Abstract
The human MxA gene belongs to the class of interferon (IFN)-stimulated genes (ISGs) involved in antiviral resistance against influenza viruses. Here, we studied the requirements for MxA induction by influenza A virus infection. MxA is transcriptionally upregulated by type I (alpha and beta) and type III (lambda) IFNs. Therefore, MxA is widely used in gene expression studies as a reliable marker for IFN bioactivity. It is not known, however, whether viruses can directly activate MxA expression in the absence of secreted IFN. By using an NS1-deficient influenza A virus and human cells with defects in IFN production or the STAT1 gene, we studied the induction profile of MxA by real-time reverse transcriptase PCR. The NS1-deficient virus is known to be a strong activator of the IFN system because NS1 acts as a viral IFN-antagonistic protein. Nevertheless, MxA gene expression was not inducible by this virus upon infection of IFN nonproducer cells and STAT1-null cells. Likewise, neither IFN-α nor IFN-λ had a sizeable effect on the STAT1-null cells, indicating that MxA expression requires STAT1 signaling and cannot be triggered directly by virus infection. In contrast, the expression of the IFN-stimulated gene ISG56 was induced by influenza virus in these cells, confirming that ISG56 differs from MxA in being directly inducible by viral triggers in an IFN-independent way. In summary, our study reveals that MxA is a unique marker for the detection of type I and type III IFN activity during virus infections and IFN therapy.
The expression of interferon (IFN)-stimulated genes (ISGs) is widely used as a surrogate marker for type I IFN activity in various experimental and clinical settings (5, 9, 13, 19, 18, 60, 72). However, in many instances, the expression of ISGs results from a direct response to virus infection, in addition to induction by IFNs (2, 15, 23). Therefore, often it is not clear whether the observed expression of ISGs is indeed induced by IFN or rather by other pathways, such as Toll-like receptor signaling or direct activation by specific virus components (14, 38, 81). Type I and type III IFNs are induced by viral infection and act through the JAK/STAT pathway (17, 74). The activation of receptor-associated JAK kinases induces the assembly of heterotrimeric ISG factor 3 (ISGF3), which consists of STAT1, STAT2, and IFN regulatory factor 9 (IRF9) (17). ISGF3 binds to IFN-stimulated response elements (ISREs) present in the promoter region of ISGs and stimulates the expression of the respective genes. Knockout mice lacking STAT1 (22, 52, 55) or STAT1-deficient humans (21) cannot respond to type I and III IFNs and do not upregulate genes that are strictly IFN dependent. Nevertheless, some ISGs are induced by virus infection or by other appropriate stimuli, even in these nonresponsive cells (6). For example, ISG56 can be directly induced by double-stranded RNA (dsRNA) or virus infection (15, 28, 64). This IFN-independent induction occurs via an alternative pathway involving the crucial transcription factor IRF3 (27, 57). IRF3 has been shown to bind to the ISRE site of the ISG56 promoter (78). In contrast, other genes, such as Mx, show at best a poor primary response to viral infection (8) but are highly responsive to type I and III IFNs (40, 48, 67).
Mx proteins belong to the superfamily of dynamin-like large GTPases (33). They were originally identified as factors conferring resistance to lethal influenza A virus infections in mice (32, 35, 44, 69). The human Mx homologue MxA is able to inhibit the replication of orthomyxoviruses and other RNA viruses (33). The exclusive induction of murine Mx1 by type I IFN was demonstrated by the prevention of Mx1 resistance in virus-infected mice treated with IFN-neutralizing antibodies (31). However, the situation of human MxA induction is less clear, although MxA gene expression is used as a marker for the detection of biologically active type I IFN generated during viral infections (26, 60, 75) or administered during IFN therapy (3, 4, 12, 39).
Transcriptional profiles by cDNA analysis are increasingly used in biomedical research to better characterize human infectious diseases or to evaluate therapeutic regimens. In this context, reliable markers of IFN activity are needed. Here, we used influenza A virus and human cells defective in IFN synthesis or IFN signaling to analyze MxA expression. We found that in sharp contrast to ISG56, MxA is induced selectively by type I IFNs but not directly by virus infection. We further demonstrate that MxA expression depends on STAT1 and provide evidence that the weak MxA activation induced by virus infection in type I IFN-defective cell systems is most likely due to the action of type III IFNs.
(This work was conducted by D. Holzinger in partial fulfillment of the requirements for an M.D. degree from the Medical Faculty of the University of Freiburg, Freiburg, Germany.)
MATERIALS AND METHODS
Cells, viruses, IFNs, and antibodies.
A549 human lung adenocarcinoma cells and GRE human glioblastoma cells (kindly provided by Ganes C. Sen, Cleveland Clinical Foundation, Cleveland, OH) (6, 51) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS). Epstein-Barr virus-transformed B cells from two individuals with different mutations in the STAT1 gene and a cell line from a healthy individual (21) were cultivated in RPMI medium with 20% FCS.
Human peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-treated blood from healthy blood donors by centrifugation through a Ficoll gradient (Amersham Pharmacia, Freiburg, Germany) as described previously (10) and cultivated in RPMI medium with 10% FCS. A total of 2 × 106 cells were cultivated in 1 ml RPMI medium and stimulated with different concentrations of IFN for 6 and 16 h.
Influenza A virus (FLUAV) infections were performed with influenza A/PR/8/34 virus (FLUAVwt) and the recombinant influenza A/PR/8/34 virus with a deletion in the NS1 gene (FLUAVdNS1) (kindly provided by Adolpho Garcia-Sastre, Mount Sinai Hospital, New York, NY) (25). FLUAV stocks were diluted in phosphate-buffered saline supplemented with 0.3% bovine serum albumin. The cells were washed with phosphate-buffered saline and incubated with 1 or 5 PFU/cell of the appropriate virus stock for 1 h. Then the cells were further incubated in the normal growth medium until harvest.
Human IFN-α2a (Roche, Mannheim, Germany) (3 × 107 U/ml) was used for induction experiments; human IFN-γ (106 U/ml) was a gift from Peter Staeheli, Freiburg, Germany; and human IFN-λ1 (10 μg/ml) was kindly provided by Sergei V. Kotenko, UMDNJ, Newark, NJ (40).
A mouse monoclonal antibody (M143) directed against human MxA (24), a polyclonal rabbit antibody directed against p56 (kindly provided by Ganes C. Sen) (28), a polyclonal rabbit antibody directed against the nucleoprotein (NP) of FLUAV (29), and a monoclonal β-tubulin antibody (Sigma, Munich, Germany) were used for the immunodetection of the proteins by Western blotting.
Plasmid construction.
pGL3-MxAP-FF-Luc carrying the firefly luciferase (FF-Luc) gene under the control of the human MxA promoter was constructed by inserting a 1,800-nucleotide (nt) NdeI/HindIII fragment of the MxA promoter, pMxA-CAT (kindly provided by Ilkka Julkunen, Department of Microbiology, NPHI, Helsinki, Finland) (61), into pGL3 basic (Promega, Mannheim, Germany). A reporter plasmid carrying the FF-Luc gene under the control of the ISG54 promoter (pISG54-Luc) was kindly provided by David Levy, New York University, New York, NY (56).
The expression construct for a constitutively activated, hemagglutinin (HA)-tagged, human IRF3(5D) (43) in pCAGGS was kindly provided by Luis Martinez-Sobrido, Department of Microbiology, Mount Sinai School of Medicine, New York, NY (7). The expression construct for the dominant negative, HA-tagged, human IRF3(58-427) was kindly provided by Takashi Fujita, Department of Tumor Cell Biology, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan (82).
