Abstract
The human immunodeficiency virus type 1 (HIV-1) RNA genome contains a terminal repeat (R) region that encodes the transacting responsive (TAR) hairpin, which is essential for Tat-mediated activation of gene expression. TAR has also been implicated in several other processes during viral replication, including translation, dimerization, packaging, and reverse transcription. However, most studies in which replication of TAR-mutated viruses was analyzed were complicated by the dominant negative effect of the mutations on transcription. We therefore used an HIV-1 variant that does not require TAR for transcription to reinvestigate the role of TAR in HIV-1 replication. We demonstrate that this virus can replicate efficiently upon complete deletion of TAR. Furthermore, evolution of a TAR-deleted variant in long-term cultures indicates that HIV-1 requires a stable stem-loop structure at the start of the viral transcripts in which the 5′-terminal nucleotides are base paired. This prerequisite for efficient replication can be fulfilled by the TAR hairpin but also by unrelated stem-loop structures. We therefore conclude that TAR has no essential function in HIV-1 replication other than to accommodate Tat-mediated activation of transcription.
Retroviral RNA genomes contain a sequence repeat (R) that forms the extreme 5′ and 3′ ends of the viral transcripts. The transacting responsive (TAR) region of the 97-nucleotide (97-nt) R region of human immunodeficiency virus type 1 (HIV-1) RNA can fold a stable hairpin structure (8) (Fig. 1) which has been suggested to play multiple important roles in viral replication. The best-studied function of this TAR hairpin is its essential role in the activation of transcription from the promoter in the 5′ long terminal repeat (LTR) of the proviral genome (reviewed in references 2 and 11). Important features in TAR are the highly conserved 3-nt pyrimidine bulge in the stem, which binds the viral Tat transactivator protein (32), and the apical 6-nt loop to which the cyclin T1 subunit of the positive transcriptional elongation factor (pTEFb) binds in a Tat-dependent manner (27, 35). Upon binding, the kinase component of pTEFb, cyclin-dependent kinase 9 (CDK9), can phosphorylate the C-terminal domain of RNA polymerase II, which enhances the processivity of the elongating polymerase (10, 24). Furthermore, it was recently demonstrated that pTEFb also directs the recruitment of TATA-box-binding protein to the LTR promoter and thus stimulates the assembly of new transcription complexes (26).
FIG. 1.
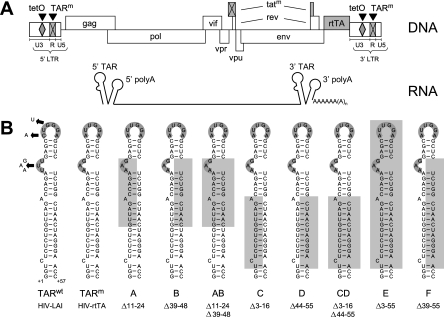
TAR mutations introduced in the dox-dependent HIV-rtTA. (A) A schematic of the HIV-rtTA genome is shown, with the LTR region subdivided into the U3, R, and U5 domains. Transcription starts at the first nucleotide of the 5′ R region, and the RNA transcripts are polyadenylated at the last nucleotide of the 3′ R region. Both the 5′ and the 3′ ends of the RNA molecule can fold a TAR and poly(A) hairpin. In the HIV-rtTA virus, the Tat-TAR axis of transcription regulation has been inactivated by mutation of both Tat and TAR (tatm and TARm; crossed boxes). Transcription and replication of the virus was made dox dependent by the introduction of tetO elements in the U3 promoter region and by replacing the Nef gene with the rtTA gene. (B) HIV-rtTA is based on the HIV-1 molecular clone LAI, which contains a 57-nt wild-type TAR hairpin (TARwt). In HIV-rtTA, TAR was inactivated by nucleotide substitutions in both the bulge and loop motifs (TARm). The TARm sequence was partially or nearly completely deleted (mutants A to F). The deleted nucleotides are boxed in gray. All mutations were introduced in both the 5′ and 3′ LTR of HIV-rtTA.
There have been numerous reports in which additional functions of TAR in translation, dimerization, packaging, and reverse transcription of the viral transcripts have been proposed (reviewed in references 1, 2, 6, and 16). More recently, it has been suggested that TAR may also affect the cellular RNA interference process (3, 4). Thus, a pleiotropy of functions has been attributed to the TAR motif in a variety of experimental systems. The most biologically relevant assay system is that of the replicating virus, and the importance of TAR is underlined by the observation that mutations within TAR cause severe replication defects. However, these studies are complicated by the fact that nearly all mutations in TAR affect the transcription process. This dominant effect on transcription makes it difficult or impossible to distinguish the effect of such mutations on other processes during virus replication.
