Abstract
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections lead to AIDS in humans and rhesus macaques (RM), while they are asymptomatic in species naturally infected with SIV, such as chimpanzees, sooty mangabeys (SM), and African green monkeys (AGM). Differential CD4+ T-cell apoptosis may be responsible for these species-specific differences in susceptibility to disease. To identify factors that influence the different apoptotic responses of these species, we analyzed virus-infected human and nonhuman primate peripheral blood mononuclear cells (PBMC). We found that the apoptotic factor TRAIL was present at higher levels in human and RM PBMC cultures and was mediating, at least in part, CD4+ T-cell apoptosis in these cultures. The species-specific increase in TRAIL and death receptor expression observed with cultures also occurred in vivo in SIV-infected RM but not in SIV-infected SM. In human and RM myeloid immature dendritic cells and macrophages, the virus-induced expression of TRAIL and other interferon-inducible genes, which did not occur in the same cells from chimpanzee, SM, and AGM, was Tat dependent. Our results link the differential induction of TRAIL in human and nonhuman primate cells to species-specific differences in disease susceptibility.
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections lead to a progressive loss of CD4+ T cells and AIDS in some species but not in others. The hallmark of the infection in disease-susceptible species is the progressive decline of the CD4+ T-cell population. CD8+ T cells also show increased sensitivity to HIV-mediated apoptosis (8, 12, 13, 22, 35, 39), but this population remains more stable during the course of the infection in vivo. Why CD4+ T cells are progressively lost in infected humans and rhesus macaques (RM) yet remain stable in chimpanzees, sooty mangabeys (SM), and African green monkeys (AGM) is not well understood. The range of plasma viremia in naturally infected SM and AGM is similar to that in humans during the progression to AIDS (14, 41). Therefore, containment of viremia is an unlikely explanation for the lack of pathogenicity of SIVagm and SIVsm in their natural host species.
There are at least two mechanisms that may account for the loss of CD4+ T cells: direct cytopathic effect and activation of programmed cell death. The Env-mediated cytopathic effect can be observed with HIV type 1 (HIV-1)-infected CD4+ T cells from both humans and chimpanzees. HIV-1 can also eliminate both infected and uninfected CD4+ T cells via activation of apoptosis (8, 16, 43). Programmed cell death can be induced by viral proteins and as a result of persistent enhanced immune activation. Abnormal T-cell proliferation, which leads to immune exhaustion and the progressive deterioration of the immune system, has been observed with susceptible species but not in naturally infected hosts (4, 5, 8, 9, 16, 43, 49). A model for CD4+ T-cell depletion by the induction of apoptosis has been suggested (1, 24).
Susceptibility to HIV- and SIV-mediated apoptosis varies within and across species. Apoptosis is substantially reduced in HIV-infected individuals that are long-term nonprogressors (28, 33). Additional evidence suggests that apoptosis occurs significantly less in species such as chimpanzees and SM that do not develop AIDS (10, 17, 49). In the rare cases when chimpanzees do progress to AIDS, the loss of CD4+ T cells and increased apoptosis have been observed previously (9, 48).
Several HIV-1 proteins have been shown to interfere with cellular proteins implicated in the control of cell death (reviewed in reference 45). It is not known if one or more of these apoptotic pathways are regulated differently in infected humans or RM, species which develop AIDS, relative to those in infected chimpanzees or AGM and SM, which do not. These differences among species and the roles played by different cell types in the progression toward AIDS may result, in part, from differences in the response of host cells to viral proteins.
HIV Tat has been shown to increase transcription of tumor necrosis factor-related-apoptosis-inducing-ligand/Apo-2 ligand (TRAIL) in human cells (23, 54, 55). TRAIL has been shown to trigger apoptosis in T cells from HIV-infected individuals, while T cells from uninfected controls are completely resistant to TRAIL-induced apoptosis (26). There are at least four receptors for TRAIL, some of which are critical for mediating TRAIL signaling (death receptors DR4 and DR5) (44, 56). The differential sensitivity to TRAIL observed with HIV-infected individuals (19, 25, 26, 30, 32) may result from the upregulation of death-mediating receptors, which can be induced by gp120 in HIV-exposed cultures (19, 25, 26, 30, 32). Whether SIV-mediated TRAIL apoptosis has a different impact on different infected nonhuman primate species is not known.
To gain insights into the reasons for the different outcomes of HIV infection in primate species, we investigated differences in human and nonhuman primate cells infected with HIV and SIV or expressing Tat. Our results reveal a link between the differential induction of TRAIL and the species-specific differences in AIDS susceptibility.
MATERIALS AND METHODS
Cells, antibodies, and enzyme-linked immunosorbent assays.
Samples of fresh human peripheral blood mononuclear cells (PBMC) from the Children's Hospital Boston blood bank and primate PBMC of RM and AGM (Chlorocebus sabaeus) from the New England Regional Primate Research Center or PBMC of chimpanzee and SM shipped overnight from Yerkes National Primate Research Center were used in the experiments. PBMC were stained with antibodies against CD3, CD4, CD8, CD11c, HLA-DR, CD123, CD95, and CD95L (BD Biosciences, San Jose, CA), CD14 (Immunotech, Beckman Coulter, Miami, FL), TRAIL, DR4, DR5, DcR1, and DcR2 (eBioscience, San Diego, CA).
Human immature dendritic cells (iDC) were generated from monocytes purified by elutriation. For nonhuman primates, negative selection (Miltenyi Biotec, Auburn, CA) and/or plastic adhesion was used to enrich the population for iDC precursors. CD14+ cells, as determined by staining with an anti-CD14 antibody (Miltenyi Biotec, Auburn, CA) were seeded at a density of 106 cells/ml with interleukin 4 (100 ng/ml) and granulocyte-macrophage colony-stimulating factor (GM-CSF; 200 ng/ml) (3). Seven days after plating, approximately 95% of cells were DC+, CD11chigh, CD1a−/low, CD14low, CD83−, CD86low, and HLA-DRlow by flow cytometry analysis, consistent with the profile for iDC (29, 34, 38, 53). The characterization of these cells was done with human iDC, with multiple markers as described in reference 23. The most common contaminating cells were CD14high monocytes, varying between 5 and 10% of the different cultures. Macrophages were generated from elutriated human cells or from monocytes purified by negative selection (Miltenyi Biotec, Auburn, CA) and/or by plastic adhesion, depending on the species (36, 40). Monocytes were seeded at a density of 106 cells/ml and analyzed by flow cytometry 7 days after seeding. Before infection, 95% or more of the populations were CD68+ CD11b+.
For phenotypic analysis, differentiated cells were incubated with monoclonal antibodies including CD1a, CD11b, CD11c, CD25, CD40, and HLA-DR (BD Bioscience, San Diego, CA), and CD14 and CD83 (Immunotech, Beckman Coulter, Miami, FL), and CD80 and CD86 (BD Bioscience, San Diego, CA) and were analyzed by flow cytometry. Other antibodies used in the studies were those against membrane TRAIL (mTRAIL) (eBioscience, San Diego, CA), DR4, DR5 (Alexis, San Diego, CA), and type I alpha interferon receptor chain 2 (IFN-αR) and IFN-α (U.S. Biological, Swampscott, MA).
Enzyme-linked immunosorbent assays (ELISA) for CD95L, soluble TRAIL (sTRAIL), IP-10, MIG, MCP-2, MCP-3, and IFN-α were obtained from R&D Systems (Minneapolis, MN) and Biosource (Camarillo, CA), and from PBL Biomedical Lab (Piscataway, NJ) for IFN-β, and from Cell Sciences (Canton, MA) for IFN-γ. Positive controls for sTRAIL and for the detection of other cytokines reported in Fig. 5 and Fig. 6 in each species were provided by supernatants from monocytes treated with recombinant human IFN-α (rhIFN-α). Nonhuman primate PBMC treated with rhIFN-α or with RM rIFN-α provided the positive control for mTRAIL. Purified human TRAIL was used as a standard in all experiments. Influenza virus-treated nonhuman primate PBMC from each of the primate species were used as a source of IFN-γ- and IFN-α-positive controls. Human IFN-α and RM rIFN-α were used as standards for detection of IFN-α in human and nonhuman primate assays. Human cytokine antibody array kits were obtained from RayBiotech (Norcross, GA). Anti-annexin V antibody and an Annexin V apoptosis detection kit were obtained from BD Biosciences (San Jose, CA). Positive controls for the detection of DR4 and DR5 in nonhuman primate species were obtained by treating PBMC with doxorubicin, which induces expression of these receptors in approximately 80 to 90% of the T cells present in the culture (27, 52).
FIG. 5.