18S rRNA (accession no. X03205) in pGEM, hypoxanthine phosphoribosyltransferase (HPRT) (accession no. M31642) in pBSK, and IP-10 (accession no. NM_001565) in pBSK were amplified from a cDNA reverse transcribed using RNA isolated from human PBMCs. For the construction of pGEM-18SrRNA, the primers 18SrRNA(+) (5′-ACT GTG GTA ATT CTA GAG CTA) and 18SrRNA(−) (5′-GGC ACT TAC TGG GAA TTC CTC) were used. The 1,450-nt-long PCR product was cloned into the EcoRI and XbaI sites of pGEM-4 (Promega). For the construction of pBSK-HPRT, the primers HPRT(+) (5′-GAC AGA ATT CCC TCC TGA GCA GTC AGC C) and HPRT(−) (5′-GAC AGG ATC CAC TGC TGA CAA AGA TTC ACT GG) were used. The 1,250-nt-long PCR product was cloned into the EcoRI and BamHI sites of pBSK (Stratagene, Heidelberg, Germany). For the construction of pBSK-IP-10, the primers IP-10(+) (5′-GAC AAA GCT TCT CCA GTC TCA GCA CCA TGA) and IP-10(−) (5′-GAC AGG ATC CTT GAA AAC CAT TCA GCA CAT TT) were used. The 1,100-nt-long PCR product was cloned into the HindIII and BamHI sites of pBSK.
In vitro transcription of synthetic RNA standards.
For quantification by LightCycler reverse transcriptase PCR (RT-PCR), RNA standards were produced by runoff transcription. Transcription was performed from the T7 promoter by using plasmids pBS-T7-MxA (NM_002462) (79), pBSK-p56 (NM_001548; kindly provided by Ganes C. Sen) (28), pBSK-IP-10, pBSK-HPRT, and pGEM-18SrRNA. For T7 transcription, plasmids were linearized, the RNAs were transcribed by T7 RNA polymerase, and the template DNAs were digested using RNase-free DNase (Ambion, Austin, TX). The efficiency of the DNase digestion improved markedly when, prior to adding the DNase, the reaction mixture was incubated at 95°C for 1 min and then cooled in ice water for 1 min to melt down DNA-RNA hybrids. After two rounds of DNase treatment, the RNA was purified using TRIzol (Invitrogen, Karlsruhe, Germany) extraction. The quantification of RNA was performed using the fluorescent dye RiboGreen (Molecular Probes, OR), which specifically binds single-stranded RNA. Briefly, RNA samples and standard RNA in the range of 20 to 1,000 ng/ml were mixed with RiboGreen in Tris-EDTA buffer. Fluorescent emission of samples and standards at 525 nm were measured using the plate read mode of the SDS1.6.3 software of ABI-PRISM 7700 (Perkin Elmer, Weiterstadt, Germany), and the quantities of the sample RNAs were calculated. Copy numbers of the RNA transcripts were calculated from their molecular weights, and RNA standards ranging from 1010 to 101 molecules were diluted in 100 μg of tRNA/ml (Sigma, Munich, Germany).
RNA extraction.
Total cellular RNA was isolated from 2 × 106 cells by use of an RNeasy Mini RNA isolation kit (QIAGEN, Hilden, Germany). Total RNA was eluted from the RNeasy Mini columns with 50 μl of RNase-free water.
Conventional RT-PCR.
For reverse transcription, 1 μg of RNA was transcribed using SuperScript II RT (Invitrogen, Karlsruhe, Germany) and random hexamer primers (Roche) in a volume of 10 μl at 42°C for 1 h. After digestion with RNase A, 3 μl of the RT reaction mixture was used for 50-μl PCRs to amplify the IFN-λ1 (GenBank accession no. NM_172140), IFN-β (GenBank accession no. NM_002176), FLUAV-NP (GenBank accession no. M38279), and γ-actin (GenBank accession no. NM_001614) cDNAs using the specific primer pairs indicated in Table 1 and Taq DNA polymerase (Roche). Blank aliquots of reaction mixtures without reverse transcriptase were used as internal controls. IFN-λ cDNA fragments were amplified by 35 cycles (95°C, 64°C, and 72°C for 30, 60, and 30 s per step, respectively). After amplification, 10-μl aliquots of the PCR products were electrophoretically separated in a 1% agarose gel and stained with ethidium bromide. The bands were visualized under UV light.
TABLE 1.
Nucleotide sequences of the PCR primers and probes used for LightCycler real-time qRT-PCR and conventional PCRs
| Primer or probe | Nucleotide sequence |
|---|---|
| MxA forward primer | 5′-TTCAGCACCTGATGGCCTATC |
| MxA reverse primer | 5′-TGGATGATCAAAGGGATGTGG |
| MxA probe | 5′-FAM-CAGGAGGCCAGCAAGCGCCATC |
| ISG56 forward primer | 5′-CCTGGAGTACTATGAGCGGGC |
| ISG56 reverse primer | 5′-TGGGTGCCTAAGGACCTTGTC |
| ISG56 probe | 5′-FAM-ACAGAGTTCTCAAAGTCAGCAGCCAGTCTCAGT |
| 18S rRNA forward primer | 5′-GCTGGAATTACCGCGGCT |
| 18S rRNA reverse primer | 5′-CGGCTACCACATCCAAGGAA |
| 18S rRNA probe | 5′-FAM-TGCTGGCACCAGACTTGCCCTC |
| HPRT forward primer | 5′-CGGCTCCGTTATGGCG |
| HPRT reverse primer | 5′-GGTCATAACCTGGTTCATCATCAC |
| HPRT probe | 5′-FAM-CGCAGCCCTGGCGTCGTGT |
| IP-10 forward primer | 5′-GCSATTCTGATTTGCTGCCT |
| IP-10 reverse primer | 5′-GATGCAGGTACAGCGTACRGT |
| IP-10 probe | 5′-FAM-CTGACTCTAAGTGGCATTCAAGGAGTACCTCTCTC |
| IFN-λ forward primer | 5′-TGAGCTGGCCCTGACGCTGAA |
| IFN-λ reverse primer | 5′-AGGCGGAGGTTGAAGGTGA |
| IFN-β forward primer | 5′-GACGCCGCATTGACCATCTA |
| IFN-β reverse primer | 5′-CCTTAGGATTTCCACTCTGACT |
| FLUAV-NP forward primer | 5′-GCAGGTGCTGCAGTCAAAGGA |
| FLUAV-NP reverse primer | 5′-AAAGCTTCCCTCTTGGG |
| γ-Actin forward primer | 5′-GGCGGTCGCAATGGAAGAAGA |
| γ-Actin reverse primer | 5′-CATGGCCGGGGTGTTGAAGGTC |
RT-PCR amplicon design.
Primers were designed for an annealing melting temperature of 60°C using the PCR document window of the Primer Express software (Applied Biosystems), which operates using the algorithm developed as described previously (63). The melting temperature of the 5′ 6-carboxyfluorescein (FAM)-tagged and 3′ carboxytetramethylrhodamine-tagged probes was 70°C. Specific primers were designed in reference to sequences with the following accession numbers: for MxA, NM_002462; for ISG56, NM_001548; for 18S rRNA, X03205; for IP-10, NM_001565; and for HPRT, M31642. Primers and probes used for LightCycler real-time quantitative RT-PCR (qRT-PCR) are listed in Table 1. The primers were purchased at TIB-MolBiol, Berlin, Germany.
LightCycler real-time RT-PCR.