We and others previously reported the construction of an HIV-1 variant that does not depend on the Tat-TAR interaction for activation of transcription (13, 30, 34). In our HIV-rtTA variant, both the Tat protein and its TAR binding site were inactivated by mutation (Fig. 1A) and functionally replaced by the components of the Tet-On gene regulation system (33). The gene encoding the rtTA transcriptional activator protein was inserted in place of the 3′-terminal nef gene, and tet operator (tetO) binding sites were introduced in the LTR promoter. Administration of the effector doxycycline (dox) induces a conformational switch in the rtTA protein, which subsequently can bind to the tetO-LTR promoter region and activate transcription of the proviral genome. Thus, transcription and replication of HIV-rtTA are critically dependent on the presence of dox. Since this virus does not require TAR for the activation of transcription, it is the ideal tool to study nontranscriptional functions of TAR in virus replication. We here present the development of an efficiently replicating HIV-rtTA variant that completely lacks the TAR-hairpin. Our results demonstrate that TAR is not essential for processes other than transcription during HIV-1 replication.
MATERIALS AND METHODS
Construction of HIV-rtTA variants.
Construction of the HIV-rtTA molecular clone was described previously (13, 34). The HIV-rtTA variant used in this study (HIV-rtTAF86Y A209T2ΔtetO) contains the 2ΔtetO promoter configuration in both the 5′ and 3′ LTR (22) and the optimized rtTAF86Y A209T gene (14). Deletions in TAR were introduced in both the 5′ and 3′ LTRs of the HIV-rtTA plasmid in three steps as described below. Briefly, we first introduced the deletions by PCR mutagenesis in the 3′ LTR of a shuttle vector encompassing the 3′ half of the HIV-rtTA genome, subsequently introduced the mutations in the 5′ LTR of HIV-rtTA, and finally combined the 5′ and 3′ LTR mutated fragments. For the construction of the A variant, the 3′ LTR sequence was amplified with primers tTA-tetO-1 (ctccccgggtaactaagtaaggat; sense primer annealing at the 3′ end of the rtTA gene) and TAR-A (cagagagctcca-Δ-atgctccagagagacccagtacaggc [SacI site underlined, Δ indicating position of deletion]), with plasmid pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO, which includes Env, rtTA, and 3′ LTR sequences of the HIV-rtTA genome (14), as the template. The PCR product was digested with BspEI and SacI and ligated into the corresponding sites of the shuttle vector pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL, which is a derivative of pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO in which the SacI site has been removed from the vector sequence by digestion with BssHII and BamHI, blunting of the sticky ends with Klenow polymerase and deoxynucleoside triphosphates, and subsequent religation. The C variant was constructed in the same way, but PCR was performed with primers tTA-tetO-1 and TAR-C (cagagagctccaatgctcctttctgg-Δ-cccagtacaggcaaaaagcag). Similarly, for the construction of the B variant, PCR was performed with primers TAR-B (attggagctc-Δ-tagggaacccactgcttaagcc) and pLAI-3′seq (tgtctcatgagcggatacata [antisense primer annealing to vector sequences downstream of 3′ LTR]), and the PCR product was digested with SacI and AatII and subsequently ligated into the corresponding sites of pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL. The D and F variants were constructed in the same way but with primers TAR-D (attggagctctctgg-Δ-cccactgcttaagcctcaata) and TAR-F (attggagctc-Δ-cccactgcttaagcctcaata), respectively. For construction of the E variant, we used primers tTA-tetO-1 and TAR-E (aggcaagctttattgaggcttaagcagtggg-Δ-cccagtacaggcaaaaagca [HindIII site underlined]), digested the PCR product with BspEI and HindIII, and subsequently ligated this fragment into the corresponding sites of pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL. For the construction of the double-mutant AB, we combined the A and B deletions. The A variant of pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL was digested with BamHI and SacI, and the 1,463-bp env-rtTA-LTR fragment was used to replace the corresponding sequences in the B variant of this shuttle vector. Similarly, to combine the C and D deletions in the CD double mutant, the BamHI-SacI env-rtTA-LTR fragment of the C variant was used to replace the corresponding sequences in the D variant. For the introduction of the TAR mutations into the 5′ LTR of HIV-rtTA, we employed PCR to amplify the LTR regions from the pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL variants A to F with primers U3-Xba-Not (acgtctagagcggccgcactggaagggctaattcactc [positions −331 to −313]) and U5-Nar (ttcgggcgccactgctagagattttccacactg [positions +192 to +160]), digested the PCR product with NotI and NarI, and used this fragment to replace the corresponding 5′ LTR sequences in HIV-rtTA. To construct 5′-plus-3′ TAR-mutated HIV-rtTA variants, the BamHI-BglI fragments of the pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL variants were used to replace the corresponding Env-rtTA-3′ LTR sequences in the 5′ LTR-mutated HIV-rtTA variants. All mutations were verified by sequence analysis.