Expression analysis of selected IFN-inducible genes in different nonhuman primate iDC. (a) RT activity in macrophage supernatants. (b) Tat RNA levels measured by real time RT-PCR. (c) Induction of selected IFN-inducible genes based on RNA levels from human and nonhuman primate iDC after HIVBal or SIVmac316 infection (day 14) and Tat expression (30 h). Results are reported as the average (n-fold) induction relative to that of the time zero mock-infected sample. Two samples from two independent donors are reported. A positive control for the real-time RT-PCR in RNAs from each species was provided by an RNA sample from CD14+ cells treated with IFN-α. All detection values are normalized according to the GAPDH internal control. (d) Cytokine detection in primate iDC supernatants after HIVBal or SIVmac316 infection (means ± standard errors of three independent samples).
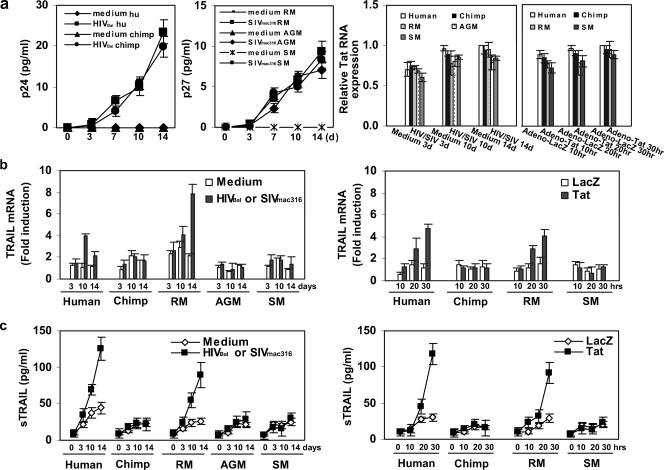
FIG. 6.
TRAIL expression in macrophages from different primate species after Tat expression or viral infection (HIVBal or SIVmac316). HIV and SIV infection in primate iDC and macrophages. (a) Virus production in infected macrophages measured by p24 (left panel) or p27 (middle panel) ELISA assay. Tat expression detected by real-time RT-PCR in the different cultures (right panel). (b) Changes in TRAIL RNA levels for each species (means ± standard errors [SE]). (c) Levels of sTRAIL measured by ELISA in the supernatant of macrophage cultures (means ± SE). Three independent experiments were carried out with human macrophages and two with macrophages from the other species.
Immunofluorescent staining and flow cytometry.
Intracellular staining (ICS) and phenotypic analysis by flow cytometry were carried out as previously described (51). Flow cytometric acquisition was performed on a Beckman Coulter XL (Beckman Coulter, Inc.) with fluorescein isothiocyanate, phycoerythrin, phycoerythrin-Texas Red, and phycocyanine 5 as the four fluorescent parameters. For cell analysis, the mean fluorescence intensity of staining for each marker was assessed on the gated live cell population using EXPO 32 ADC analysis software (Beckman Coulter, Inc.).
Viruses, viral infections, and viral replication detection.
Strain HIV-1Bal was obtained from Advanced Biotechnology, Inc. (Rockville, MD). Strains HIVHXB2, HIVSF2, SIVmac239, and SIVmac316 were all derived from the transfection of molecular clones and subsequent brief amplification in human or rhesus macaque PBMC. HIV-1Bal was used for infection of iDC and macrophages from humans and chimpanzees, and SIVmac316 was used for RM, SM, and AGM macrophages and iDC. We also evaluated PBMC from SIVSME041 (a gift of Preston Marx), SIVagm(sab-2), and SIVrcmGABI (AIDS repository) with RM, SM, AGM, and chimpanzee cells. Staphylococcal enterotoxin B (SEB) was used as an immune activator to increase levels of virus replication in the PBMC culture at the concentration of 100 ng/ml. The use of this activator resulted in a better ratio of activation and infection to toxicity in chimpanzee cells than the use of phytohemagglutinin, which produced high background, or anti-CD3 plus anti-CD28, which does not work well in this species (37). Viral stock titers were determined with human or macaque PBMC by limiting dilution, according to the 50% tissue culture infective dose (TCID50) determination protocol reported at http://aactg.s-3.com/pub/download/labmanual/43-ALM-TCID50-Determination.pdf. Viral infections were carried out with 10,000 TCID50/106 cells, with a multiplicity of infection of 0.01. The viability of iDC and macrophages over the 30-h Tat expression time course did not change. During the HIV or SIV time course, a limited number of cells became detached at the time of medium change. On day 14 after infection, when the viability was assessed by trypan blue exclusion after detaching the adherent cells in the medium, it varied between 75% and 90%, when infected cells were compared to mock-infected cells.
Virus replication was monitored in the PBMC cultures by p24 or p27 ELISA and by p24 or p27 ICS. HIVBal and SIVmac316 were used for the five (human) and three (other primates) independent experiments with independent donor cells shown in Fig. 1. In some chimpanzee PBMC, infectivity was substantially lower than that observed with human cells, and those cultures were disregarded. The apoptosis rate and additional parameters were reported for three chimpanzee cultures in which the virus production and infectivity were within twofold of that observed with the human PBMC cultures. The behavior of R5 HIVBal or R5/X4 HIVSF2 and of X4 HIVHXB2 and HIVNL4-3 was evaluated with PBMC from separate human donors. Comparison of HIVHXB2 and HIVBal was carried out with chimpanzee PBMC from three donors; comparison of SIVmac316 and SIVmac239 was carried out with RH, SM, and AGM PBMC from three independent donors. An adenovirus carrying the tat gene transfer system (Adeno-Tat) and an Adeno-LacZ system were constructed at the Vector Facility of the Harvard Gene Therapy Initiative according to established protocols (6). In iDC cultures, virus production was measured by reverse transcriptase (RT) activity in the supernatant (23). In addition, HIV tat gene expression was evaluated by real-time RT-PCR with the RNA extracted from the cultures. In macrophage cultures, virus replication was monitored by supernatant p24 or p27 ELISA. Viral infection rate in iDC and macrophages was analyzed by ICS with a serum reactive against HIV or SIV, followed by a horseradish peroxidase secondary antibody and analyzed by optical microscopy or by ICS with p24 or p27 antibodies, followed by flow cytometric analysis 14 days after the initial infection. Tat expression after Adeno-Tat infection was evaluated by real-time RT-PCR and by Tat ICS using an anti-Tat antibody.
FIG. 1.
Apoptosis and TRAIL production in lentivirus-infected primate PBMC cultures. Levels of apoptosis in (a) HIV- and (b) SIV-infected PBMC from human and nonhuman primates. Panels in the left column display the percentage of CD4+ T cells in the PBMC cultures that stain for annexin V, a marker for apoptosis. Panels in the middle column report the flow cytometric analysis of CD4+ T-cell viability in PBMC. Values at each time point are expressed as the percentage of viable CD4+ T cells present in the mock-infected sample. Panels in the right column report virus production in infected primate PBMC measured by p24 or p27 ELISA assay. The data represent the mean percentages ± standard errors (SE) of five (human) and three (other species) experiments. PBMC subpopulation mean ± SE percentages on the day of infection were as follows: for human PBMC, CD3, 71 ± 4.5; CD4, 45 ± 2.8; CD8, 24 ± 3.6; CD14, 13 ± 3.7; CD11c, 1.04 ± 0.30; CD123, 0.70 ± 0.14; for chimpanzee PBMC, CD3, 73 ± 4.1; CD4, 41 ± 2.8; CD8, 21 ± 1.2; CD14, 9.2 ± 0.7; CD11c, 1.03 ± 0.19; CD123, 0.47 ± 0.12; for RM PBMC, CD3, 67 ± 4.2; CD4, 32 ± 3.9; CD8, 33 ± 2.1; CD14, 7.9 ± 0.9; CD11c, 1.87 ± 0.34; CD123, 0.53 ± 0.12; for AGM PBMC, CD3, 68 ± 5; CD4, 19 ± 3.6; CD8, 49 ± 6.9; CD14, 7.7 ± 2.5; CD11c, 0.77 ± 0.18; CD123, 0.59 ± 0.24; for SM PBMC, CD3, 72 ± 1.6; CD4, 23 ± 1.4; CD8, 50 ± 3; CD14, 14.6 ± 1.7; CD11c, 0.81 ± 0.30; CD123, 0.54 ± 0.13. HIV or SIV infection mean ± SE percentages (determined by p24 or p27 intracellular staining on day 7 postinfection in CD4+ T cells or in CD14+ cells) for HIV-infected human PBMC were CD4, 15 ± 2.9; CD14, 10.1 ± 2; for SEB/HIV-infected human PBMC were CD4, 24.8 ± 5.3; CD14, 15.2 ± 4.5; for HIV-infected chimpanzee PBMC were CD4, 9 ± 1.5; CD14, 5 ± 1.5; for SEB/HIV-infected chimp PBMC were CD4, 14.2 ± 1.8; CD14, 9 ± 2.1; for SIV-infected RM PBMC were CD4+, 12 ± 3; CD14, 5 ± 1.9; for SEB/SIV-infected RM PBMC were CD4, 18 ± 4; CD14, 8 ± 2.6; for SIV-infected AGM PBMC were CD4, 12.6 ± 2.2; CD14, 4.8 ± 1.7; for SEB/SIV-infected AGM PBMC were CD4, 16.9 ± 3.7; CD14, 7.3 ± 3.8; for SIV-infected SM PBMC were CD4, 8.9 ± 1.9; CD14, 5.5 ± 2.1; for SEB/SIV-infected SM PBMC were CD4, 18.1 ± 5.3; CD14, 8.7 ± 3.9. (c) Levels of IFN-α, IFN-γ, and soluble TRAIL in primate PBMC culture supernatants after infection with HIVBal or SIVmac239 during 10 days of culture (means ± SE from three experiments).
