Abstract
The early events of hepatitis B virus (HBV) infection remain unclear. In 2006, Stoeckl et al. proposed a new entry mechanism involving a translocation motif (TLM) present in the pre-S2 domain of envelope proteins (L. Stoeckl, A. Funk, A. Kopitzki, B. Brandenburg, S. Oess, H. Will, H. Sirma, and E. Hildt, Proc. Natl. Acad. Sci. USA 103:6730-6734, 2006). After receptor binding and internalization into the endosomal compartment, this motif would allow the translocation of HBV particles through the endosomal membrane into the cytosol. In this study we have used two different mutated viruses containing a truncated TLM and showed their ability to infect human hepatocytes in primary culture, thus demonstrating the dispensability of the TLM for HBV infectivity.
The hepatitis B virus (HBV) envelope contains three transmembrane proteins known as hepatitis B surface (HBs) proteins: the small (S), middle (M), and large (L) polypeptides. These proteins are encoded by a single open reading frame containing three in-phase start codons. The hydrophobic S domain serves as a membrane anchor and plays important roles in virus assembly (5) and possibly membrane fusion (2). The M protein is formed by the S protein extended by the pre-S2 region (55 amino acids), while the L protein is composed of the entire M protein extended by the pre-S1 region (108 amino acids for genotype D). The N-terminal extremities of both S and M proteins are exposed on the surface of secreted particles, while the pre-S part of the L protein may be present either on the internal part of the virus, interacting with the nucleocapsid (4, 5, 8, 9, 20), or on the outer face (7, 21), available for interaction with target cells and necessary for viral infectivity (12, 17).
In 1994, I. Rodriguez-Crespo et al. (24) predicted the presence of a putative fusion peptide in the S protein of HBV. Its in vitro fusogenic properties support the hypothesis that it might be involved in the initial infective steps of hepadnavirus (22, 23). While no evidence proves its role in HBV infection, it seems to be crucial for duck HBV (DHBV) infectivity (11). In contrast, in 2006, Stoeckl et al. (25) proposed a new original entry mechanism involving a translocation motif (TLM) in the pre-S2 domain of envelope proteins. The TLM is a 12-amino-acid domain that can mediate an energy- and receptor-independent transfer of proteins, when fused to them, across the membrane without affecting their integrity (19, 25). According to this hypothesis, after virus binding at the cell surface and internalization into the endosomal compartment, the TLM would be exposed on the surface of viral particles following a conformational change of envelope proteins and would allow the translocation of viruses through the endosomal membrane into the cytosol. The role of the TLM in HBV infection steps was suggested by experiments demonstrating that artificial exposure of the TLM on the surface of viral particles leads to infection of HuH7 cells. However, no assays were conducted to analyze the capacity of TLM-deficient viruses to infect human hepatocytes, leaving some doubt about its role in the virus life cycle. Furthermore, we have already shown in 1998, the dispensability of the L-protein TLM for HBV infection (18). Indeed, virions lacking TLM in this protein, as a consequence of internal deletions, were still able to assemble and infect primary human hepatocytes (PHH). However, in these experiments TLM was still present in the M protein which, although not necessary for viral infection (13, 18), could provide in trans, a functional TLM responsible for the infectivity of chimeric viruses lacking TLM only in the L protein.
To complete these results, we decided to analyze the infectivity of virions lacking TLM both in the L and M proteins. To produce viruses with wild-type (WT) L and M proteins or viruses with the L and M proteins deleted, we cotransfected HepG2 cells (1) with three different plasmids. The first construct, named pHBV L−E−, corresponds to a viral genome competent for viral replication but deficient for envelope protein production. It is derived from the plasmid pHBV L− (18) in which we have introduced an opal mutation into codon 15 and an amber mutation into codon 94 of the S domain. The second plasmid, pSVSX, used to optimize HBV particle production, encodes the WT S protein. It contains the 1,986-bp EcoRI-BglII fragment of WT HBV DNA, bearing the entire S and X coding region, cloned downstream of the simian virus 40 early promoter in plasmid pSV-SPORT 1 (Life Technology). Finally, the third construct, pSV12SX, is an expression vector of the three surface proteins (18). Mutations consisting of contiguous deletions of 10 amino acids in the pre-S2 domain between positions 114 and 163 (Fig. 1) have been introduced in this plasmid. The five first amino acids of the pre-S2 domain (positions 109 to 113) are crucial for the assembly process (18) and therefore have not been deleted. The unaffected expression of proteins in HepG2 cells transfected with the WT or mutant protein expression plasmids was previously assessed by Western blotting (18). HBs antigen was also found to be actively produced in the culture supernatant of transfected HepG2 cells (data not shown), thus demonstrating that deletions did not alter the secretion of envelope proteins. Finally, immunolocalization studies of the mutated proteins in HuH7 cells did not show any alteration compared to the WT proteins (data not shown).
