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier, J. L., M. J. Keller, and J. J. McCoy. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for activation of viral gene expression and replication. J. Virol. 76:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier, J. L., and J. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier, J. L., and M. F. Stinski. 2006. Major immediate-early enhancer and its gene products, p. 151-166. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 35.Messerle, M., B. Bühler, G. M. Keil, and U. H. Koszinowski. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messerle, M., G. M. Keil, and U. H. Koszinowski. 1991. Structure and expression of murine cytomegalovirus immediate-early gene 2. J. Virol. 65:1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, C. O., R. Holtappels, H.-M. Tervo, V. Böhm, T. Däubner, S. A. Oehrlein-Karpi, B. Kühnapfel, A. Renzaho, D. Strand, J. Podlech, M. J. Reddehase, and N. K. A. Grzimek. 2006. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 80:10436-10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, C. O., C. K. Seckert, D. Dreis, M. J. Reddehase, and N. K. Grzimek. 2005. Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, C. O., C. K. Seckert, N. K. A. Grzimek, and M. J. Reddehase. 2006. Murine model of cytomegalovirus latency and reactivation: the silencing/desilencing and immune sensing hypothesis, p. 483-500. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 42.Sinclair, J., and P. Sissons. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763-1779. [DOI] [PubMed] [Google Scholar]
- 43.Tang, Q., and G. G. Maul. 2006. Immediate-early interactions and epigenetic defense mechanisms, p. 131-149. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 44.Trinklein, N. D., S. F. Aldred, S. J. Hartman, D. I. Schroeder, R. P. Otillar, and R. M. Myers. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uitenbroek, D. G. 1997. Binomial. SISA. http://home.clara.net/sisa/fisher.htm.