Blocking assays.
These assays were performed using antibodies for TRAIL (eBioscience, San Diego, CA), DR4, DR5 (Alexis, San Diego, CA), and IFN-αR and IFN-α (US Biological, Swampscott, MA). For these assays, PBMC were cultured for 2 h with the neutralizing antibodies against TRAIL, DR4, and DR5 at the concentration of 5 μg/ml. Cells were then exposed to HIV-1HXB2 and then grown for 7 days. A second dose of the blocking antibodies was added to the cultures after 72 h. The percentage of annexin V+ cells was determined by fluorescence-activated cell sorter analysis on day 7. The negative control was an isotype-matched unrelated mouse monoclonal antibody, used at the same dose as the total amount of the blocking antibodies (three in the case of TRAIL, two for CD95 or IFN-α). The positive control was a PBMC culture treated with IFN-α with or without the IFN-α neutralizing antibody, and the readout was the production of TRAIL. TRAIL production was reduced by approximately 60% in the presence of the neutralizing antibody compared to that of the culture that did not receive antibody treatment.
RNA isolation, labeling, and array hybridization.
At each time point of interest, 106 cells were harvested and lysed by using Trizol (Molecular Research Center). Total RNA was isolated, and 1 μg was prepared for hybridization to HG U133A 2.0 oligonucleotide arrays (Affymetrix) according to the standard Affymetrix one-cycle target-labeling protocol (Affymetrix). Hybridization was carried out overnight with 1.5 μg of labeled cRNA product, and arrays were scanned on Affymetrix scanners.
DNA array data analysis.
GeneChip arrays were scanned using a GeneArray scanner (Affymetrix). The CEL files were generated from DAT files using GeneChip Operating Software (Affymetrix). Each array was scaled to a target signal of 150. Expression ratios were calculated for each probe set relative to the intensity in the T = 0 control (chimpanzee) or the geometric mean of duplicate T = 0 controls (human). Changes in gene expression were considered significant if each of the following criteria was met: (i) expression changed at least twofold at two consecutive time points or threefold at a single time point compared with T = 0; (ii) expression changed at least twofold compared to the expression in negative controls (medium control for HIV-1 infection and adenovirus expressing the LacZ gene, for the adenovirus infection experiment) at the same time point; and (iii) increased gene expression included at least one “Present” call (Affymetrix algorithm), or both zero time points were present when gene expression decreased.
RT-PCR.
Primers for Stat-1, IRF-7, MIG, IP-10, and TRAIL were selected to match 100% of both the human and the RM sequences (www.ncbi.nlm.nih.gov; www.macaque.org). The RM MCP2 and MCP3 sequences are not available, and therefore, the primer sequence is based on the human sequence. RT-PCR was performed with 100 ng of total RNA, using a one-step RT-PCR kit (QIAGEN), according to previously published procedures (23). Quantitative real-time RT-PCR analysis was performed using a SYBR green system (catalog no. ABI7900HT; Applied Biosystems, Foster City, CA). Data were normalized to the level of β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each individual sample.
Statistical analysis.
A two-tailed, two-sample Student t test was used to calculate P values for differences in means between groups. The data were expressed as means ± standard errors (SE) of the means. For statistical inference, a P value of <0.05 was considered significant.
RESULTS
HIV-mediated apoptosis in PBMC.
We investigated whether HIV- or SIV-related apoptosis occurs in PBMC cultures from species that are not susceptible to AIDS, in the presence or absence of T-cell activation induced by exposure to SEB. Significantly lower levels of apoptosis were observed with chimpanzee CD4+ T cells exposed to HIVBal than with human CD4+ T cells (Fig. 1a). Similarly, although the difference was of a magnitude lower than that between human and chimp cells, significantly lower levels of apoptosis were observed for SEB-activated AGM and SM CD4+ T cells exposed to SIVmac316 than for SEB-activated RM CD4+ T cells (Fig. 1b). Rates of infection, measured on day 7 by p24 or p27 ICS, were within twofold when percentages of infected human PBMC were compared to those of chimp PBMC or when the comparison was carried out among the three other nonhuman primates. Levels of 24 or p27 in the supernatants, measured by ELISA, were comparable in the infected cultures (Fig. 1). These results were confirmed using viral strains with different tropism such as HIVBal or SIVmac316 in the different nonhuman primate PBMC and HIVSF2, HIVHXB2, and HIVNL4-3 in human cells (data not shown). Similar results were also observed with nonhuman primate PBMC with species-specific isolates such as SIVSME041, SIVagm(sab-2), and SIVrcmGABI (data not shown). These results indicated that the PBMC culture model of HIV- and SIV-induced CD4+ T-cell death would provide an experimental system to further investigate the mechanisms that lead to different levels of virus-mediated apoptosis in CD4+ T cells from species that are differentially susceptible to AIDS.
Because apoptosis in HIV-exposed PBMC populations has been shown to occur in both infected and uninfected CD4+ T cells (15), we reasoned that soluble factors such as cytokines could play a role in the sensitization of the uninfected cells. We used cytokine assays to determine whether there were differences between the protein levels in the supernatants of human and those in chimpanzee PBMC exposed to infectious HIV (Table 1). Of the 44 cytokines measured in these assays, only TRAIL was present at a considerably higher level in the media of HIV-exposed activated human PBMC than in HIV-exposed chimpanzee PBMC (Table 1). Other cytokines were produced at the same level in the cultures from the two species or at different levels in human and chimpanzee cultures, but these differences were not HIV specific, as they were also observed with activated cells in the absence of viral infection. Increased expression of sTRAIL was observed with at least three independent human and chimpanzee cultures (Fig. 1c) and with different viral strains (not shown). The differential TRAIL expression obtained with activated HIV-exposed human PBMC and chimpanzee PBMC was recapitulated, even if to a lesser extent, between activated SIV-exposed RM compared to that of AGM and SM PBMC (Fig. 1c). TRAIL was present at significantly higher levels in the supernatant of SIV-exposed RM PBMC than in uninfected or SEB-treated RM PBMC. The difference in TRAIL production observed for the supernatant of SIV-exposed AGM or SM PBMC compared to mock-infected or SEB-treated samples was not statistically significant (Fig. 1c).
TABLE 1.