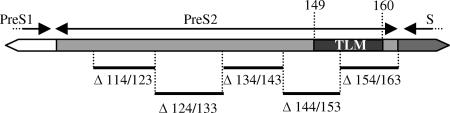
FIG. 1.
Amino acid deletions in the pre-S2 region of the S gene. Deletions (shown in the form Δx/y, where x is the position of the first deleted amino acid and y is the position of the last deleted amino acid) are indicated as thin lines. Positions are given relative to the first N-terminal amino acid of the L protein (subtype ayw, EMBL accession no. X02496).
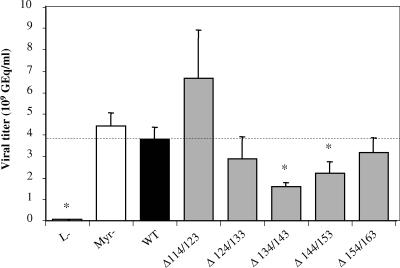
Since no changes were observed at the protein expression level, the ability of the mutated proteins to complement replication of the envelope protein-defective genome for viral particle secretion was tested. The culture supernatants of cotransfected HepG2 cells were concentrated 50-fold to obtain the diverse inocula (15). To assess the relative amount of viruses produced, particles from 50 μl of concentrated inocula were fixed on 96-well plates coated with a monoclonal anti-pre-S1 antibody (MA18/7, a generous gift from W. H. Gerlich), and viral DNA was quantified by quantitative PCR (Q-PCR) with primers that amplify the core gene (10). Cotransfections with constructs causing deletion of the L and M proteins have clearly shown that mutants with a 10-amino-acid deletion in the pre-S2 domain between positions 114 and 163 were still able to assemble into complete viral particles (Fig. 2). The two controls, L− and Myr−, consist of HBV with the L protein deleted, impairing virion production (4, 5), and of HBV with a mutated L protein defective for myristoylation at its N-terminal extremity, resulting in the production of noninfectious viruses (6, 15), respectively. Depending on the deletion, the relative amount of secreted virions was slightly different compared to that of WT virus, and as expected, no viral particles were found in the L− control (Fig. 2). The level of viral secretion was slightly reduced for the Δ134/143 (deletion from positions 134 to 143) and Δ144/153 deletions. However, we can conclude that the five deletions introduced in the pre-S2 domains of both M and L proteins, two of which impair the TLM (Δ144/153 and Δ154/163), allow the production of viruses.
FIG. 2.
Viral titers of inocula determined by quantitative PCR analysis of viral DNA. Results are expressed in GEq per ml of inoculum. The two controls, L− and Myr−, consist of HBV with the L protein deleted, impairing virion production, and of HBV with an unmyristoylated L protein, resulting in the production of noninfectious viruses, respectively. Mutant viruses, with deletions in the pre-S2 domain, are identified as Δx/y. Statistical analysis was performed by the Mann-Whitney test; significant differences compared to the WT condition are indicated by asterisks (P < 0.05). The dotted horizontal line indicates the level of the WT condition.