Proviral DNA analysis and cloning of evolved sequences.
Virus-infected cells were pelleted by centrifugation at 4,000 rpm for 4 min and washed with phosphate-buffered saline (PBS). DNA was solubilized by resuspending the cells in 10 mM Tris-HCl (pH 8.0)-0.1 mM EDTA-0.5% Tween 20 followed by incubation with 200 μg of proteinase K per ml at 56°C for 30 min and at 95°C for 10 min. Proviral DNA sequences were PCR amplified from total cellular DNA with primers U3-Xba-Not (acgtctagagcggccgcactggaagggctaattcactc [positions −331 to −313]) and AD-GAG (atggatccgttctagctccctgcttgccc [positions +463 to +442]), ligated into pCR2.1-TOPO TA-cloning vector (Invitrogen), and sequenced with primer BB3 (gagtcctgcgtcgagagagctcctctggtt [positions +245 to +216]). For the cloning of the evolved ER1-3 sequences into the HIV-rtTA provirus, the U3-R sequences were PCR amplified from the corresponding TA clones with primers U3-Xba-Not and AD-GAG. The PCR product was digested with NarI and NotI and used to replace the corresponding 5′ LTR sequences in HIV-rtTA. Moreover, the PCR product was digested with SalI and HindIII and used to replace the corresponding fragment in pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL. The BamHI-BglI fragments of these 3′ LTR-mutated plasmids were used to replace the corresponding sequences in the 5′ LTR-mutated HIV-rtTA variants, which resulted in HIV-rtTA variants with ER1-3 sequences in both the 5′ and 3′ LTRs.
rtTA activity assay.
In the plasmid pLTR-2ΔtetO-X/H-lucff the expression of firefly luciferase is under the control of the LTR-2ΔtetO promoter of HIV-rtTA. For the construction of this plasmid, we isolated the XbaI-HindIII fragment encompassing the LTR-2ΔtetO promoter sequences (−333 to +82) of HIV-rtTAF86Y A209T2ΔtetO and ligated this fragment into the compatible NheI and HindIII sites of the luciferase reporter construct pGL3-Basic (Promega). For cloning purposes, the SalI site in pGL3-basic had been removed by digestion with SalI, blunting of the sticky ends with Klenow polymerase and deoxynucleoside triphosphates, and subsequent religation.
For the introduction of the TAR mutations into pLTR-2ΔtetO-X/H-lucff, we PCR amplified the LTR region from the pBlue3′LTRext-ΔU3-rtTAF86Y A209T-2ΔtetO-mPL variants A to F with primers U3-Xba-Not and U5-Nar, digested the PCR product with SalI and HindIII, and used this fragment to replace the corresponding sequences in pLTR-2ΔtetO-X/H-lucff. Plasmid pBlue3′LTR-lucff contains the complete U3 region and R sequences up to position +82 of the wild-type HIV-1 LAI proviral DNA coupled to the firefly luciferase reporter gene (19). The plasmid pRL-CMV (Promega), in which the expression of Renilla luciferase is controlled by a cytomegalovirus promoter, is cotransfected into the C33A cells to allow correction for differences in transfection efficiency.
C33A cells were cultured in 2-cm2 wells and transfected with 20 ng pLTR-2ΔtetO-lucff construct (TARm and mutants A to F) or 20 ng pBlue3′LTR-lucff (TARwt), 0.4 ng rtTA-expression plasmid pCMV-rtTAF86Y A209T (14), and 0.5 ng pRL-CMV. pBluescript was added to the transfection mix to achieve a total of 1 μg of DNA. The cells were cultured after transfection for 48 h with 0 to 1,000 ng/ml dox (Sigma D-9891). Cells were lysed in passive lysis buffer, and firefly and Renilla luciferase activities were determined with a dual-luciferase assay (Promega). The expression values for firefly and Renilla luciferase were within the linear range, and no squelching effects were observed. The promoter activity was calculated as the ratio between the firefly and Renilla luciferase activities and was corrected for between-session variation (29).
Cells and viruses.
SupT1 T cells were cultured and transfected by electroporation, as previously described (14). To assay virus replication, 5 × 106 SupT1 cells were transfected with 1 μg of the proviral constructs and cultured in 5 ml medium with 1 μg/ml dox. For the selection of variants with improved replication capacity, the viruses were cultured for up to 168 days. When virus-induced cytopathic effects were observed, high-level virus replication was maintained by passage of the cell-free culture supernatant onto uninfected SupT1 cells. Cell and supernatant samples were stored at −80°C for subsequent analysis. C33A cervix carcinoma cells (ATCC HTB31) were cultured in 2-cm2 wells and transfected with 1 μg HIV-rtTA construct by calcium phosphate precipitation, as previously described (14). Virus production was measured using a CA-p24 enzyme-linked immunosorbent assay (ELISA) and culture medium samples (20).