Changes in cytokine induction in the supernatant of human and chimpanzee PBMC infected with strains HIVHXB2 and HIVBala
| Gene product | Fold-induction (X4-tropic HIV)
|
Fold-induction (R5-tropic HIV)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human
|
Chimpanzee
|
Human
|
Chimpanzee
|
|||||||||
| HIV | SEB | HIV/SEB | HIV | SEB | HIV/SEB | HIV | SEB | HIV/SEB | HIV | SEB | HIV/SEB | |
| GCSF | 1.3 | 1.6 | 2.0 | 1.1 | 1.3 | 0.9 | 1.1 | 1.2 | 1.0 | 1.3 | 1.8 | 1.5 |
| ICAM1 | 1.3 | 1.7 | 1.6 | 0.8 | 1.2 | 0.7 | 1.0 | 1.0 | 1.3 | 1.3 | 2.1 | 1.8 |
| IL-1β | 2.1 | 4.3 | 4.6 | 2.2 | 1.2 | 1.7 | 2.0 | 1.8 | 2.3 | 2.1 | 2.6 | 3.1 |
| IL-3 | 1.0 | 6.3 | 4.6 | 1.1 | 1.3 | 1.7 | 1.1 | 1.2 | 1.3 | 1.1 | 1.3 | 1.3 |
| IL-6 | 1.3 | 2.2 | 2.3 | 1.0 | 1.0 | 1.3 | 1.2 | 1.4 | 1.4 | 1.2 | 1.1 | 1.2 |
| IL-7 | 1.8 | 1.3 | 1.4 | 1.1 | 1.1 | 1.1 | 1.8 | 2.3 | 2.3 | 1.3 | 1.1 | 1.3 |
| IL-8 | 1.1 | 1.4 | 1.5 | 1.0 | 1.0 | 1.0 | 1.6 | 2.2 | 2.1 | 1.1 | 1.1 | 1.2 |
| IL-10 | 1.4 | 1.2 | 1.9 | 2.2 | 1.8 | 2.3 | 1.6 | 1.8 | 1.9 | 1.0 | 1.6 | 1.7 |
| IL-11 | 1.1 | 1.5 | 2.1 | 1.2 | 1.1 | 0.9 | 1.1 | 0.9 | 1.1 | 1.2 | 1.6 | 1.6 |
| IL-13 | 1.2 | 6.3 | 7.3 | 1.0 | 1.1 | 1.0 | 2.4 | 3.2 | 3.3 | 1.2 | 1.7 | 1.4 |
| IL-15 | 1.1 | 1.9 | 2.4 | 0.9 | 1.2 | 1.4 | 1.1 | 1.0 | 1.1 | 1.4 | 1.9 | 1.7 |
| IL-17 | 1.0 | 1.9 | 2.5 | 1.0 | 0.8 | 0.7 | 1.0 | 1.1 | 1.2 | 1.3 | 2.0 | 1.9 |
| IP10 | 0.9 | 2.1 | 2.5 | 1.7 | 1.3 | 1.7 | 1.1 | 1.3 | 1.4 | 0.6 | 1.1 | 0.8 |
| MCP2 | 2.0 | 2.2 | 2.6 | 1.1 | 0.8 | 0.7 | 1.0 | 0.8 | 0.8 | 1.0 | 1.0 | 0.9 |
| MIG | 1.0 | 1.4 | 2.5 | 1.1 | 1.1 | 0.7 | 0.9 | 1.0 | 1.2 | 1.5 | 1.6 | 1.5 |
| MIP1α | 2.3 | 3.5 | 4.1 | 0.8 | 0.9 | 0.6 | 2.6 | 2.0 | 2.5 | 1.4 | 2.0 | 2.2 |
| MIP1δ | 1.0 | 1.9 | 2.6 | 0.7 | 1.0 | 0.4 | 1.5 | 1.4 | 1.4 | 1.0 | 1.6 | 1.4 |
| RANTES | 2.8 | 3.9 | 4.1 | 1.0 | 1.1 | 1.0 | 1.0 | 1.1 | 1.1 | 1.1 | 1.8 | 1.4 |
| TGFβ1 | 0.6 | 1.3 | 1.4 | 1.1 | 1.2 | 1.3 | 1.2 | 1.1 | 1.2 | 1.3 | 2.0 | 2.0 |
| TNF-α | 2.5 | 2.4 | 2.7 | 2.0 | 2.5 | 2.0 | 1.7 | 2.4 | 2.1 | 0.9 | 2.1 | 1.7 |
| TNFβ | 1.1 | 1.3 | 1.8 | 1.8 | 5.6 | 4.1 | 1.3 | 2.3 | 2.5 | 1.3 | 2.4 | 2.1 |
| sTNFRII | 1.7 | 1.4 | 1.8 | 1.5 | 0.8 | 1.4 | 1.2 | 2.1 | 2.1 | 1.0 | 1.3 | 1.0 |
| PDGF-BB | 1.1 | 1.0 | 1.6 | 1.7 | 1.0 | 1.5 | 1.1 | 1.1 | 1.1 | 1.6 | 2.0 | 1.9 |
| TIMP2 | 1.2 | 0.7 | 0.9 | 5.4 | 0.9 | 4.5 | 2.0 | 1.3 | 2.1 | 0.8 | 1.1 | 1.1 |
| CD95L | 1.7 | 2.5 | 2.5 | 1.5 | 1.7 | 2.0 | 1.8 | 2.0 | 2.2 | 1.3 | 1.3 | 1.4 |
| IFNα | 7.4 | 3.1 | 9.4 | 4.4 | 3.2 | 9.0 | 6.2 | 2.5 | 6.9 | 6.9 | 3.2 | 5.0 |
| TRAIL | 3.2 | 1.5 | 4.3 | 1.3 | 0.9 | 1.1 | 4.7 | 1.9 | 6.7 | 1.7 | 1.6 | 2.0 |
The n-fold induction is calculated using the values obtained from supernatants from mock-infected cultures as the denominator. The cytokine amount in each sample was determined by evaluating the intensity of the antibody signal produced on the membranes with a scanner. Supernatants from two independent donors were evaluated. Cytokines for which a twofold or higher induction was observed in any of the experimental samples are reported. The following cytokines were also screened and their induction was lower than twofold in all the experimental conditions, as follows: eotaxin, eotaxin 2, GM-CSF, IFN-γ, I309, interleukin 1α (IL-1α), IL-2, IL-4, IL-6sR, IL-12p40, IL-12p70, IL-16, MCP1, MCSF, soluble tumor necrosis factor R1 (sTNFRI). The last four cytokines were measured by ELISA in four independent samples, and the number represents the change in induction of the average of the four samples. IFN-β was undetectable in all cultures. The differences in the production of TRAIL (see also Fig. 1c) were statistically significant (P < 0.05 and P < 0.041 for the HIVHXB2 infection experiment; P < 0.045 and P < 0.01 for the HIVBal infection experiment). The differences in IFN-α production were not statistically significant. Averages ± standard deviations (SD) for values of IFN-α production in the HIVHXB2 experiment were 3.3 ± 3 pg/ml (mock), 10.1 ± 1.8 (SEB); 24 ± 10 (HIV), 30.5 ± 12 (SEB plus HIV) for human PBMC; 3.3 ± 3 pg/ml (mock), 13.9 ± 5 (SEB); 27.3 ± 10.8 (HIV), 38.9 ± 11.8 (SEB plus HIV) for chimpanzee PBMC. Averages ± SD for values of IFN-α production in the HIVBal experiment were 4.4 ± 3 pg/ml (mock), 11 ± 1.4 (SEB); 28.6 ± 10 (HIV), 30.4 ± 6.2 (SEB plus HIV) for human PBMC; 4.3 ± 3 pg/ml (mock), 13.9 ± 5 (SEB); 29.9 ± 7.3 (HIV), 21.3 ± 6.9 (SEB plus HIV) for chimpanzee PBMC. It is possible that the reactivity of human monoclonal antibodies against the chimpanzee cytokines may be lower than against the human cytokines, and this may result in an underestimate of the chimpanzee cytokines, with some of the above chimpanzee cytokines being more abundant than we observed. Additional differences may be detected when chimpanzee reagents become available.
TRAIL can be present both as a soluble and as a membrane-bound factor, so we measured the percentage of human PBMC expressing mTRAIL and identified their phenotype. We found that the percentage of mTRAIL+ cells was considerably higher in SEB/HIV-exposed human PBMC than in SEB-exposed human PBMC (P < 0.05) (Fig. 2a). The percentage of mTRAIL+ cells was also considerably higher in SEB/HIV-exposed human PBMC than in chimpanzee PBMC (P < 0.04) (Fig. 2a). Increased numbers of mTRAIL+ cells could be found among human SEB/HIV-exposed CD4+ T cells, CD14+ monocytes, and CD11c/HLA-DR+ iDC, and the differences between these numbers and those observed after SEB stimulation were statistically significant (on day 4, P < 0.03, P < 0.017, and P < 0.0037, respectively; on day 10, P < 0.045, P < 0.037, and P < 0.01, respectively) (Fig. 2b).
FIG. 2.
TRAIL and TRAIL death receptor (DR4 and DR5) expression in HIV- or SIV-exposed PBMC. (a) Percentage of mTRAIL+ PBMC in PBMC from human (closed symbols) and chimpanzee (open symbols) PBMC during 10 days of culture. The data represent the means ± standard errors (SE) of five experiments. (b) Percentage of human CD4+ T cells, monocytes (CD14+), and iDC (CD11c/HLA-DR+) expressing mTRAIL after HIV infection within each population in the total PBMC cultures. The data represent the means ± SE of three experiments. (c) Analysis of TRAIL receptors DR4 and DR5 in human and chimpanzee PBMC infected with HIV. Mean percentages ± SE of cells expressing the receptor are reported for total PBMC and for CD4+ T cells present in the PBMC culture (data derived from three independent donors). (d) Effect of antibodies against TRAIL and TRAIL receptors on HIV-induced PBMC apoptosis, showing the percentage of apoptotic human CD4+ T cells present on day 7 of culture under different conditions. Five experiments were carried out, and the average percentages of apoptosis observed for the mock-treated samples were subtracted from the other experimental values. (e) Effect of antibodies against CD95, CD95L, IFN-α, and IFN-αR on HIV-induced CD4+ T-cell apoptosis (mean percentages of three independent samples). Rates of HIV or SIV infection shown by ICS on day 7 in all the virus-infected cultures were comparable.
Linking TRAIL expression to CD4+ T-cell apoptosis.