Five different real-time RT-PCR assays, based on TaqMan chemistry to detect MxA, ISG56, IP-10, HPRT, and 18S rRNA expression, were developed. All qRT-PCR assays were performed in the same format and were run on a LightCycler (Roche, Germany) with identical cycle conditions: RT at 61°C for 20 min, activation at 95°C for 5 min, and 45 cycles of PCR at 95°C for 15 s and 60°C for 60 s. We used the RNA master hybridization probe kit (Roche) with 500-nM concentrations of primers, 200-nM concentrations of probes, and 3.25 mM manganese acetate. The kit includes an aptamer-blocked Tth DNA polymerase which performs both RT and hot-start PCR amplification. One microliter of the synthetic RNA standards or of RNA prepared from cells was added to 19 μl of LightCycler RNA master hybridization probe reaction mix (Roche). DNA contamination was controlled in the same run by using a PCR Fast Start DNA master kit (Roche). One microliter of the RNA prepared from cells was added to 19 μl Fast Start DNA master hybridization probe reaction mix (Roche) supplemented with 5 mM MgCl2, 500-nM concentrations of primers, and 200-nM concentrations of probes. The cycle conditions were identical to those for the RT-PCR assay. Standard curves were obtained using dilutions of the synthetic RNA preparations and used to determine the concentration of the target transcripts. Results were expressed as the ratio of the target sequence concentration relative to 18S rRNA or HPRT transcripts. The results are presented as mean values with standard deviations calculated from at least three experiments.
Reporter gene assays.
Transient transfection of GRE cells was performed by using 2 μl Dac-30 (Eurogentec, Seraing, Belgium) per μg DNA in 200 μl OptiMEM (Gibco-BRL) as described previously (30). Transient transfection of A549 cells was performed by using 3 μl FuGENE 6 (Roche) per μg DNA in 100 μl OptiMEM according to the manufacturer's protocol. Cells were transfected with 1.0 μg of FF-Luc reporter plasmid under the control of the different IFN-responsive promoters, together with 0.1 μg of a Renilla luciferase (REN-Luc) plasmid under the control of the constitutive simian virus 40 (SV40) promoter (pRL-SV40, Promega). For the IRF induction experiments, cells were additionally transfected with 0.5 μg of pCAGGS expression constructs encoding HA-tagged IRF3(5D) and IRF3(58-427). At 6 h posttransfection, cells were treated with recombinant IFN-α2a, infected with 1 PFU per cell of FLUAV, or left untreated. At 24 h posttransfection, cells were harvested and lysed in 200 μl of passive lysis buffer (Promega). An aliquot of 20 μl was used to measure FF-Luc and REN-Luc activities as described previously by the manufacturer (dual-luciferase reporter assay system; Promega). FF-Luc activities were normalized to REN-Luc activities.
Oligonucleotide binding assay.
Binding of IRF3 to elements of the ISG56 and MxA promoter was analyzed as previously described (49). A549 cells were infected with 1 multiplicity of infection of FLUAVdNS1 for 5 h and lysed afterwards. Cleared nuclear extracts were incubated with streptavidin-agarose beads (Neutravidin; Pierce) coupled to 5′-biotinylated oligonucleotides. The oligonucleotides used were the NF-κB elements of the IFN-β promoter (49), 5′-biotin (bio)-GGATCCGGAATTTCCCGGAATTTCCC, and the reverse strain, 5′-GGGAAATTCCGGGAAATTCC; the IRE sites of ISG56 (76), 5′-bio-GGATCCTTACAAAAGGGAAAGTGAAACTAGAA and 5′-TTCTAGTTTCACTTTCCCTTTTGTAA; ISRE1 of MxA (61), 5′-bio-GGATCCGCGAAGAAATGAAACTCAC and 5′-GTGAGTTTCATTTCTTCGC; and ISRE2 of MxA (61), 5′-bio-GGATCCGGGAGCAGAAACGAAACCTAGC and 5′-GTGAGTTTCATTTCTTCGC. Binding was performed for 2 h at 4°C, and then the beads were washed and oligonucleotide-bound proteins were detected by Western blotting using rabbit antibodies directed against IRF3 (Santa Cruz; catalog no. FL425) and against the p50 subunit of NF-κB (Santa Cruz; catalog no. H119).
Western blot analysis.
Cells were lysed in 50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.25% Na-deoxycholate, 1 mM dithiothreitol, complete protease inhibitor mix (Roche), 1 μg/ml Benzonase (Novagen, Merck, Bad Soden, Germany). The proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred on a polyvinylidene fluoride membrane (Millipore, Schwalbach, Germany). The Western blot was probed with the monoclonal mouse antibody M143 directed against MxA, a polyclonal rabbit antibody directed against p56, a polyclonal rabbit antibody directed against FLUAV nucleoprotein, and a mouse monoclonal antibody against β-tubulin (Sigma). Horseradish peroxidase-labeled secondary antibodies and the ECL detection system (Amersham) were used to detect the proteins.
RESULTS
MxA expression is tightly controlled by IFN-α.
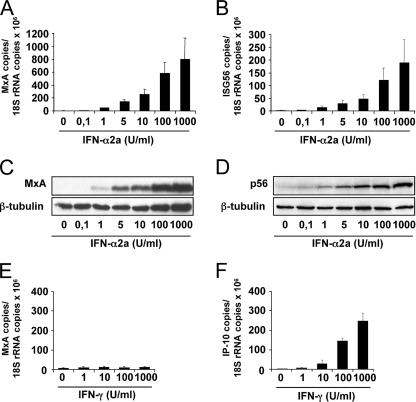
To compare the expression of MxA with that of ISG56 with regard to inducibility by IFN-α, we established a real-time RT-PCR (qRT-PCR) protocol. PBMCs from healthy donors were treated with increasing amounts of human IFN-α2a for 6 h. Total RNA was extracted, and the amounts of MxA and ISG56 transcripts were determined using LightCycler qRT-PCR technology. Ribosomal 18S rRNA was quantified in parallel and used to normalize the expression levels of the two induced genes. As expected, MxA and ISG56 expression were barely detectable in the absence of IFN but were stimulated by low concentrations of IFN-α2a (Fig. 1A and B). Gene expression increased with increasing amounts of IFN. Likewise, the respective gene products accumulated in a dose-dependent manner, as demonstrated by Western blot analysis (Fig. 1C and D). A weak but significant induction of MxA by IFN-γ has previously been reported (1, 61, 75). To verify these findings, PBMCs were treated with increasing amounts of IFN-γ for 6 h and were then analyzed for the presence of MxA and IP-10 transcripts by qRT-PCR. IP-10 is an IFN-γ-regulated chemokine (37, 45) and served as a positive control. Clearly, IFN-γ treatment strongly induced IP-10 expression in PBMCs but failed to stimulate MxA expression to any sizeable degree (Fig. 1E and F). The difference in MxA-inducing capacity between IFN-α and IFN-γ was striking (compare Fig. 1A and E). The present findings indicate that the expression of MxA is strongly induced by type I IFN but negligibly, if at all, by IFN-γ. We therefore conclude that MxA expression is a marker predominantly for type I IFN action.
FIG. 1.
Induction of MxA expression by IFN-α and IFN-γ. PBMCs from healthy donors (2 × 106 cells) were treated with increasing concentrations of IFN-α2a (A to D) or IFN-γ (E and F). After 6 h, the amounts of MxA (A and E), ISG56 (B), and IP-10 (F) transcripts were determined by qRT-PCR and normalized to the amount of 18S rRNA transcripts. After 16 h, MxA protein (C) and p56 protein (D) were analyzed by Western blotting using specific antibodies. β-Tubulin was used as an internal standard. The error bars represent standard deviations calculated from three experiments.
MxA gene expression is not directly induced by virus but requires IFN production.