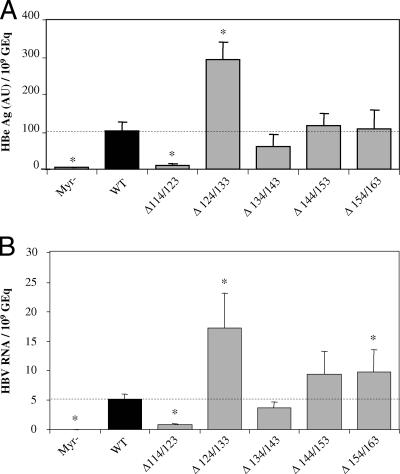
To determine whether the pre-S2 region of both M and L proteins, where the TLM is found, was involved in viral infectivity, we performed in vitro infections of PHH (14) with both mutant and WT viruses. During infection, performed without enhancer of viral infection, hepatocytes (4.16 × 105 per 1.9-cm2 well) were covered with 250 μl of serum-free culture medium (14) containing 50 μl of inocula with titers between 1.6 × 109 and 6.7 × 109 genome equivalents (GEq) per ml (Fig. 2). After infection, cells were washed three times, and the medium was renewed every 2 days. We have assessed infection of human hepatocytes 10 days postinfection, by measuring both secreted core antigen (HBeAg), a sensitive and specific marker of HBV infection, in the culture supernatant of infected cells and intracellular HBV RNA. HBeAg was measured in culture medium with the Bio-Rad kit Monolisa HbeAg-Ab Plus, and viral RNAs were measured by Q-PCR after reverse transcription with primers that amplify the core gene (10). The infectivity was expressed as a ratio between the level of infection markers (HBe or HBV RNA) and the number of GEq used for infection. As expected, no infection was detected with the control inoculum Myr−, which corresponds to a noninfectious mutant (Fig. 3), confirming the specificity of our infection assays. Depending on the deletion, the infectious ability of mutants was modulated compared to that of the WT virions. While the first deletion (Δ114/123) slightly increased viral assembly and strongly decreased the infectivity of the virus, the contiguous one (Δ124/133) slightly decreased assembly and increased infectivity about three times (Fig. 3). We can assume that the region of the pre-S2 domain between positions 114 and 133, although not crucial for infectivity, could nevertheless act as a strong modulator. Finally, concerning the three last mutants, two of which bear a truncated TLM, we clearly noticed that they were able to infect PHH as efficiently as the WT virus. We can therefore conclude that the TLM, present in the pre-S2 domain of HBV envelope proteins, is dispensable for both particle assembly and infectivity. Furthermore, while the manuscript was in preparation, a study using hepatitis delta virus as a surrogate model for HBV infection showed the dispensability of the TLM for infectivity (16).
FIG. 3.
Infectivity of mutant and WT viruses. The infectivity is expressed as a ratio between the level of infection markers (HBe or HBV RNA) and the number of GEq used for infection. The control Myr− is described in the legend to Fig. 2. Mutant viruses with deletions in the pre-S2 domain are identified as Δx/y. Statistical analysis was performed using the Mann-Whitney test; significant differences compared to the WT condition are indicated by asterisks (P < 0.05). The dotted horizontal line indicates the level of the WT condition. (A) Infection was assessed by measuring HBeAg in the culture supernatant of infected cells 10 days postinfection. One arbitrary unit (AU) corresponds to the HBe concentration in the positive control of the assay kit. (B) Infection was assessed by measuring intracellular viral RNA by Q-PCR after reverse transcription 10 days postinfection. Expression data are normalized to 18S rRNA, and the relative viral RNA expression levels are calculated according to the ΔΔCt (comparative Ct) method (Applied Biosystems user bulletin 2 on relative quantification of gene expression) in which the reference corresponds to the WT condition.
Thus, while infectivity assays seem to prove that the TLM of the DHBV L protein is involved in the infection process (25), this motif seems to be dispensable for HBV infection. Therefore, although our data tend to exclude a role of TLM in HBV entry, we cannot rule out the possibility that some still unidentified functional TLM located elsewhere in the viral surface proteins could substitute for the impaired pre-S2 TLM, thus allowing infection by the corresponding virions. Nevertheless, involvement of a fusion peptide in a regular fusion process, in agreement with data obtained in DHBV system (11), should be considered and is currently under investigation.
While this article was in revision, a paper by Blanchet and Sureau also showed that the pre-S2 TLM was dispensable for HBV infectivity in another model where the infection was enhanced by the use of polyethylene glycol (3).