It is possible that increased expression of TRAIL death receptors in human PBMC compared to that of chimpanzee PBMC may account for the higher levels of CD4+ T-cell apoptosis observed with the human PBMC culture model presented here (Fig. 1). We sought to determine whether there is an increase in the number of CD4+ T cells expressing death receptors in HIV-infected PBMC cultures. Ten days after infection, we found that CD4+ T cells expressing both DR4 and DR5 receptors were present in activated, HIV-infected PBMC cultures at substantially higher levels than that observed with activated, uninfected PBMC (Fig. 2c). No significant differences were observed 4 days after infection. Significant expression of DR4 and DR5 TRAIL receptors was not detected under similar conditions in chimpanzee cultures. DR4 and DR5 expression, possibly induced by immunoactivation, exposure to viral products, noninfectious virions, or soluble factors released by infected cells, was detected on both p24+ and p24− cells, suggesting that TRAIL could also mediate apoptosis in uninfected cells. These results provide an explanation for the increased susceptibility of HIV-infected PBMC to TRAIL-mediated apoptosis.
We evaluated the impact of TRAIL-mediated apoptosis on the human PBMC cultures by using antibodies against sTRAIL and DR4 and DR5, the TRAIL receptors that mediate apoptosis. When human PBMC cultures were exposed to HIV-1HXB2 and simultaneously treated with anti-TRAIL, anti-DR4, and anti-DR5 antibodies, the levels of observed apoptosis decreased substantially (Fig. 2d). The differences between the levels of apoptosis observed for HIV-infected PBMC compared to those of the same culture treated with the three antibodies were statistically significant (P < 0.03). When a similar experiment was carried out in the presence of antibodies against CD95 and CD95L, we observed some reduction of apoptosis for HIV-infected human PBMC (approximately 30%; Fig. 2e), but the effect was not as impressive as the treatment with antibodies against TRAIL and TRAIL receptors, which reduced the apoptosis observed with PBMC treated with SEB and infected by HIV by 50 to 60%. A similar reduction was observed for the SEB-treated sample. Furthermore, CD95L was produced at similar levels in human and chimpanzee infected PBMC and after cell activation (Table 1). Therefore, it is possible that CD95-mediated apoptosis is more directly linked to cell activation than to HIV infection.
It has been suggested that interferons, and more specifically IFN-α produced by plasmacytoid DC (pDC), are critical to the more pronounced and chronic immunoactivation observed for species that develop AIDS (46, 47). In this culture model of HIV or SIV infection, we found that the levels of IFN-α produced within the first 10 days of infection were not significantly different among AIDS-susceptible and non-AIDS-susceptible species (Table 1 and Fig. 1c), yet the rates of TRAIL production and apoptosis were. Furthermore, when antibodies against IFN-α and type I IFN receptor were used in human PBMC cultures since the day of infection, no significant reduction in apoptosis was observed (Fig. 2e). TRAIL production was present at similar levels in both treated cultures and cultures not treated with antibodies against IFN-α and type I IFN receptor (data not shown). Because neither IFN-β RNA nor protein was detectable in these PBMC cultures, its inhibition was not pursued. IFN-γ was produced at similar levels in stimulated and/or infected cultures from all species, suggesting that this cytokine may not be responsible for the significantly higher production of TRAIL observed after HIV infection for human and RM cultures (Fig. 1c). As IFN-α is present in these cultures, these results do not exclude the possibility of some contribution of IFN-α to TRAIL induction and associated apoptosis. However, the presence of significantly different levels of TRAIL in human and rhesus macaque infected cultures and similar levels of IFN-α in all primate cultures indicate that IFN-α alone is not the only factor contributing to the differential TRAIL expression.
Taken together, these results indicate that TRAIL-mediated apoptosis occurs in activated, HIV-exposed human PBMC or activated, SIV-exposed RM PBMC but is significantly lower in chimpanzee, AGM, and SM PBMC. Thus, enhanced TRAIL production is associated with virus infection in cells from AIDS-susceptible primates and may play a significant role in the decline of CD4+T cells observed with HIV-infected human cells.
In vivo evaluation of TRAIL, DR4, and DR5 expression.
Having observed increased expression of TRAIL after HIV infection of human PBMC and SIV infection of macaque PBMC, we investigated whether increased TRAIL expression also occurs in vivo. We measured TRAIL expression in uninfected and SIV-infected RM and SM (Fig. 3b). We found significant differences between the serum TRAIL levels of SIV-infected RM relative to those of the uninfected animals (234 ± 43 pg/ml in SIV-infected compared to 41 ± 23 pg/ml in noninfected animals; P = 1.72 × 10−8). These differences were not apparent in SM (68 ± 29 pg/ml in SIV-infected compared to 84 ± 33 pg/ml in noninfected animals). The differences between the levels of TRAIL in the serum of SIV-infected RM and those in SIV-infected SM were statistically significant (P = 3.35 × 10−5) (Fig. 3b). Furthermore, when we evaluated mTRAIL expression in PBMC derived directly from the animals, the percentages of mTRAIL+ PBMC and mTRAIL+ CD4+ T cells were significantly higher in SIV-infected RM than in noninfected RM (P = 0.033 for PBMC, P = 0.0087 for CD4+ T cells) or SIV-infected SM (P = 0.0046 for PBMC, P = 0.0017 for CD4+ T cells) (Fig. 3c). SIV-infected RM also showed a higher percentage of DR4+ CD4+ T cells than noninfected RM (P = 0.0084) or than SIV-infected SM (P = 0.0011) (Fig. 3d). The percentage of DR5+ CD4+ T cells was also higher in SIV-infected RM than in the other groups, but this difference compared to that of noninfected RM was not significant. The expression of these death receptors was almost entirely restricted to the CD4+ T cells in the PBMC. A positive correlation could be found between TRAIL levels in the serum and RNA viral loads (R2 = 0.364; P < 0.01) (Fig. 3e) but not between TRAIL levels and CD4+ T-cell counts (not shown). TRAIL has been shown to trigger apoptosis in T cells from HIV-infected individuals, while T cells from uninfected controls are resistant to TRAIL-induced apoptosis (26). Combined with similar evaluations carried out in humans (18, 26) and in the SCID-hu mouse system (32), these data confirm that differential expression of TRAIL and its death receptors occurs during infection in vivo, when a species that progresses to AIDS is compared to a species that is resistant to disease. These results support a model where increased expression of TRAIL in vivo causes the accelerated depletion of CD4+ T cells that is the hallmark of AIDS.
FIG. 3.
In vivo TRAIL, DR4, and DR5 expression analysis. (a) Detection of soluble TRAIL in supernatants of nonhuman primate CD14+ cells by a human TRAIL ELISA, showing means ± standard errors of supernatant levels after rIFN-α stimulation of three PBMC-derived CD14+monocyte samples. (b) sTRAIL serum levels in SIV-infected and noninfected RM and SM samples. These samples were obtained from 20 macaques sampled multiple times before and after infection with SIVmac251 and from 74 SIV-positive and 35 SIV-negative SM. The SIV-positive SM were naturally infected, with the exception of two animals experimentally infected with SIVmac239. (c) Percentages of mTRAIL+ cells in PBMC and CD4+ T cells obtained from SIV-infected and noninfected RM and SM. (d) Percentages of DR4 and DR5+ cells in CD4+ T cells obtained from SIV-infected and noninfected RM and SM. (e) Correlation between SIV RNA viral loads in RM and serum TRAIL levels.
Response to viral infection and Tat expression in primate iDC.
As TRAIL expression was elevated among CD14+ and CD11c/HLA-DR+ cells in the PBMC cultures, we further studied the differential response to infection in primate iDC and macrophages from various species. We recently studied the response of human iDC to HIV infection in some detail and found that TRAIL is among a small set of genes induced by HIV-1 and its Tat transactivator in these cells (23), so we focused our initial comparative studies on human and chimpanzee iDC.
We used DNA arrays to determine the effects of HIV-1 and Tat on the human and chimpanzee iDC gene expression program. Affymetrix DNA microarrays based on human sequences were used for these experiments. The high level of identity between human and chimpanzee gene sequences (99.5% identity) is sufficient to permit the use of human arrays for analysis of gene expression in chimpanzee RNA (11). Primary human and chimpanzee iDC were generated from precursors in peripheral blood by culturing CD14+ monocytes with interleukin 4 and GM-CSF for 6 to 7 days (3, 29, 42). To identify iDC genes whose expression was affected by HIV-1 Tat with high confidence, we studied the effects of both Tat expression and HIV-1 infection independently, and we determined these effects at multiple points over a time course.
Primary iDC were infected with HIV-1Bal or exposed to control medium, aliquots were taken at 0, 10, and 14 days, and target RNA was prepared for microarray analysis. The progress of the infection in the iDC population was monitored by reverse transcriptase activity in the supernatant and the Tat mRNA levels (Fig. 4a and b). Furthermore, p24 ICS was carried out on a separate aliquot of cells on day 14 and results were comparable (55% and 59% in infected human and chimp cells, respectively). Aliquots of cells exposed to uninfected macrophage-derived medium (the HIV-1Bal stock used in the infection was produced in macrophages) were also taken during a similar time course, and their corresponding RNAs provided negative controls for each time point.
FIG. 4.