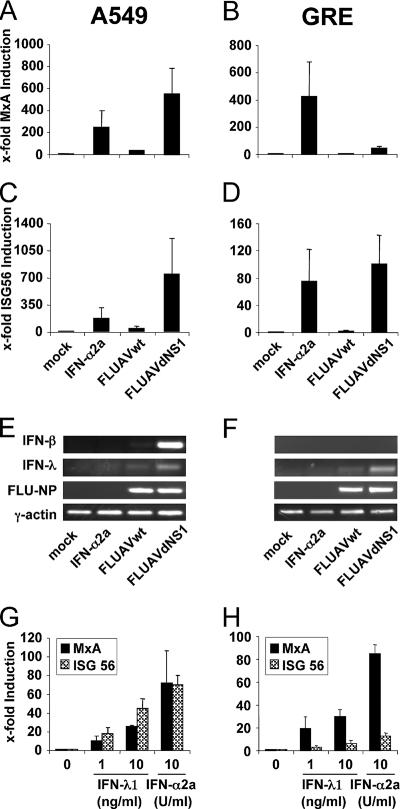
Several ISGs, such as ISG56, can be stimulated directly by virus infection in the absence of IFN action (6, 15, 28). We asked whether virus infection could also directly stimulate MxA expression in the absence of type I IFN. To distinguish between direct (virus-mediated) and indirect (IFN-mediated) induction pathways, we compared the induction of MxA gene expression in A549 human lung adenocarcinoma cells with that in the human glioblastoma GRE cell line. GRE cells lack the region of chromosome 9 that encodes the IFN-α and IFN-β genes and are therefore unable to synthesize type I IFNs (51). ISG56 expression served as a positive control. FLUAV was used as a viral inducer. FLUAVdNS1, a recombinant mutant virus of FLUAV that is defective in the expression of the viral IFN-antagonistic NS1 protein, is known to be a particularly strong activator of type I IFN genes and ISG expression (25). The cells were infected with wild-type FLUAV or mutant FLUAVdNS1 and analyzed for MxA and ISG56 transcripts 16 h later. Control cells were treated with IFN-α2a or were left untreated. FLUAVdNS1 was an excellent inducer of MxA transcription in IFN-competent A549 cells but not in IFN-defective GRE cells (Fig. 2A and B). In contrast, the same virus was a good inducer of ISG56 transcription in both cell lines. FLUAVdNS1 was able to establish infection in A549 and GRE cells, as demonstrated by equal accumulation of newly synthesized viral nucleoprotein transcripts in both cell lines by conventional RT-PCR (Fig. 2E and F). A549 cells responded with IFN-β transcription, while GRE cells were unable to do so because of their genetic defect (Fig. 2E and F). In summary, ISG56 expression was greatly induced in the GRE cell line in the absence of IFN gene expression, whereas MxA gene expression was not induced to substantial levels. IFN-α treatment, however, induced MxA expression in GRE cells to the same degree as that in A549 cells (Fig. 2A and B). These results show that MxA gene expression requires the synthesis of virally induced IFNs, but in contrast to ISG56, MxA is not directly triggered by viral replication.
FIG. 2.
MxA is induced by type I or type III IFNs in influenza A virus-infected cells. A549 (A, C, E, and G) and GRE (B, D, F, and H) cells were infected with 1 PFU per cell of either FLUAVwt or FLUAVdNS1. Parallel cultures were treated with 1,000 U/ml of IFN-α2a or were left untreated (mock). To monitor induction by IFN-λ (G and H), cells were incubated with IFN-λ1 (1 ng/ml or 10 ng/ml) or IFN-α2a (10 U/ml). After 16 h, the amounts of MxA (A, B, G, and H) and ISG56 (C, D, G, and H) were determined by qRT-PCR and normalized to the amount of HPRT transcripts. The induction in untreated cells was defined as onefold. Error bars represent standard deviations calculated from three experiments. The induction of IFN-β and IFN-λ (E and F) was monitored by conventional RT-PCR using specific primers. Transcripts of FLUAV-NP and γ-actin were analyzed to monitor virus infection and expression of a housekeeping gene, respectively (E and F).
Interestingly, infection with wild-type FLUAV had only a weak stimulatory effect (Fig. 2A to D), presumably due to the antagonistic activity of the viral NS1 protein. However, infection of A549 cells with FLUAVdNS1 led to a much stronger expression of MxA and ISG56 than did treatment with 1,000 U/ml of IFN-α (Fig. 2A and C). This indicates the high-inducing capacity of FLUAVdNS1, which is almost completely blocked by the expression of the viral NS1 protein in wild-type-infected cells.
The weak approximately 40-fold induction of MxA gene expression observed in FLUAVdNS1-infected GRE cells (Fig. 2B) must have been due to additional virus-induced activators of MxA expression. Virus-induced type III IFNs (also called IFN-λ) are known activators of MxA expression (40, 66). An analysis of the FLUAVdNS1-infected cells by RT-PCR showed an induction of IFN-λ in both A549 and GRE cells, whereas the expression of IFN-β was not detectable in GRE cells (Fig. 2E and F). To confirm that IFN-λ is indeed able to induce MxA expression in our cell culture systems, A549 and GRE cells were treated with recombinant IFN-λ1, in parallel with IFN-α2a as a positive control. Both types of IFN were able to induce MxA as well as ISG56 expression (Fig. 2G and H), leading to the assumption that IFN-λ is responsible for the weak induction of MxA detected in the FLUAVdNS1-infected GRE cells (Fig. 2B).
Lack of MxA promoter activation by virus infection.
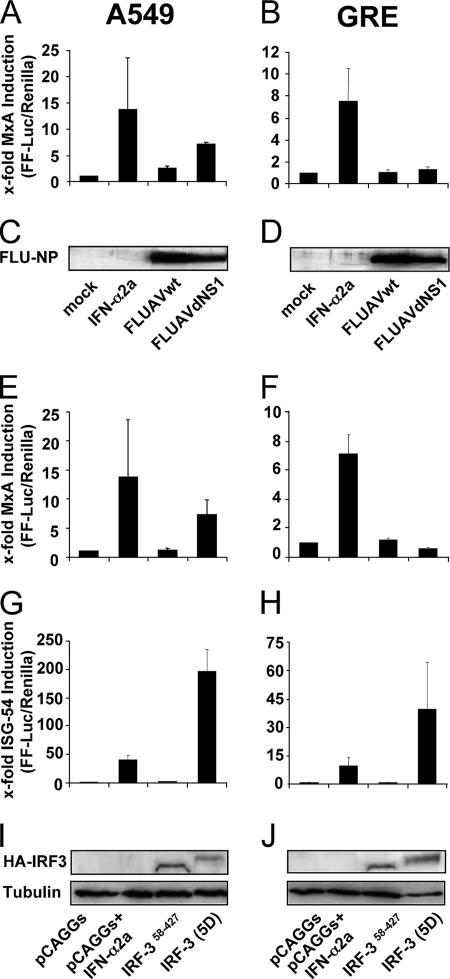
To further characterize the induction of MxA gene expression in virus-infected cells, we generated a reporter plasmid encoding FF-Luc under the control of the MxA promoter. A549 and GRE cells were transfected with the reporter construct, together with a plasmid encoding Renilla luciferase under the control of the constitutive SV40 promoter that was used as an internal transfection control. Treatment of the transfected cells with IFN-α showed that the activity of FF-Luc determined for the cell lysates correlates with the activation of the MxA promoter (Fig. 3A and B). The infection of A549 cells with FLUAVdNS1, but not with wild-type FLUAV, led to a significant activation of the MxA promoter (Fig. 3A). However, in GRE cells, the promoter was not activated by virus infection (Fig. 3B). Comparable virus replication in the cells was monitored by Western blot analysis of the viral nucleoprotein (Fig. 3C and D).
FIG. 3.
MxA promoter activation in response to virus infection occurs via type I IFN. A549 and GRE cells were transfected with pMxAP-FF-Luc (A, B, E, and F) or pISG54-FF-Luc (G and H) reporter plasmids. As an internal control, the cells were cotransfected with the SV40-REN-Luc reporter plasmid. (A to D) Cells were infected 6 h later with 1 PFU per cell of FLUAVwt or FLUAVdNS1. Parallel cultures were treated with 1,000 U/ml IFN-α2a or were left untreated (mock). (E to J) Cells were cotransfected with pCAGGS or pCAGGS expression constructs coding for IRF3(58-427) or IRF3(5D). At 24 h postinfection (A to D) or posttransfection (E to J), cells were lysed and FF-Luc and REN-Luc activities were measured. The FF-Luc activities were normalized to that of REN-Luc. The induction in untreated cells was defined as onefold. Bars represent standard deviations calculated from three experiments. The expression of viral NP (C and D) or HA-tagged IRF3 (I and J) was monitored by Western blotting using NP- or HA-specific antibodies, respectively. β-Tubulin (I and J) was used as a loading control.