Acknowledgments
This work was supported by funding from INSERM (Institut National de la Santé et de la Recherche Médicale), ARC (Association pour la Recherche sur le Cancer), and ANRS (Agence Nationale de Recherche contre le Sida). C. Lepère is the recipient of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie.
We thank the Biological Resource Centre of Rennes, France, for the isolated human hepatocytes. Finally, we gratefully acknowledge Isabelle Cannie for helpful technical support.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Aden, D. P., A. Fogel, S. Plotkin, I. Damjanov, and B. B. Knowles. 1979. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 282:615-616. [DOI] [PubMed] [Google Scholar]
- 2.Berting, A., C. Fischer, S. Schaefer, W. Garten, H. D. Klenk, and W. H. Gerlich. 2000. Hemifusion activity of a chimeric influenza virus hemagglutinin with a putative fusion peptide from hepatitis B virus. Virus Res. 68:35-49. [DOI] [PubMed] [Google Scholar]
- 3.Blanchet, M., and C. Sureau. 2007. The infectivity determinants of the large hepatitis B virus envelope protein pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81:5841-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V., J. Hagelsten, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. [DOI] [PubMed] [Google Scholar]
- 7.Bruss, V., X. Y. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruss, V., and R. Thomssen. 1994. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 68:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruss, V., and K. Vieluf. 1995. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 69:6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R. W., H. Piiparinen, M. Seppanen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65:250-256. [DOI] [PubMed] [Google Scholar]
- 11.Chojnacki, J., D. A. Anderson, and E. V. Grgacic. 2005. A hydrophobic domain in the large envelope protein is essential for fusion of duck hepatitis B virus at the late endosome. J. Virol. 79:14945-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouteau, P., J. Le Seyec, I. Cannie, M. Nassal, C. Guguen-Guillouzo, and P. Gripon. 2001. A short N-proximal region in the large envelope protein harbors a determinant that contributes to the species specificity of human hepatitis B virus. J. Virol. 75:11565-11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernholz, D., P. R. Galle, M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1993. Infectious hepatitis-B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194:137-148. [DOI] [PubMed] [Google Scholar]
- 14.Gripon, P., C. Diot, N. Theze, I. Fourel, O. Loreal, C. Brechot, and C. Guguen-Guillouzo. 1988. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol. 62:4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213:292-299. [DOI] [PubMed] [Google Scholar]
- 16.Gudima, S., A. Meier, R. Dunbrack, J. Taylor, and V. Bruss. 2007. Two potentially important elements of the hepatitis B virus large envelope protein are dispensable for the infectivity of hepatitis delta virus. J. Virol. 81:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oess, S., and E. Hildt. 2000. Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther. 7:750-758. [DOI] [PubMed] [Google Scholar]
- 20.Poisson, F., A. Severac, C. Hourioux, A. Goudeau, and P. Roingeard. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228:115-120. [DOI] [PubMed] [Google Scholar]
- 21.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Crespo, I., E. Nunez, J. Gomez-Gutierrez, B. Yelamos, J. P. Albar, D. L. Peterson, and F. Gavilanes. 1995. Phospholipid interactions of the putative fusion peptide of hepatitis B virus surface antigen S protein. J. Gen. Virol. 76:301-308. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Crespo, I., E. Nunez, B. Yelamos, J. Gomez-Gutierrez, J. P. Albar, D. L. Peterson, and F. Gavilanes. 1999. Fusogenic activity of hepadnavirus peptides corresponding to sequences downstream of the putative cleavage site. Virology 261:133-142. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Crespo, I., J. Gomez-Gutierrez, D. L. Peterson, and F. Gavilanes. 1994. Interaction of a peptide corresponding to the amino terminus region of the S protein of hepatitis B virus with liposomes. Biochem. Soc. Trans. 22:S365. [DOI] [PubMed] [Google Scholar]
- 25.Stoeckl, L., A. Funk, A. Kopitzki, B. Brandenburg, S. Oess, H. Will, H. Sirma, and E. Hildt. 2006. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc. Natl. Acad. Sci. USA 103:6730-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]