Gene expression profiles of human and chimpanzee iDC infected with HIVBal or expressing Tat. (a) Virus production in infected iDC measured by RT activity. (b) Detection of Tat expression by real-time RT-PCR in HIV-infected, SIV-infected, and Adeno-Tat-expressing primate iDC. Cycle threshold values in all samples were normalized by GAPDH gene expression, and the Tat amount in the day-14 human iDC HIV-infected sample was assigned an arbitrary value of 1. The Tat amount in other samples is expressed as a fraction of the human iDC HIV-infected sample. (c) Venn diagrams illustrate the number of genes modulated in different culture conditions and how the numbers overlap in the two species. (d) The left panel shows profiles of genes upregulated in human iDC infected with Adeno-Tat and with HIV (see Table S2a in the supplemental material). The gene expression profile of the same genes in chimpanzee cells is also reported. IFN-inducible genes are listed in red. The right panel shows gene expression profiles of genes downregulated in human iDC infected with HIVBal and Adeno-Tat (see Table S2b in the supplemental material). The gene expression profile of the same genes in chimpanzee cells is also reported. The color scale is shown below the panels.
We used an algorithm for identifying differentially expressed genes (23) to compare the responses of human and chimpanzee iDCs to HIV infection (Fig. 4c). Of the 1,071 genes induced by HIV infection of human iDCs, only 32 were also induced in chimpanzee iDCs. TRAIL RNA induction was observed in the infected human but not in the infected chimpanzee iDCs. The numbers of genes downregulated by infection in cells from both species were also small. (For the list of genes whose expression was modified in these experiments, see Tables S1a to S1d in the supplemental material.) We next investigated what portions of the modified gene expression programs in iDCs might be induced by Tat alone by using an Adeno-Tat gene transfer system (6) to achieve Tat gene expression. Aliquots of cells infected with Adeno-LacZ (control) or Adeno-Tat were harvested at 0, 10, 20, and 30 h, and RNA was isolated, labeled, and prepared for microarray analysis. Tat mRNA could be detected in iDC in all Adeno-Tat-infected samples (not shown). Based on ICS, approximately 61% of the human cells and 57% of the chimp cells expressed Tat at 30 h after Adeno-Tat infection. Of the 57 genes induced by both HIV infection and Tat expression in human iDCs, none was similarly induced in chimpanzee iDCs (Fig. 4c and d). TRAIL was among the genes that were induced both by HIV infection and Tat expression in human iDCs, together with a spectrum of IFN-inducible genes (indicated in red in Fig. 4d), confirming results of our previous study (23). The expression of most of these genes was not modulated in chimp iDC. The use of DNA arrays with a larger number of probes accounts for the additional genes found modulated in this study compared to those reported in reference 23. Among the 107 genes downregulated by both HIV infection and Tat expression in human iDCs, only 6 were also downregulated in chimpanzee iDCs (Fig. 4c and d). (For the list of genes whose expression was similarly modified in these experiments by HIV and Adeno-Tat, see Tables S2a to S2d in the supplemental material.)
If TRAIL induction plays a role in the progression to AIDS, we would expect to observe TRAIL induction in RM iDCs but not in SM or AGM iDCs exposed to SIV. We used real-time RT-PCR to investigate the expression profiles of TRAIL and six additional interferon-inducible gene products (IRF-7, STAT1, MCP-2, MCP3, IP-10, and MIG) in at least two independent donors from chimpanzees, SM, RM, and AGM iDCs over a time course of SIV infection or Tat expression (Fig. 5). Virus production and Tat mRNA expression were comparable in all the cultures (Fig. 5a and b). ICS to detect p24 or p27 on day 14 postinfection varied between 50 and 67% of cells among all reported samples (samples below 50% were excluded from further analysis). ICS to detect Tat on samples obtained 30 h after Adeno-Tat infection ranged between 58 and 71%. We found that each of these genes was induced in human and RM iDCs but not in iDCs from chimpanzees, SM, and AGM exposed to SIV (Fig. 5c). Similar results were obtained when Tat was expressed in iDCs from humans, chimpanzees, SM, and RM (Fig. 5c). The induction of the chemokines and TRAIL in humans and RM but the lack of it in the other nonhuman primate species was also confirmed with three donors at the protein level (Fig. 5d). In the iDC cultures infected with HIV or Adeno-Tat, type I IFN RNAs and proteins were below the level of detection (not shown), suggesting the possibility that the induction of these genes was IFN independent.
We conclude that TRAIL induction is a common feature of iDCs exposed to HIV or SIV when these cells are derived from AIDS-susceptible but not AIDS-resistant primate species. Furthermore, Tat expression alone is capable of stimulating this response in cells from AIDS-susceptible primate species.
Response to viral infection and Tat expression in primate macrophages.
Because both monocytes and myeloid iDC cells were among the mTRAIL-positive cells in HIV-infected human PBMC cultures (Fig. 2b), we next investigated the expression of TRAIL in HIV- or SIV-infected macrophages derived from CD14+ cells from AIDS-susceptible and AIDS-resistant primates. Virus production and Tat expression were comparable among the different cultures (Fig. 6a). The percentage of p24 or p27 positive cells detected by ICS on samples obtained on day 14 postinfection varied between 55 and 64% among all samples. The steady-state RNA levels of some of the IFN-inducible genes that were activated in iDC were evaluated by RT-PCR in RNA from HIV-infected and uninfected human macrophages, and the results obtained were similar to those described for iDCs (not shown). In particular, both TRAIL and chemokine genes were among those induced by HIV infection. We confirmed by RT-PCR and ELISA that TRAIL RNA and protein levels were induced in HIV-infected human macrophages derived from multiple independent human PBMC samples (Fig. 6b and c, left panels).
We then compared the effects of viral exposure and Tat expression on TRAIL induction in macrophages from humans, chimpanzees, RM, AGM, and SM. Tat ICS in samples obtained 30 h after Adeno-Tat infection ranged between 65 and 76%. We found that viral exposure or Tat expression led to increased TRAIL mRNA and protein levels in macrophages from humans and RM but not from chimpanzees, AGM, and SM (Fig. 6b and c, right panels). No IFN-α expression could be detected in these cells by real-time RT-PCR. Thus, TRAIL induction is a common feature of both macrophages and iDCs exposed to virus when these cells are derived from AIDS-susceptible, but not AIDS-resistant primate species.
Taken together, our results indicate that IFN-related genes, and among these the apoptotic factor TRAIL, are upregulated in iDC and macrophages from primates that progress to AIDS after infection with HIV or SIV or after Tat expression. This gene modulation does not occur in chimpanzee, SM, and AGM myeloid antigen-presenting cells (APC).
DISCUSSION
We have explored the effects of HIV-1 and its Tat transactivator on PBMC and on antigen-presenting cells such as myeloid iDC and macrophages from AIDS-susceptible and AIDS-resistant species. We found that expression of a subset of IFN-responsive genes, and among these TRAIL, occurs in APC from human and RM cells but not from chimpanzee, SM, and AGM cells. Differential TRAIL expression was confirmed in vivo when infected SM were compared to infected RM.
The mechanisms that have been proposed to account for HIV-mediated destruction of CD4+ T cells include direct cytopathic effects and apoptosis. The viral gp120 protein and the host proteins CD4 and CXCR4 contribute to cell death via syncytium formation. The viral and host factors that contribute to AIDS pathogenesis in humans and RM through their effects on apoptosis are numerous. Tat, Vpr, Vpu, Nef, and gp120 have all been reported as possible mediators of apoptosis. The intrinsic and extrinsic apoptotic pathways are engaged by these HIV proteins (15). Both the Fas/Fas ligand and the TRAIL pathways have been shown to be activated during HIV-induced apoptosis, although evidence for the involvement of the Fas/Fas ligand pathway appears to be controversial (2, 13, 26, 50). We did not observe higher levels of Fas/Fas ligand-mediated apoptosis for human and RM cells than for cells from AIDS-resistant species. Instead, our evidence implicates TRAIL as an important mediator of apoptosis in cells from AIDS-susceptible species.
TRAIL has been shown to trigger apoptosis in T cells from HIV-infected individuals, while T cells from uninfected controls are resistant to TRAIL-induced apoptosis (26). Elevated levels of TRAIL have been observed in serum samples from HIV-infected patients compared to those of uninfected subjects (18), and higher numbers of TRAIL+ cells were observed with the tonsils of HIV+ progressors than with those of HIV-infected nonprogressors (21), providing in vivo evidence consistent with our results in the human PBMC culture model. Blocking of TRAIL in a SCID mouse model reconstituted with human PBMC and infected with HIV also resulted in significantly reduced apoptosis (32). These and additional studies linked TRAIL to HIV-induced apoptosis in vivo and in vitro in humans (18, 26, 31, 32, 55). We found that the selective induction of TRAIL and its DR4 and DR5 receptors also occurs in vivo in RM, which like humans, are susceptible to AIDS, but not in SM, an AIDS-resistant species.