The direct induction of ISG56 in virus-infected cells is mediated via the activation of cellular IRF3, a transcription factor that is also critical for the virus-induced expression of IFN (57, 65). IRF3 is activated through the phosphorylation of serine and threonine residues in its C-terminal part (43, 82). Accordingly, we used a cDNA construct encoding constitutively activated IRF3. IRF3(5D) is active because of the conversion of five critical phosphorylation sites into phosphomimetic aspartic acid residues in its C-terminal autoinhibitory domain (43). As a negative control, we used a plasmid encoding an N-terminally truncated IRF3, IRF3(58-427), which lacks a DNA-binding domain and is therefore inactive (82). To study a direct stimulation of the MxA promoter by IRF3, cells were cotransfected with the reporter plasmids under the control of the MxA promoter and with the expression plasmids for IRF3 or the empty vector (pCAGGS). We used a reporter construct containing the promoter of ISG54, which is induced in a fashion similar to that of ISG56, as a control to monitor activation by IRF3 (56, 76). After 24 h, the accumulation of the reporter activity was determined in the cell lysates. The treatment of the cells with IFN-α2a was used as a positive control and showed the activation of the respective promoter constructs (Fig. 3E to H). Interestingly, the MxA promoter was activated by IRF3(5D) in A549 cells, but its induction was completely absent in GRE cells (Fig. 3E and F). In contrast and as expected, the ISG54 promoter was strongly activated by IRF3(5D) in both cell lines, whereas IRF3(58-427) was completely inactive (Fig. 3G and H). Western blot analysis confirmed the expression of the recombinant IRF3s in the transfected cells (Fig. 3I and J). These data indicate that in contrast to that of ISG54, the promoter of human MxA is not directly activated by virus infection or by virus-induced activation of IRF3 but requires the preceding induction of IFN.
IRF3 binding to the IFN-stimulated elements of the MxA promoter.
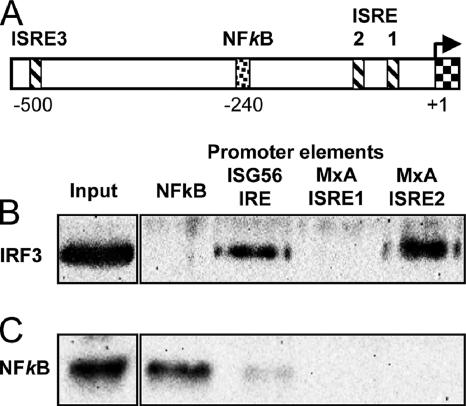
To further analyze the response of the MxA promoter compared to that of the ISG56 promoter, we analyzed putative regulatory elements of the MxA promoter. There are two functional ISREs near the transcriptional start site, one more distal ISRE-like element and a putative NF-κB binding site (Fig. 4A) (61). Recently, a negative regulatory role of NF-κB was postulated for murine Mx1 gene expression (80). Therefore, we mutated the putative NF-κB binding site (nucleotide positions −234 to −244) in the MxA promoter-driven reporter construct to a nonfunctional sequence according to the information given by Ping et al. (58). However, the mutation of the NF-κB site neither changed the responsiveness of the promoter to IFN-α nor stimulated activation by cotransfected IRF3(5D) in transfected GRE cells (data not shown). This result correlates with a previous analysis showing no binding of NF-κB to the respective binding site in the MxA promoter (61).
FIG. 4.
IRF3 binding to the MxA promoter. (A) Schematic presentation of the MxA promoter region (−1 to −530). The transcriptional start site is marked with an arrow (+1, exon 1). ISREs ISRE1 (−45 to −57), ISRE2 (−88 to −103), and ISRE3 (−490 to −505) are indicated with striped boxes, and one NF-κB site is indicated with a punctated box (−234 to −244) according to the data in reference 61. Binding of activated IRF3 (B) and NF-κB (C) to the Sepharose-immobilized NF-κB promoter element of IFN-β PRDII, IRE of the ISG56 promoter, or ISRE1 and ISRE2 of the MxA promoter. Nuclear extracts of FLUAVdNS1-infected A549 cells were incubated with the immobilized oligonucleotides, and DNA-bound proteins were detected by Western blot analysis using IRF3 and NF-κB p50-specific antibodies. The left panels show the respective protein signals in the nuclear extract. The results are representative of two individual experiments.
Next, we tested the ISREs for IRF3 binding. Previous studies showed two functional elements, ISRE1 and ISRE2, whereas ISRE3 lacked regulatory activity. ISRE1 is necessary for IFN-induced activation of the MxA promoter, whereas ISRE2 has an enhancing effect in the presence of an activated ISRE1 (61). We compared the IRF3 binding capacity of the two elements with that of the IRE of the ISG56 promoter (positions −82 to −100) (76) by using an oligonucleotide-binding assay. As a negative control, we used a consensus NF-κB-binding element. As a source of activated IRF3, nuclear extracts from A549 cells infected with FLUAVdNS1 were prepared and incubated with the short dsDNA elements as described previously (49). As expected, IRF3 bound to the IRE of ISG56 but not to the NF-κB response element (Fig. 4B). Interestingly, IRF3 also recognized ISRE2 of the MxA promoter with an efficiency comparable to that of the ISG56 IRE but was not found to interact with the ISRE1 binding site. NF-κB was used as a specificity control and showed binding only to the NF-κB response element, not to the ISREs (Fig. 4C). Our results indicate that the critical ISRE1 is not recognized by activated IRF3, explaining why the MxA promoter is not responsive to direct stimulation by IRF3 in contrast to the ISG56 or ISG54 promoter.
Virus-mediated MxA gene expression requires IFN signaling.
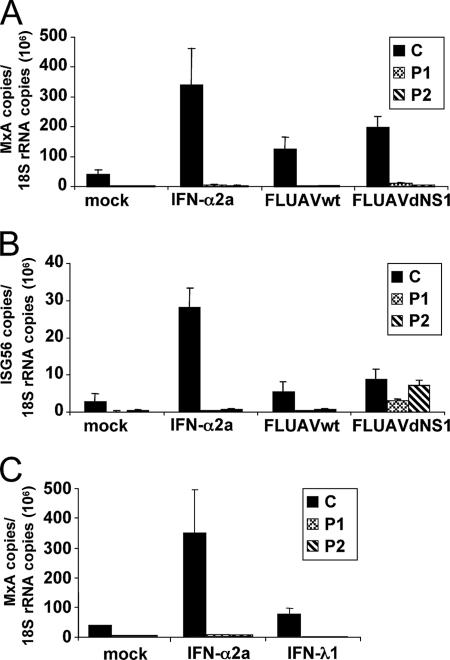
The induction of ISGs by IFN is dependent on the activation of ISGF3. Binding of type I or III IFNs to their receptors leads to the activation of STATs, which assemble to form the ISGF3 heterotrimeric protein complex (40, 71). To demonstrate the exclusive induction of MxA by IFN and by the IFN-stimulated signal transduction pathway, we took advantage of human cell lines with defects in the STAT1 function (21). The Epstein-Barr virus-transformed B cells originated from one patient (P1) with a 2-nt deletion from positions 1757 and 1758, generating a premature stop in the STAT1 open reading frame, and a second patient (P2) with a single amino acid exchange from leucine to proline at position 600. In contrast to the GRE cells, these cells were functional in the synthesis of type I IFN but were defective in IFN signaling (21). A control B-cell line from a healthy individual (C) was fully responsive to IFN. The cells were infected with the two FLUAV strains used before or were treated with IFN-α2a. Subsequently, the induction of MxA and ISG56 expression was analyzed by qRT-PCR. The control cells showed the induction of MxA and ISG56 gene expression by these stimuli (Fig. 5A and B). However, the expression of MxA was absent in cells that lack functional STAT1 (Fig. 5A). In contrast, ISG56 was also induced by FLUAVdNS1 in the STAT1-deficient cell lines (Fig. 5B), indicating that virus infection leads to a direct stimulation of ISG56 expression independent of IFN signaling. Furthermore, exogenous IFN-λ1 did not stimulate MxA expression in the STAT1-defective cell lines (Fig. 5C). These data clearly exclude a direct induction of MxA by virus infection and confirm that MxA gene expression is exclusively dependent on the signaling pathway that is linked to the IFN-α and IFN-λ receptors.