RNA analysis.
For the isolation of viral transcripts, C33A cells were cultured in 10-cm2 wells and transfected with 5 μg HIV-rtTA construct. After 48 h, cells were washed with PBS, briefly incubated with 0.5 ml 0.05% trypsin-EDTA (Invitrogen) until cells detached, resuspended in 1 ml 10% fetal bovine serum-containing medium to inactivate trypsin, and subsequently centrifuged at 2,750 × g for 5 min. Cells were washed in 1 ml PBS, centrifuged at 2,750 × g for 5 min, lysed in 0.6 ml RLT buffer (QIAGEN), and homogenized with a QIAshredder column (QIAGEN). Total cellular RNA was isolated with an RNeasy kit (QIAGEN), and contaminating DNA was removed with RNase-free DNase (QIAGEN) during isolation. When indicated, RNA was decapped with tobacco acid pyrophosphatase (BIOzymTC; Epicenter Biotechnologies, Landgraaf, The Netherlands).
We used 5′ rapid amplification of cDNA ends (RACE) system version 2.0 (Invitrogen) to analyze the 5′ ends of the RNA transcripts. Briefly, the primer AD-GAG was annealed to the viral RNA at 85°C for 2 min and 70°C for 10 min. The RNA was reverse transcribed with SuperScript II reverse transcriptase (RT; Invitrogen) at 50°C for 50 min. After inactivation of RT at 70°C for 15 min, RNA was degraded with RNase H and RNase T1. The cDNA product was purified with a QIAquick PCR purification kit (QIAGEN) and dA-tailed with terminal deoxynucleotidyl transferase and dATP. After inactivation of terminal deoxynucleotidyl transferase at 65°C for 10 min, the dA-tailed cDNA was amplified by PCR with primers AD-SD (catggatccagtcgcctcccctcgcctc [positions +290 to +270]) and 3′RACE Abridged Primer (ggccacgcgtcgactagtac[t]17) and ligated into pCR2.1 TA-cloning vector. Cloned cDNA fragments were sequenced with primer BB3.
RESULTS
Deletions in TAR do not affect gene expression.
HIV-rtTA carries multiple nucleotide substitutions in TAR (TARm in Fig. 1B) that completely abolish transactivation of the viral promoter by Tat. This virus no longer requires the Tat-TAR axis for the activation of gene expression, but the TAR stem-loop structure may still have other roles in viral replication. Previous attempts to identify such secondary functions of TAR were complicated by the indispensability of the wild-type TAR structure for activation of transcription. We therefore set out to determine whether the TAR structure in the HIV-rtTA variant is required for viral replication and introduced more-rigorous mutations in the hairpin motif. Partial deletion of the left-hand side (mutants A and C) or the right-hand side (mutants B, D, and F) of the TAR stem severely reduces the stability of this hairpin (Fig. 1B). Combination of these deletions in the double mutants AB and CD results in truncated stem-loop structures. In mutant E, the most rigorous mutant, we deleted nearly the complete TAR structure except for the bottom two base pairs, which were left in place to preserve the G residues at the transcription initiation site.
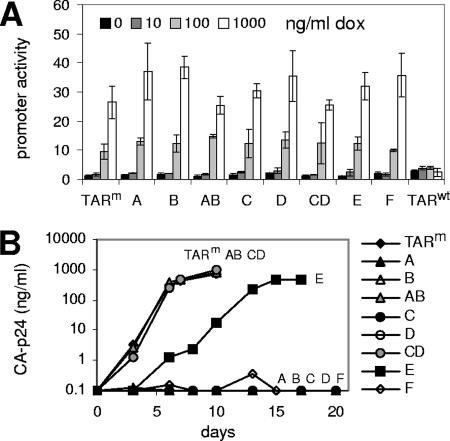
To determine the effect of these TAR deletions on gene expression, we made plasmid reporter constructs in which the LTR-tetO promoter of HIV-rtTA, carrying the original TAR sequence (TARm) or a mutated TAR sequence (A to F), was coupled to the luciferase reporter gene. We assayed dox responsiveness of these constructs upon cotransfection with an rtTA-expression plasmid into C33A cervix carcinoma cells (Fig. 2A). The TARm construct demonstrates low basal luciferase expression in the absence of dox, and the activity level gradually rises with increases in dox concentrations. Similar dox-dependent expression levels are observed for mutants A to F, demonstrating that the TAR deletions do not affect gene expression in this promoter context. A control construct with the wild-type HIV-1 LTR promoter (TARwt) obviously does not respond to dox. We also assayed Tat responsiveness of these promoter luciferase constructs upon cotransfection with a Tat expression plasmid. As anticipated, TARm and all TAR deletion constructs (A to F) were not activated by Tat, and only the control TARwt promoter construct was greatly responsive to Tat (data not shown).