Both HIV-1 infection and Tat expression in myeloid iDC and macrophages caused increased expression of a subset of the IFN-inducible genes whose expression is usually affected by interferons. mTRAIL expression was also observed with CD4+ T cells exposed to HIV. Other reports indicated that TRAIL production during HIV infection is IFN-α dependent (20). In our experiments, IFN-α or IFN-β could not be detected in the supernatants of infected iDC and macrophages, indicating that activation of the IFN-inducible genes in these cells might be type I IFN independent. The ability of HIV and SIV proteins like Tat to stimulate TRAIL production in the cells of some species but not others, as our data suggest, argues that increased TRAIL expression in infected myeloid cells may contribute to mediating elevated levels of CD4+ T-cell apoptosis in humans and RM but not in the other species. This finding does not rule out the contribution of additional host factors and viral proteins in this process, and the in vivo contribution of TRAIL-mediated apoptosis to the CD4+ T-cell decline relative to that induced by other factors remains to be determined.
More TRAIL+ and IFN-α+ cells have been detected in tonsils from HIV-infected progressors than in nonprogressors (21, 28). There are substantial virological differences between human nonprogressors and infected AIDS-resistant species (49). AIDS-resistant species have viral loads comparable to progressors, but in these species, TRAIL levels are low and do not correlate with their viral loads. It is possible that their infected pDC produce less IFN-α than humans or RM, but the finding that their purified iDC and macrophages do not produce TRAIL after infection, while human and rhesus MDC and macrophages do so, points to an important difference between AIDS-resistant and AIDS-susceptible species, one that is independent of possible differences in the production of IFN-α among these species. Myeloid DC are abundant in skin and mucosal tissues where the influence of pDC, usually found in lymphoid tissues, and IFN-α may be limited. Considering the high levels of viral replication and CD4+ T-cell depletion in the gastrointestinal mucosa and the significant number of iDC and macrophages residing there that can also be targeted by the infection, the host-pathogen interactions occurring in myeloid APCs may contribute to AIDS progression in a significant way.
Enhanced immunoactivation has been reported as one of the major differences between AIDS-susceptible and AIDS-resistant species (8, 16, 43, 47). The interferon mimicry produced by Tat in human and RM monocytes, macrophages, and myeloid iDC, which activates a subset of IFN-inducible genes that include specific chemokines, could contribute to the higher levels of immunoactivation in these species. In addition to its role in inducing apoptosis, TRAIL itself can enhance T-cell proliferation after T-cell receptor engagement and signal the augmentation of IFN-γ secretion (7).
Our evidence that TRAIL can be induced by HIV Tat suggests that pharmacological inhibition of Tat might prevent disease progression in HIV-infected individuals by multiple mechanisms. In addition to inhibiting this essential viral transactivator, reduced TRAIL production may facilitate preservation of CD4+ T cells, as has been shown in the hu-SCID mouse (32).
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI74011 and AI 72803-01.
We thank Cristian Apetrei (Tulane University), Amintider Kaur (NERPRC), H. M. McClure, Jim Else, and other personnel of the Yerkes Primate Center for providing the nonhuman primate sera and PBMC, Preston A. Marx (Tulane National Primate Research Center, grant RR00164) for providing the SIVSME041 viral strain, and Jennifer Love (Whitehead Institute) for technical assistance with the array hybridization experiments.
Footnotes
Published ahead of print on 9 May 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ameisen, J. C., and A. Capron. 1991. Cell dysfunction and depletion in AIDS: the programmed cell death hypothesis. Immunol. Today 12:102-105. [DOI] [PubMed] [Google Scholar]
- 2.Baumler, C. B., T. Bohler, I. Herr, A. Benner, P. H. Krammer, and K. M. Debatin. 1996. Activation of the CD95 (APO-1/Fas) system in T cells from human immunodeficiency virus type-1-infected children. Blood 88:1741-1746. [PubMed] [Google Scholar]
- 3.Bender, A., M. Sapp, G. Schuler, R. M. Steinman, and N. Bhardwaj. 1996. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J. Immunol. Methods 196:121-135. [DOI] [PubMed] [Google Scholar]
- 4.Bostik, P., A. E. Mayne, F. Villinger, K. P. Greenberg, J. D. Powell, and A. A. Ansari. 2001. Relative resistance in the development of T cell anergy in CD4+ T cells from simian immunodeficiency virus disease-resistant sooty mangabeys. J. Immunol. 166:506-516. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, A. H., H. F. Tsai, L. L. Lin, S. L. Hsieh, P. I. Hsu, and P. N. Hsu. 2001. Enhanced proliferation and increased IFN-gamma production in T cells by signal transduced through TNF-related apoptosis-inducing ligand. J. Immunol. 167:1347-1352. [DOI] [PubMed] [Google Scholar]
- 8.Cotton, M. F., D. N. Ikle, E. L. Rapaport, S. Marschner, P. O. Tseng, R. Kurrle, and T. H. Finkel. 1997. Apoptosis of CD4+ and CD8+ T cells isolated immediately ex vivo correlates with disease severity in human immunodeficiency virus type 1 infection. Pediatr. Res. 42:656-664. [DOI] [PubMed] [Google Scholar]
- 9.Davis, I. C., M. Girard, and P. N. Fultz. 1998. Loss of CD4+ T cells in human immunodeficiency virus type 1-infected chimpanzees is associated with increased lymphocyte apoptosis. J. Virol. 72:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehret, A., M. O. Westendorp, I. Herr, K. M. Debatin, J. L. Heeney, R. Frank, and P. H. Krammer. 1996. Resistance of chimpanzee T cells to human immunodeficiency virus type 1 Tat-enhanced oxidative stress and apoptosis. J. Virol. 70:6502-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enard, W., P. Khaitovich, J. Klose, S. Zollner, F. Heissig, P. Giavalisco, K. Nieselt-Struwe, E. Muchmore, A. Varki, R. Ravid, G. M. Doxiadis, R. E. Bontrop, and S. Paabo. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296:340-343. [DOI] [PubMed] [Google Scholar]
- 12.Estaquier, J., T. Idziorek, F. de Bels, F. Barre-Sinoussi, B. Hurtrel, A. M. Aubertin, A. Venet, M. Mehtali, E. Muchmore, P. Michel, et al. 1994. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc. Natl. Acad. Sci. USA 91:9431-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estaquier, J., M. Tanaka, T. Suda, S. Nagata, P. Golstein, and J. C. Ameisen. 1996. Fas-mediated apoptosis of CD4+ and CD8+ T cells from human immunodeficiency virus-infected persons: differential in vitro preventive effect of cytokines and protease antagonists. Blood 87:4959-4966. [PubMed] [Google Scholar]
- 14.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gougeon, M. L. 2003. Apoptosis as an HIV strategy to escape immune attack. Nat. Rev. Immunol. 3:392-404. [DOI] [PubMed] [Google Scholar]
- 16.Gougeon, M. L., H. Lecoeur, A. Dulioust, M. G. Enouf, M. Crouvoiser, C. Goujard, T. Debord, and L. Montagnier. 1996. Programmed cell death in peripheral lymphocytes from HIV-infected persons: increased susceptibility to apoptosis of CD4 and CD8 T cells correlates with lymphocyte activation and with disease progression. J. Immunol. 156:3509-3520. [PubMed] [Google Scholar]
- 17.Heeney, J., W. Bogers, L. Buijs, R. Dubbes, P. ten Haaft, W. Koornstra, H. Niphuis, P. Nara, and V. Teeuwsen. 1996. Immune strategies utilized by lentivirus infected chimpanzees to resist progression to AIDS. Immunol. Lett. 51:45-52. [DOI] [PubMed] [Google Scholar]
- 18.Herbeuval, J. P., A. Boasso, J. C. Grivel, A. W. Hardy, S. A. Anderson, M. J. Dolan, C. Chougnet, J. D. Lifson, and G. M. Shearer. 2005. TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected patients and its in vitro production by antigen-presenting cells. Blood 105:2458-2464. [DOI] [PubMed] [Google Scholar]