FIG. 5.
MxA induction depends on STAT1 signaling. Control cells, C, and two STAT1-deficient cell lines, P1 (STAT1/1757-1758delAG) and P2 (STAT1/L600P), were infected with 5 PFU per cell of FLUAVwt or FLUAVdNS1 or treated with 1,000 U/ml of IFN-α2a or left untreated. After 16 h, the amounts of MxA (A) and ISG56 (B) transcripts were determined by qRT-PCR and normalized to the amount of 18S rRNA transcripts. Error bars represent standard deviations calculated from three experiments. (C) Induction of MxA by IFN-λ. Cells were treated with 1,000 U/ml of IFN-α2a or 10 ng/ml IFN-λ1. The amounts of MxA and 18S rRNA transcripts were determined as described above.
DISCUSSION
Here we demonstrate that MxA gene expression is inducible by type I and III IFNs but not directly by virus infection, using different cell lines defective for IFN synthesis or signaling. Therefore, MxA can be used as a specific and reliable marker to detect type I IFN activity even in the context of virus infection.
In the present study, we used a FLUAV that is known to strongly stimulate IFN production, due to a deletion of the NS1 gene (25). FLUAVdNS1, a recombinant FLUAV that lacks the viral interferon antagonistic NS1 protein, was shown to cause the activation of IRF3 and the induction of type I IFN (73). Accordingly, we found a strong induction of MxA and ISG56 in human A549 lung adenocarcinoma cells after infection with FLUAVdNS1 but almost no induction after infection with wild-type FLUAV. Using human glioblastoma GRE cells defective in type I IFN synthesis (6), we showed that MxA is not directly induced by FLUAVdNS1 infection, in contrast to ISG56. MxA is also not induced by activated IRF3. Its induction requires the action of IFN. Our findings are in contrast to those of previous reports that have indicated an IFN-independent expression of MxA by virus infection (1, 26, 38, 59, 62). According to our data, we would also exclude a direct induction of MxA gene expression by the Toll-like receptor agonists lipopolysaccharide and poly(I:C), as reported recently, because they are known to efficiently induce type I IFNs (11, 20, 46, 81).
Interestingly, we observed a weak activation of the MxA promoter in virus-infected GRE cells. Indeed, a previous report has demonstrated a low induction of Mx gene expression in virus-infected mouse cells lacking a functional type I IFN receptor (8). In that report, Bazzigher et al. speculated that the murine Mx1 promoter might be directly activated upon virus infection albeit at low levels. However, it is conceivable that infected GRE cells produce type III IFN, which has recently been shown to induce MxA expression (40). Accordingly, we found an induction of IFN-λ in FLUAVdNS1-infected GRE cells and MxA gene expression in GRE cells treated with recombinant IFN-λ. Interestingly, in the cell types tested in our study and in accordance with a recent study by Meager et al. (48), IFN-λ showed a much weaker induction of MxA than did IFN-α, although the amounts of recombinant IFN-λ1 and IFN-α used in these experiments were difficult to compare because they were isolated from different sources.
Induction of ISGs by type I and III IFNs depends on the activation of the Jak/STAT signal transduction pathway and formation of an active ISGF3 (40, 71). To show the exclusive dependency of MxA gene induction on the presence of IFN, we used human cell lines with a genetic defect in IFN signaling based on nonfunctional STAT1 (21). Infection of these cells with FLUAVdNS1 caused an induction of ISG56, most likely by activated IRF3, but failed to stimulate MxA gene expression. These data indicate that the MxA promoter requires the formation of ISGF3 by the IFN-activated Jak/STAT pathway and that the expression of activated IRF3 or other IFN regulatory factors, like IRF1 and IRF7 (data not shown), is not sufficient for activation of the MxA promoter.
It is still unclear, however, why some ISGs are exclusively induced by IFNs, as described previously for the Mx genes (this study and see references 8 and 68), IRF7 (47), and some other ISGs (27, 72), while ISGs like ISG56, ISG54, and ISG15 can also be induced directly by virus infection or dsRNA, most likely via activated IRFs (6, 23, 27). The expression of ISGs is induced via ISREs located in their promoter regions. These elements share common sequence similarities (34, 36, 76). In general, ISRE sites are not used exclusively by ISGF3 but also by activated IRFs (27, 28). Using an oligonucleotide precipitation assay, we tested the ISREs of MxA for IRF3 binding. The MxA promoter contains two functional ISRE sites and one nonfunctional ISRE site (11, 34). The two active ISRE sites are essential and sufficient for the induction of MxA expression by type I IFN and show direct binding of activated ISGF3 (53, 61). An interferon regulatory element was also characterized for the ISG56 promoter (77) and was shown to be directly activated by virus infection or activated IRF3 (this study and see reference 27). Therefore, it was surprising that the MxA promoter is quite sensitive to activation by type I and III IFNs but unresponsive to virus infection and IRF3 stimulation. Interestingly, our analysis revealed that the distal ISRE2 was recognized by activated IRF3 similarly to the regulatory element of the ISG56 promoter, whereas ISRE1 showed no IRF3 binding. It has been shown that activated IRF3 needs additional surrounding sequences to bind to ISRE-containing promoter regions (16, 42, 54). Therefore, sequence variations in the ISREs or differences in the surrounding sequences might be responsible for the differences in IRF3-binding specificity. A previous analysis showed that ISRE1 was absolutely necessary for MxA promoter activation, whereas ISRE2 had only a modulatory enhancing function (61). Therefore, we propose the model that the binding of IFN-activated ISGF3 to ISRE1 is absolutely required for a basal stimulation of the MxA promoter. However, the status of promoter activation can be enhanced either by the binding of IFN-stimulated ISGF3 to ISRE2, as has previously been shown (61), or by the binding of virus-activated IRF3 to ISRE2. This possible synergistic route of MxA promoter activation by the IFN-α-activated as well as the IRF3-dependent pathway would explain the strong expression of the MxA gene by FLUAVdNS1 compared to IFN-α2a treatment that we observed in some experimental settings in A549 cells (Fig. 2A). Furthermore, cis-acting, negative regulatory elements that are yet unknown might suppress IFN-independent activation of the MxA promoter, as has been suggested previously by Chang et al. (11).
The exclusive induction of Mx protein synthesis by IFN suggests a strong restriction of Mx gene expression. Negative effects on the normal homoeostasis of the cell could be provoked by Mx accumulation. Accordingly, MxA overexpression has been shown to increase the sensitivity of cells to apoptotic stimuli (41, 50). Thus, it has been suggested that the upregulation of MxA is one factor that promotes the development of Fanconi anemia, a disorder that predisposes patients for skeletal malformation, progression to bone marrow failure, and acute leukemia (41).