FIG. 2.
Effect of TAR mutations on gene expression and virus replication. (A) The LTR-tetO promoter region of HIV-rtTA with the original (TARm) or mutant (A to F) TAR sequence was placed upstream of the firefly luciferase gene. To determine dox responsiveness, C33A cells transfected with these plasmid reporter constructs and an rtTA-expression plasmid were cultured at different dox levels (0 to 1,000 ng/ml). A plasmid constitutively expressing Renilla luciferase was cotransfected to correct for differences in transfection efficiency, and the ratio of the firefly and Renilla luciferase activities measured two days after transfection reflects the promoter activity. A construct with the wild-type HIV-1 LTR promoter (TARwt) was included as a control. Average values obtained in three transfections are shown, with the error bars indicating standard deviations. (B) SupT1 T cells were transfected with the original HIV-rtTA (TARm) and TAR-mutated variants (A to F) and cultured with 1 μg/ml dox for several weeks. CA-p24 levels in the culture supernatant were measured by ELISA. No virus replication was observed in the absence of dox (not shown). This experiment was repeated three times with similar results.
Replication of TAR-deleted variants.
To reveal additional roles of TAR in viral replication, we introduced TAR deletions A to F into the HIV-rtTA genome. The TAR sequence is part of the 97-nt R region that is present at both ends of the viral RNA genome and plays an important role in first-strand transfer during reverse transcription (9). Since sequence differences between the 5′ and 3′ R regions may hamper the reverse transcription process, we introduced the TAR mutations at both ends of the proviral genome. We transfected these HIV-rtTA plasmids into C33A cells, which support HIV-1 gene expression and virion production but not HIV-1 infection due to lack of a CD4 receptor. Cells were cultured for 48 h with dox, and we subsequently measured CA-p24 production in the culture supernatant. The TARm and TAR deletion variants all produced a high level of CA-p24 (data not shown), which confirms that viral gene expression is not significantly affected by partial or complete deletion of TAR.
We transfected the HIV-rtTA plasmids into the HIV-1-susceptible SupT1 T-cell line to determine the effect of the TAR deletions on viral replication. These cells support efficient replication of the original HIV-rtTA in the presence of dox (Fig. 2B; TARm). While replication of the variants with a deletion in either the left- or right-hand side of the TAR stem (A, B, C, D, and F) was below the detection level, the double mutants AB and CD replicated as efficiently as the original HIV-rtTA, and the TAR-deleted E mutant showed delayed replication kinetics. These results demonstrate that whereas destabilization of TAR abolishes viral replication, truncation or complete removal of TAR does not affect or only moderately affects viral replication.
Improved replication of TAR-deleted HIV-rtTA upon viral evolution.
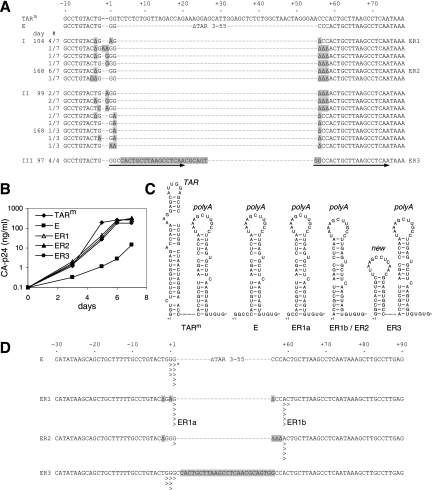
Although the complete removal of the TAR structure does not abolish replication, the E mutant replicates much slower than the original HIV-rtTA (TARm), and we anticipated that this mutant could evolve to a better-replicating variant when cultured for a prolonged period. We therefore started three long-term cultures of the E mutant and passaged the virus onto fresh cells at the peak of infection when massive syncytia were observed. We noticed that the time interval between infection and the appearance of syncytia became shorter upon prolonged culturing, suggesting that the viral replication capacity was improved. Sequence analysis of the proviral genome present in these long-term cultures revealed that the E mutant had acquired multiple nucleotide substitutions, deletions, and insertions at the U3-R boundary in cultures I and II, while part of the R sequence had been duplicated in culture III (Fig. 3A).
FIG. 3.