- 19.Herbeuval, J. P., J. C. Grivel, A. Boasso, A. W. Hardy, C. Chougnet, M. J. Dolan, H. Yagita, J. D. Lifson, and G. M. Shearer. 2005. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood 106:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbeuval, J. P., A. W. Hardy, A. Boasso, S. A. Anderson, M. J. Dolan, M. Dy, and G. M. Shearer. 2005. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: role of type I IFN-producing plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA 102:13974-13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbeuval, J. P., J. Nilsson, A. Boasso, A. W. Hardy, M. J. Kruhlak, S. A. Anderson, M. J. Dolan, M. Dy, J. Andersson, and G. M. Shearer. 2006. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc. Natl. Acad. Sci. USA 103:7000-7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm, G. H., and D. Gabuzda. 2005. Distinct mechanisms of CD4+ and CD8+ T-cell activation and bystander apoptosis induced by human immunodeficiency virus type 1 virions. J. Virol. 79:6299-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izmailova, E., F. M. Bertley, Q. Huang, N. Makori, C. J. Miller, R. A. Young, and A. Aldovini. 2003. HIV-1 Tat reprograms immature dendritic cells to express chemoattactants for activated T cells and macrophages. Nat. Med. 9:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Jaworowski, A., and S. M. Crowe. 1999. Does HIV cause depletion of CD4+ T cells in vivo by the induction of apoptosis? Immunol. Cell Biol. 77:90-98. [DOI] [PubMed] [Google Scholar]
- 25.Jeremias, I., I. Herr, T. Boehler, and K. M. Debatin. 1998. TRAIL/Apo-2-ligand-induced apoptosis in human T cells. Eur. J. Immunol. 28:143-152. [DOI] [PubMed] [Google Scholar]
- 26.Katsikis, P. D., M. E. Garcia-Ojeda, J. F. Torres-Roca, I. M. Tijoe, C. A. Smith, and L. A. Herzenberg. 1997. Interleukin-1 beta converting enzyme-like protease involvement in Fas-induced and activation-induced peripheral blood T cell apoptosis in HIV infection. TNF-related apoptosis-inducing ligand can mediate activation-induced T cell death in HIV infection. J. Exp. Med. 186:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, N. S., H. J. Cheong, S. J. Kim, S. E. Kim, C. K. Kim, K. T. Lee, S. K. Park, S. H. Baick, D. S. Hong, H. S. Park, and J. H. Won. 2003. Ex vivo purging of leukemia cells using tumor-necrosis-factor-related apoptosis-inducing ligand in hematopoietic stem cell transplantation. Leukemia 17:1375-1383. [DOI] [PubMed] [Google Scholar]
- 28.Liegler, T. J., W. Yonemoto, T. Elbeik, E. Vittinghoff, S. P. Buchbinder, and W. C. Greene. 1998. Diminished spontaneous apoptosis in lymphocytes from human immunodeficiency virus-infected long-term nonprogressors. J. Infect. Dis. 178:669-679. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y. J. 2001. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell 106:259-262. [DOI] [PubMed] [Google Scholar]
- 30.Lum, J. J., D. J. Schnepple, and A. D. Badley. 2005. Acquired T-cell sensitivity to TRAIL mediated killing during HIV infection is regulated by CXCR4-gp120 interactions. AIDS 19:1125-1133. [DOI] [PubMed] [Google Scholar]
- 31.Miura, Y., and Y. Koyanagi. 2005. Death ligand-mediated apoptosis in HIV infection. Rev. Med. Virol. 15:169-178. [DOI] [PubMed] [Google Scholar]
- 32.Miura, Y., N. Misawa, N. Maeda, Y. Inagaki, Y. Tanaka, M. Ito, N. Kayagaki, N. Yamamoto, H. Yagita, H. Mizusawa, and Y. Koyanagi. 2001. Critical contribution of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) to apoptosis of human CD4+ T cells in HIV-1-infected hu-PBL-NOD-SCID mice. J. Exp. Med. 193:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moretti, S., S. Marcellini, A. Boschini, G. Famularo, G. Santini, E. Alesse, S. M. Steinberg, M. G. Cifone, G. Kroemer, and C. De Simone. 2000. Apoptosis and apoptosis-associated perturbations of peripheral blood lymphocytes during HIV infection: comparison between AIDS patients and asymptomatic long-term non-progressors. Clin. Exp. Immunol. 122:364-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortara, L., M. J. Ploquin, A. Faye, D. Scott-Algara, B. Vaslin, C. Butor, A. Hosmalin, F. Barre-Sinoussi, O. M. Diop, and M. C. Muller-Trutwin. 2006. Phenotype and function of myeloid dendritic cells derived from African green monkey blood monocytes. J. Immunol. Methods 308:138-155. [DOI] [PubMed] [Google Scholar]
- 35.Mueller, Y. M., S. C. De Rosa, J. A. Hutton, J. Witek, M. Roederer, J. D. Altman, and P. D. Katsikis. 2001. Increased CD95/Fas-induced apoptosis of HIV-specific CD8(+) T cells. Immunity 15:871-882. [DOI] [PubMed] [Google Scholar]
- 36.Mwaengo, D. M., and F. J. Novembre. 1998. Molecular cloning and characterization of viruses isolated from chimpanzees with pathogenic human immunodeficiency virus type 1 infections. J. Virol. 72:8976-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, D. H., N. Hurtado-Ziola, P. Gagneux, and A. Varki. 2006. Loss of Siglec expression on T lymphocytes during human evolution. Proc. Natl. Acad. Sci. USA 103:7765-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Doherty, U., R. Ignatius, N. Bhardwaj, and M. Pope. 1997. Generation of monocyte-derived dendritic cells from precursors in rhesus macaque blood. J. Immunol. Methods 207:185-194. [DOI] [PubMed] [Google Scholar]
- 39.Petrovas, C., Y. M. Mueller, I. D. Dimitriou, S. R. Altork, A. Banerjee, P. A. Sklar, K. C. Mounzer, J. D. Altman, and P. D. Katsikis. 2006. Increased mitochondrial mass characterizes the survival defect of HIV-specific CD8+ T cells. Blood 109:2505-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poaty-Mavoungou, V., R. Onanga, I. Bedjabaga, and E. Mavoungou. 2001. Simian immunodeficiency virus from mandrill (Mandrillus sphinx) SIVmnd experimentally infects human and nonhuman primate cells. Microbes Infect. 3:599-610. [DOI] [PubMed] [Google Scholar]
- 41.Rey-Cuillé, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romani, N., D. Reider, M. Heuer, S. Ebner, E. Kampgen, B. Eibl, D. Niederwieser, and G. Schuler. 1996. Generation of mature dendritic cells from human blood. An improved method with special regard to clinical applicability. J. Immunol. Methods 196:137-151. [DOI] [PubMed] [Google Scholar]
- 43.Ross, T. M. 2001. Using death to one's advantage: HIV modulation of apoptosis. Leukemia 15:332-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu, H. S., K. H. Chang, S. J. Chang, M. S. Kim, H. J. Joo, and K. S. Oh. 2000. Expression of TRAIL (TNF-related apoptosis-inducing ligand) receptors in cervical cancer. Int. J. Gynecol. Cancer 10:417-424. [DOI] [PubMed] [Google Scholar]
- 45.Selliah, N., and T. H. Finkel. 2001. Biochemical mechanisms of HIV induced T cell apoptosis. Cell Death Differ. 8:127-136. [DOI] [PubMed] [Google Scholar]
- 46.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 48.Villinger, F., S. S. Brar, G. T. Brice, N. F. Chikkala, F. J. Novembre, A. E. Mayne, S. Bucur, C. D. Hillyer, and A. A. Ansari. 1997. Immune and hematopoietic parameters in HIV-1-infected chimpanzees during clinical progression toward AIDS. J. Med. Primatol. 26:11-18. [DOI] [PubMed] [Google Scholar]
- 49.Villinger, F., T. M. Folks, S. Lauro, J. D. Powell, J. B. Sundstrom, A. Mayne, and A. A. Ansari. 1996. Immunological and virological studies of natural SIV infection of disease-resistant nonhuman primates. Immunol. Lett. 51:59-68. [DOI] [PubMed] [Google Scholar]
- 50.Viollet, L., V. Monceaux, F. Petit, R. Ho Tsong Fang, M. C. Cumont, B. Hurtrel, and J. Estaquier. 2006. Death of CD4+ T cells from lymph nodes during primary SIVmac251 infection predicts the rate of AIDS progression. J. Immunol. 177:6685-6694. [DOI] [PubMed] [Google Scholar]
- 51.Waldrop, S. L., C. J. Pitcher, D. M. Peterson, V. C. Maino, and L. J. Picker. 1997. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. J. Clin. Investig. 99:1739-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen, J., N. Ramadevi, D. Nguyen, C. Perkins, E. Worthington, and K. Bhalla. 2000. Antileukemic drugs increase death receptor 5 levels and enhance Apo-2L-induced apoptosis of human acute leukemia cells. Blood 96:3900-3906. [PubMed] [Google Scholar]
- 53.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, Y., I. Tikhonov, T. J. Ruckwardt, M. Djavani, J. C. Zapata, C. D. Pauza, and M. S. Salvato. 2003. Monocytes treated with human immunodeficiency virus Tat kill uninfected CD4+ cells by a tumor necrosis factor-related apoptosis-induced ligand-mediated mechanism. J. Virol. 77:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, M., X. Li, X. Pang, L. Ding, O. Wood, K. Clouse, I. Hewlett, and A. I. Dayton. 2001. Identification of a potential HIV-induced source of bystander-mediated apoptosis in T cells: upregulation of trail in primary human macrophages by HIV-1 tat. J. Biomed. Sci. 8:290-296. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, X. D., T. Nguyen, W. D. Thomas, J. E. Sanders, and P. Hersey. 2000. Mechanisms of resistance of normal cells to TRAIL induced apoptosis vary between different cell types. FEBS Lett. 482:193-199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.