We also studied the induction of MxA by IFN-γ. A weak but significant induction of MxA by IFN-γ has been reported for human macrophages (61). However, in our study, IFN-γ levels of up to 1,000 U/ml did not show any induction of MxA expression in human PBMCs, whereas IP-10 expression was induced over 300-fold. This result is consistent with results obtained with mouse Mx1, which also is not induced by IFN-γ (70). Along these lines, von Wussow et al. reported that an induction of MxA by IFN-γ in mononuclear cells could be prevented by IFN-α-neutralizing antibodies, indicating that the effect of IFN-γ is indirectly mediated by IFN-γ-induced IFN-α activity (75). An alternative explanation could be that IFN-γ-activated Stat1/Stat1/IRF9 trimers bind to ISRE sites, leading to the weak induction of MxA observed in human primary macrophages (61). In contrast to results for macrophages, an IFN-independent induction of MxA has been observed in lipopolysaccharide-stimulated human neutrophils (46). This induction was found against a background of low IFN-α induction and was dependent on the activation of the p38 member of the MAP kinase family of protein kinases, suggesting that in neutrophils, MxA can also be induced via a novel MAP kinase-dependent mechanism.
In conclusion, our data demonstrate that MxA induction is dependent on type I and III IFNs and a functional IFN-stimulated signaling pathway. In contrast to ISG56, a direct induction of MxA by virus infection or activated IFN regulatory factors is suppressed due to the restrictive control of promoter activation by the proximal ISRE1 site that seems to be exclusively responsive to ISGF3. Since IFNs are produced only locally and transiently after virus infection, they are difficult to detect. In this situation, MxA expression can be used as an exclusive marker for IFN type I and III activity.
Acknowledgments
This work was supported by grants from the Wissenschaftliche Gesellschaft in Freiburg to G.K. and from the Deutsche Forschungsgemeinschaft to G.K. (Ko1579/1-8) and R.T. (TH719-2-2 Emmy-Noether-Programm). J.-L.C. is an International Scholar of the Howard Hughes Medical Institute.
We thank Simone Gruber and Nadine Kersting for excellent technical assistance and Peter Staeheli and Friedemann Weber for discussions and critical comments on the manuscript. We are grateful to Takashi Fujita, Adolpho Garcia-Sastre, Ilkka Julkunen, Sergei V. Kotenko, David Levy, Luis Martinez-Sobrido, Jesper Melchjorsen, Soeren R. Paludan, Ganes C. Sen, and Peter Staeheli for expression constructs, reporter plasmids, antibodies, viruses, cell lines, and protocols.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Aebi, M., J. Fäh, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Antonelli, G., E. Simeoni, O. Turriziani, R. Tesoro, A. Redaelli, L. Roffi, L. Antonelli, M. Pistello, and F. Dianzani. 1999. Correlation of interferon-induced expression of MxA mRNA in peripheral blood mononuclear cells with the response of patients with chronic active hepatitis C to IFN-α therapy. J. Interferon Cytokine Res. 19:243-251. [DOI] [PubMed] [Google Scholar]
- 4.Asahina, Y., N. Izumi, M. Uchihara, O. Noguchi, Y. Nishimura, K. Inoue, K. Ueda, K. Tsuchiya, K. Hamano, J. Itakura, and S. Miyake. 2003. Interferon-stimulated gene expression and hepatitis C viral dynamics during different interferon regimens. J. Hepatol. 39:421-427. [DOI] [PubMed] [Google Scholar]
- 5.Baechler, E. C., F. M. Batliwalla, G. Karypis, P. M. Gaffney, W. A. Ortmann, K. J. Espe, K. B. Shark, W. J. Grande, K. M. Hughes, V. Kapur, P. K. Gregersen, and T. W. Behrens. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA 100:2610-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandyopadhyay, S. K., G. T. Leonard, T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 7.Basler, C., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Mühlberger, M. Bray, H.-D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazzigher, L., J. Pavlovic, O. Haller, and P. Staeheli. 1992. Mx genes show weaker primary response to virus than other interferon-regulated genes. Virology 186:154-160. [DOI] [PubMed] [Google Scholar]
- 9.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyum, A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 97:77-89. [PubMed] [Google Scholar]
- 11.Chang, K. C., E. Hansen, L. Foroni, J. Lida, and G. Goldspink. 1991. Molecular and functional analysis of the virus- and interferon-inducible human MxA promoter. Arch. Virol. 117:1-15. [DOI] [PubMed] [Google Scholar]
- 12.Chen, L., I. Borozan, J. Feld, J. Sun, L. L. Tannis, C. Coltescu, J. Heathcote, A. M. Edwards, and I. D. McGilvray. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437-1444. [DOI] [PubMed] [Google Scholar]
- 13.Cheney, I. W., V. C. Lai, W. Zhong, T. Brodhag, S. Dempsey, C. Lim, Z. Hong, J. Y. Lau, and R. C. Tam. 2002. Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons. J. Virol. 76:11148-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinatl, J., Jr., G. Hoever, B. Morgenstern, W. Preiser, J. U. Vogel, W. K. Hofmann, G. Bauer, M. Michaelis, H. F. Rabenau, and H. W. Doerr. 2004. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 61:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78:1706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly, C., and N. C. Reich. 1995. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J. Biol. Chem. 270:23739-23746. [DOI] [PubMed] [Google Scholar]
- 17.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 18.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 20.Doyle, S., S. Vaidya, R. O'Connell, H. Dadgostar, P. Dempsey, T. Wu, G. Rao, R. Sun, M. Haberland, R. Modlin, and G. Cheng. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17:251-263. [DOI] [PubMed] [Google Scholar]
- 21.Dupuis, S., E. Jouanguy, S. Al-Hajjar, C. Fieschi, I. Z. Al-Mohsen, S. Al-Jumaah, K. Yang, A. Chapgier, C. Eidenschenk, P. Eid, A. Al-Ghonaium, H. Tufenkeji, H. Frayha, S. Al-Gazlan, H. Al-Rayes, R. D. Schreiber, I. Gresser, and J. L. Casanova. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388-391. [DOI] [PubMed] [Google Scholar]
- 22.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 23.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not Toll-like receptor 3. J. Virol. 79:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flohr, F., S. Schneider-Schaulies, O. Haller, and G. Kochs. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463:24-28. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 26.Goetschy, J. F., H. Zeller, J. Content, and M. A. Horisberger. 1989. Regulation of the interferon-inducible IFI-78K gene, the human equivalent of the murine Mx gene, by interferons, double-stranded RNA, certain cytokines, and viruses. J. Virol. 63:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 29.Hagmaier, K., H. R. Gelderblom, and G. Kochs. 2004. Functional comparison of the two gene products of Thogoto virus segment 6. J. Gen. Virol. 85:3699-3708. [DOI] [PubMed] [Google Scholar]
- 30.Hagmaier, K., S. Jennings, J. Buse, F. Weber, and G. Kochs. 2003. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J. Virol. 77:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haller, O., H. Arnheiter, I. Gresser, and J. Lindenmann. 1979. Genetically determined, interferon-dependent resistance to influenza virus in mice. J. Exp. Med. 149:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 33.Haller, O., and G. Kochs. 2002. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 34.Horisberger, M. A., G. K. McMaster, H. Zeller, M. G. Wathelet, J. Dellis, and J. Content. 1990. Cloning and sequence analyses of cDNAs for interferon- and virus-induced human Mx proteins reveal that they contain putative guanine nucleotide-binding sites: functional study of the corresponding gene promoter. J. Virol. 64:1171-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horisberger, M. A., P. Staeheli, and O. Haller. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. USA 80:1910-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hug, H., M. Costas, P. Staeheli, M. Aebi, and C. Weissmann. 1988. Organization of the murine Mx gene and characterization of its interferon- and virus-inducible promoter. Mol. Cell. Biol. 8:3065-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito, S., P. Ansari, M. Sakatsume, H. Dickensheets, N. Vazquez, R. P. Donnelly, A. C. Larner, and D. S. Finbloom. 1999. Interleukin-10 inhibits expression of both interferon alpha- and interferon gamma-induced genes by suppressing tyrosine phosphorylation of STAT1. Blood 93:1456-1463. [PubMed] [Google Scholar]