Evolutionary repair of a hairpin structure at the 5′ end of the viral RNA. (A) The TAR-deleted E virus was cultured for up to 168 days (cultures I and II) or 97 days (culture III). Cellular proviral DNA was isolated at 104 and 168 days (culture I), at 99 and 168 days (culture II), or at 97 days (culture III), and the LTR region was subsequently PCR amplified and cloned into the TA-cloning vector. The nucleotide sequence of the TAR region was determined for three to seven clones for each sample. The −10-to-+78 region (with +1 indicating the transcription initiation site) is shown for the original HIV-rtTA (TARm), the E mutant, and the evolved viruses (with the frequency at which each sequence is observed [#] indicated on the left). Nucleotide substitutions, insertions, and deletions (Δ) are boxed in gray. Arrows indicate the duplicated R sequence observed in culture III. At the right, ER1, ER2, and ER3 indicate the evolved sequences recloned into the HIV-rtTA virus. (B) To assay replication of the original HIV-rtTA (TARm), the E mutant, and the evolved variants (ER1, ER2, and ER3), SupT1 T cells were transfected with the proviral constructs and cultured with 1 μg/ml dox. This experiment was repeated two times with similar results. (C) Secondary structure of the 5′ end of the viral RNA transcripts. (D) Determination of the transcription initiation site. C33A cells were transfected with HIV-rtTA proviral clones carrying the mutant E or the evolved ER1, ER2, or ER3 sequence. After 2 days of culturing with 1 μg/ml dox, intracellular RNA was isolated and decapped, and the 5′-terminal sequence of the viral RNA transcripts was analyzed by 5′ RACE. The cDNA fragments were cloned in the TA-cloning vector, and 5 to 11 clones were sequenced for each sample. The transcription start site (>) observed for each clone is shown on the corresponding proviral U3-R sequence (*; transcription of the E mutant may have started one nucleotide downstream of the indicated position, because the corresponding RNA sample had not been decapped prior to reverse transcription and RT may have copied the cap-G nucleotide into cDNA) (17).
To demonstrate that these mutations improve viral replication, the sequences that were most abundant in culture I at days 104 and 168 were recloned into the HIV-rtTA proviral genome (variants ER1 and ER2, respectively). Similarly, we recloned the duplicated R sequence observed in culture III at day 97 (variant ER3). Upon transfection of these plasmids into C33A cells, similarly high levels of CA-p24 production were observed with the original construct (TARm), the TAR-deleted E mutant, and the evolved ER1, ER2, and ER3 variants (data not shown). Replication of these HIV-rtTA variants was assayed using the SupT1 T-cell line. The ER1, ER2, and ER3 variants replicated much more efficiently than the E mutant and almost as efficiently as the original HIV-rtTA (TARm) (Fig. 3B). These results demonstrate that the mutations selected during evolution at the U3-R boundary of the E mutant significantly improved replication of this virus.
Evolutionary repair of a hairpin structure at the 5′ end of the viral RNA.
Transcription of the proviral genome starts at the U3-R boundary in the 5′ LTR promoter. In both wild-type HIV-1 and HIV-rtTA, the 5′ end of the RNA transcript folds the TAR and poly(A) hairpin structures (7), with the 5′ terminal nucleotides being included in the base-paired TAR stem (Fig. 3C). It has previously been shown that the sequence GG+1GTCT (with +1 indicating the major transcription start site) is an important element for initiation of HIV-1 transcription (28). Deletion of TAR created the GGGCCC sequence at this position in the E variant, which may have affected the transcription process. The multiple mutations selected near the U3-R border do indeed suggest an evolutionary adaptation of the transcription start site. We therefore set out to identify the transcription initiation site for the E mutant and the evolved ER1, ER2, and ER3 variants.
We transfected the proviral clones into C33A cells and analyzed the 5′ terminal sequence of the RNA transcripts by 5′ RACE. As a control, we identified the transcription initiation site for the original HIV-rtTA clone (TARm), which corresponds to the wild-type G+1 start (results not shown). Transcripts of the E mutant were also found to initiate at this G+1 residue or at the adjacent G+2 residue (Fig. 3D). The 5′ end of the E transcript is predicted to fold the poly(A) hairpin structure, but the 5′ terminal nucleotides (GGCC or GCC) remain single stranded (Fig. 3C). Transcription initiation of the ER1 variant was more diffuse. This variant used the original start site (ER1a) and the position three nucleotides further downstream (ER1b), corresponding to nucleotide +59 in the original HIV-rtTA, with similar levels of efficiency (Fig. 3D). The ER2 variant predominantly initiated transcription at this A+59 position. Whereas the ER1a transcript is predicted to fold the poly(A) hairpin with two single-stranded nucleotides at the 5′ end, the ER1b/ER2 transcript was able to fold the nearly complete poly(A) hairpin but effectively removed the 5′ dangling end (Fig. 3C).
The ER3 variant did not change the transcription initiation site (Fig. 3D). Nevertheless, this variant was able to remove the 5′ dangling end by alternative means, as the sequence insert triggers the formation of a novel minihairpin immediately upstream of the poly(A) hairpin (Fig. 3C). These results demonstrate that the nucleotide substitutions, deletions, and insertions at the U3-R boundary observed during the evolution of the E mutant either cause a reallocation of the transcription initiation site (culture I; ER1 and ER2 variants) or introduce a new hairpin structure at the 5′ end of the RNA transcripts (culture III; ER3 variant). Both evolutionary routes effectively result in inclusion of the 5′ terminal nucleotides of the transcripts in a base-paired stem structure.
DISCUSSION
The TAR hairpin, which is present at the 5′ and 3′ ends of the HIV-1 genomic and messenger RNAs, has been extensively studied in recent decades. The best-studied function of TAR is its essential role in the activation of transcription from the 5′ LTR promoter. Multiple studies in which replication of TAR-mutated HIV-1 variants in T-cell lines was analyzed suggested several additional functions of TAR in other processes during viral replication, including translation, dimerization, packaging, and reverse transcription. However, most of these studies are hampered by the fact that mutation of TAR significantly reduced Tat-mediated activation of transcription, which made it difficult to distinguish effects on other replication processes. In this study, we demonstrate that complete deletion of TAR does not abolish replication of an HIV-1 variant that does not require TAR for the activation of transcription. This result demonstrates that TAR has no essential function in HIV-1 replication other than to accommodate Tat-mediated activation of transcription. However, our studies focused on replication in T-cells and we cannot exclude the possibility that TAR may have an accessory function under specific conditions or in specific cell types in vivo.
Our results suggest that efficient HIV-1 replication requires a stable stem-loop structure at the start of the viral transcripts, in which the 5′ terminal nucleotides are base paired. This structure can be the wild-type or truncated TAR hairpin (AB and CD mutants), the poly(A) hairpin (ER1 and ER2 variants), or a new stem-loop structure (ER3 variant), indicating that the nucleotide sequence of this 5′ hairpin is not important. These results are in agreement with a previous forced-evolution study in which HIV-1 replication was significantly reduced by opening of the lower TAR stem and strong evolutionary pressure restored the base pairing of this TAR region (21). Furthermore, structure analysis of multiple HIV-1, HIV-2, and simian immunodeficiency virus isolates revealed that the 5′ end of the RNA is always base paired, despite variations in overall leader and TAR structures (5).
Whereas truncation of TAR in the double-mutated variants (AB and CD) had no effect on virus replication, destabilization of this stem-loop structure in the single-mutated variants (A, B, C, D, and F) blocked replication. The untranslated RNA leader region can fold either an extended duplex through long-distance base pairing (long-distance interaction) or a branched conformation in which the RNA locally folds into hairpin structures (branched multiple hairpin) (18). Although both conformations have the TAR hairpin, the unpaired nucleotides in the destabilized structure may interact with other regions of the leader and alter the long-distance interaction-branched multiple hairpin equilibrium and thus indirectly affect viral replication (18, 23). Similarly, the unpaired nucleotides present at the 5′ end of the transcripts of the E mutant may affect the leader structure and this may explain the reduced replication capacity of this variant. Alternatively, the presence of a stable stem-loop structure at the 5′ end of the transcripts may be important for RNA longevity, as previously described for bacterial and organelle mRNAs (12, 15, 31, 36).
The TAR DNA sequence in the proviral 5′ LTR promoter region has been shown to bind various cellular transcription factors (reviewed in reference 25). We observed efficient replication of the evolved E variants in which TAR was replaced by nonrelated sequences or completely removed. Thus, although the binding of transcription factors to TAR DNA may be important for Tat-controlled transcription, these interactions are apparently not essential for virus replication in T cells when transcription is controlled by rtTA.
During reverse transcription the sequence complementarity between the 5′ and 3′ R regions facilitates the first-strand transfer in which a cDNA copy of the 5′ R-U5 region (strong-stop minus-strand DNA) is translocated to the 3′ end of the viral RNA genome. Our results demonstrate that for this function the R region can be significantly shorter than the 97-nt wild-type element. The shortest R region, only 39 nt, is observed in the efficiently replicating ER2 variant. These results are in agreement with a previous study that demonstrated that strand transfer can be efficient with a minimal R-overlap region of approximately 30 nt (9).
The dox-dependent HIV-rtTA variant was proposed as part of a novel strategy for development of a safe live attenuated HIV vaccine. In this study, we used HIV-rtTA to clarify the role of TAR in virus replication; the results demonstrated that this variant can also be a powerful tool to study HIV-1 biology.