- 38.Izmailova, E., F. M. Bertley, Q. Huang, N. Makori, C. J. Miller, R. A. Young, and A. Aldovini. 2003. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 9:191-197. [DOI] [PubMed] [Google Scholar]
- 39.Jorns, C., D. Holzinger, R. Thimme, H. C. Spangenberg, M. Weidmann, J. Rasenack, H. E. Blum, O. Haller, and G. Kochs. 2006. Rapid and simple detection of IFN-neutralizing antibodies in chronic hepatitis C non-responsive to IFN-alpha. J. Med. Virol. 78:74-82. [DOI] [PubMed] [Google Scholar]
- 40.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 41.Li, Y., and H. Youssoufian. 1997. MxA overexpression reveals a common genetic link in four Fanconi anemia complementation groups. J. Clin. Investig. 100:2873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20:6342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindenmann, J. 1962. Resistance of mouse to mice adapted influenza A virus. Virology 16:203-204. [DOI] [PubMed] [Google Scholar]
- 45.Mackay, C. R. 2001. Chemokines: immunology's high impact factors. Nat. Immunol. 2:95-101. [DOI] [PubMed] [Google Scholar]
- 46.Malcolm, K. C., and G. S. Worthen. 2003. Lipopolysaccharide stimulates p38-dependent induction of antiviral genes in neutrophils independently of paracrine factors. J. Biol. Chem. 278:15693-15701. [DOI] [PubMed] [Google Scholar]
- 47.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meager, A., K. Visvalingam, P. Dilger, D. Bryan, and M. Wadhwa. 2005. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine 31:109-118. [DOI] [PubMed] [Google Scholar]
- 49.Melchjorsen, J., J. Siren, I. Julkunen, S. R. Paludan, and S. Matikainen. 2006. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-κB and IRF-3. J. Gen. Virol. 87:1099-1108. [DOI] [PubMed] [Google Scholar]
- 50.Mibayashi, M., K. Nakad, and K. Nagata. 2002. Promoted cell death of cells expressing human MxA by influenza virus infection. Microbiol. Immunol. 46:29-36. [DOI] [PubMed] [Google Scholar]
- 51.Miyakoshi, J., K. D. Dobler, J. Allalunis-Turner, J. D. McKean, K. Petruk, P. B. Allen, K. N. Aronyk, B. Weir, D. Huyser-Wierenga, D. Fulton, et al. 1990. Absence of IFNA and IFNB genes from human malignant glioma cell lines and lack of correlation with cellular sensitivity to interferons. Cancer Res. 50:278-283. [PubMed] [Google Scholar]
- 52.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 53.Nakade, K., H. Handa, and K. Nagata. 1997. Promoter structure of the MxA gene that confers resistance to influenza virus. FEBS Lett. 418:315-318. [DOI] [PubMed] [Google Scholar]
- 54.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 55.Park, C., S. Li, E. Cha, and C. Schindler. 2000. Immune response in Stat2 knockout mice. Immunity 13:795-804. [DOI] [PubMed] [Google Scholar]
- 56.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140-147. [DOI] [PubMed] [Google Scholar]
- 57.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NF-κB- and JNK-independent activation of the 561 and IFN-β genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ping, D., G. H. Boekhoudt, E. M. Rogers, and J. M. Boss. 1999. Nuclear factor-kappa B p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J. Immunol. 162:727-734. [PubMed] [Google Scholar]
- 59.Prescott, J., C. Ye, G. Sen, and B. Hjelle. 2005. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J. Virol. 79:15007-15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roers, A., H. K. Hochkeppel, M. A. Horisberger, A. Hovanessian, and O. Haller. 1994. MxA gene expression after live virus vaccination: a sensitive marker for endogenous type I interferon. J. Infect. Dis. 169:807-813. [DOI] [PubMed] [Google Scholar]
- 61.Ronni, T., S. Matikainen, A. Lehtonen, J. Palvimo, J. Dellis, F. Van Eylen, J. F. Goetschy, M. Horisberger, J. Content, and I. Julkunen. 1998. The proximal interferon-stimulated response elements are essential for interferon responsiveness: a promoter analysis of the antiviral MxA gene. J. Interferon Cytokine Res. 18:773-781. [DOI] [PubMed] [Google Scholar]
- 62.Ronni, T., S. Matikainen, T. Sareneva, K. Melen, J. Pirhonen, P. Keskinen, and I. Julkunen. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J. Immunol. 158:2363-2374. [PubMed] [Google Scholar]
- 63.Rychlik, W., W. J. Spencer, and R. E. Rhoads. 1990. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 18:6409-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 65.Servant, M. J., N. Grandvaux, and J. Hiscott. 2002. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem. Pharmacol. 64:985-992. [DOI] [PubMed] [Google Scholar]
- 66.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 67.Simon, A., J. Fäh, O. Haller, and P. Staeheli. 1991. Interferon-regulated Mx genes are not responsive to interleukin-1, tumor necrosis factor, and other cytokines. J. Virol. 65:968-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Staeheli, P., P. Danielson, O. Haller, and J. G. Sutcliffe. 1986. Transcriptional activation of the mouse Mx gene by type I interferon. Mol. Cell. Biol. 6:4770-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staeheli, P., O. Haller, W. Boll, J. Lindenmann, and C. Weissmann. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147-158. [DOI] [PubMed] [Google Scholar]
- 70.Staeheli, P., M. A. Horisberger, and O. Haller. 1984. Mx-dependent resistance to influenza viruses is induced by mouse interferons alpha and beta but not gamma. Virology 132:456-461. [DOI] [PubMed] [Google Scholar]
- 71.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 72.Stojdl, D. F., B. D. Lichty, B. R. tenOever, J. M. Paterson, A. T. Power, S. Knowles, R. Marius, J. Reynard, L. Poliquin, H. Atkins, E. G. Brown, R. K. Durbin, J. E. Durbin, J. Hiscott, and J. C. Bell. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263-275. [DOI] [PubMed] [Google Scholar]
- 73.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vilcek, J. 2003. Novel interferons. Nat. Immunol. 4:8-9. [DOI] [PubMed] [Google Scholar]
- 75.von Wussow, P., D. Jakschies, H. K. Hochkeppel, C. Fibich, L. Penner, and H. Deicher. 1990. The human intracellular Mx-homologous protein is specifically induced by type I interferons. Eur. J. Immunol. 20:2015-2019. [DOI] [PubMed] [Google Scholar]
- 76.Wathelet, M. G., I. M. Clauss, J. Content, and G. A. Huez. 1988. Regulation of two interferon-inducible human genes by interferon, poly(rI).poly(rC) and viruses. Eur. J. Biochem. 174:323-329. [DOI] [PubMed] [Google Scholar]
- 77.Wathelet, M. G., I. M. Clauss, C. B. Nols, J. Content, and G. A. Huez. 1987. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur. J. Biochem. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 78.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 79.Weber, F., O. Haller, and G. Kochs. 2000. MxA GTPase blocks reporter gene expression of reconstituted Thogoto virus ribonucleoprotein complexes. J. Virol. 74:560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei, L., M. R. Sandbulte, P. G. Thomas, R. J. Webby, R. Homayouni, and L. M. Pfeffer. 2006. NF-κB negatively regulates interferon-induced gene expression and anti-influenza activity. J. Biol. Chem. 281:11678-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weighardt, H., G. Jusek, J. Mages, R. Lang, K. Hoebe, B. Beutler, and B. Holzmann. 2004. Identification of a TLR4- and TRIF-dependent activation program of dendritic cells. Eur. J. Immunol. 34:558-564. [DOI] [PubMed] [Google Scholar]
- 82.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the IFN-alpha/beta system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]