Acknowledgments
We thank Stephan Heynen for CA-p24 ELISA.
This research was sponsored by the Dutch AIDS Foundation (Aids Fonds grant 2005022), Technology Foundation STW (applied science division of NWO and the technology program of the Ministry of Economic Affairs, Utrecht, The Netherlands), ZON-Medical Sciences (MW; VICI grant), and NWO-Chemical Sciences (CW; TOP grant).
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Andersen, E. S., S. A. Contera, B. Knudsen, C. K. Damgaard, F. Besenbacher, and J. Kjems. 2004. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 279:22243-22249. [DOI] [PubMed] [Google Scholar]
- 2.Bannwarth, S., and A. Gatignol. 2005. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 3:61-71. [DOI] [PubMed] [Google Scholar]
- 3.Bennasser, Y., S. Y. Le, M. L. Yeung, and K. T. Jeang. 2004. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 1:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennasser, Y., M. L. Yeung, and K. T. Jeang. 2006. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 281:27674-27678. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 20:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout, B. 2000. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 48:29-73. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout, B., B. Klaver, and A. T. Das. 1995. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology 207:276-281. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout, B., J. van Wamel, and B. Klaver. 1995. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J. Mol. Biol. 252:59-69. [DOI] [PubMed] [Google Scholar]
- 10.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96:7791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady, J., and F. Kashanchi. 2005. Tat gets the “green” light on transcription initiation. Retrovirology 2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bricker, A. L., and J. G. Belasco. 1999. Importance of a 5′ stem-loop for longevity of papA mRNA in Escherichia coli. J. Bacteriol. 181:3587-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, A. T., K. Verhoef, and B. Berkhout. 2004. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 388:359-379. [DOI] [PubMed] [Google Scholar]
- 14.Das, A. T., X. Zhou, M. Vink, B. Klaver, K. Verhoef, G. Marzio, and B. Berkhout. 2004. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279:18776-18782. [DOI] [PubMed] [Google Scholar]
- 15.Hambraeus, G., K. Karhumaa, and B. Rutberg. 2002. A 5′ stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology 148:1795-1803. [DOI] [PubMed] [Google Scholar]
- 16.Harrich, D., C. W. Hooker, and E. Parry. 2000. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 74:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirzmann, J., D. Luo, J. Hahnen, and G. Hobom. 1993. Determination of messenger RNA 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res. 21:3597-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huthoff, H., and B. Berkhout. 2001. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 29:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeeninga, R. E., B. Jan, H. Van den Berg, and B. Berkhout. 2006. Construction of doxycyline-dependent mini-HIV-1 variants for the development of a virotherapy against leukemias. Retrovirology 3:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaver, B., and B. Berkhout. 1994. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 13:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzio, G., K. Verhoef, M. Vink, and B. Berkhout. 2001. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA 98:6342-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooms, M., H. Huthoff, R. Russell, C. Liang, and B. Berkhout. 2004. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 25.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raha, T., S. W. Cheng, and M. R. Green. 2005. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter, S., Y. H. Ping, and T. M. Rana. 2002. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rittner, K., M. J. Churcher, M. J. Gait, and J. Karn. 1995. The human immunodeficiency virus long terminal repeat includes a specialised initiator element which is required for Tat-responsive transcription. J. Mol. Biol. 248:562-580. [DOI] [PubMed] [Google Scholar]
- 29.Ruijter, J. M., H. H. Thygesen, O. J. Schoneveld, A. T. Das, B. Berkhout, and W. H. Lamers. 2006. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, S. M., M. Khoroshev, P. A. Marx, J. Orenstein, and K. T. Jeang. 2001. Constitutively dead, conditionally live HIV-1 genomes. Ex vivo implications for a live virus vaccine. J. Biol. Chem. 276:32184-32190. [DOI] [PubMed] [Google Scholar]
- 31.Suay, L., M. L. Salvador, E. Abesha, and U. Klein. 2005. Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 33:4754-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, R., A. Brodsky, J. R. Williamson, and A. D. Frankel. 1997. RNA recognition by HIV-1 Tat and Rev. Semin. Virol. 8:186-193. [Google Scholar]
- 33.Urlinger, S., U. Baron, M. Thellmann, M. T. Hasan, H. Bujard, and W. Hillen. 2000. Exploring the sequence space for tetracycline dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA 97:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhoef, K., G. Marzio, W. Hillen, H. Bujard, and B. Berkhout. 2001. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 75:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fisher, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 36.Zou, Z., C. Eibl, and H. U. Koop. 2003. The stem-loop region of the tobacco psbA 5′UTR is an important determinant of mRNA stability and translation efficiency. Mol. Genet. Genomics 269:340-349. [DOI] [PubMed] [Google Scholar]