Abstract
Direct functional screening of a cDNA expression library derived from primary porcine alveolar macrophages (PAM) revealed that CD163 is capable of conferring a porcine reproductive and respiratory syndrome virus (PRRSV)-permissive phenotype when introduced into nonpermissive cells. Transient-transfection experiments showed that full-length CD163 cDNAs from PAM, human U937 cells (histiocytic lymphoma), African green monkey kidney cells (MARC-145 and Vero), primary mouse peritoneal macrophages, and canine DH82 (histocytosis) cells encode functional virus receptors. In contrast, CD163 splice variants without the C-terminal transmembrane anchor domain do not provide PRRSV receptor function. We established several stable cell lines expressing CD163 cDNAs from pig, human, and monkey, using porcine kidney (PK 032495), feline kidney (NLFK), or baby hamster kidney (BHK-21) as the parental cell lines. These stable cell lines were susceptible to PRRSV infection and yielded high titers of progeny virus. Cell lines were phenotypically stable over 80 cell passages, and PRRSV could be serially passed at least 60 times, yielding in excess of 105 50% tissue culture infective doses/ml.
Porcine reproductive and respiratory syndrome (PRRS) is one of the most economically important diseases of swine (23). The syndrome appeared almost simultaneously in Western Europe and North America in the late 1980s and has since spread to become endemic in the major swine-producing nations of Europe, Asia, and the Americas. The etiologic agent of PRRS is a member of the family Arteriviridae in the order Nidovirales (1). Two distinct genotypes of PRRS virus (PRRSV), designated “European” (type I) and “North American” (type II), have been described. These two genotypes share only about 60% identity at the nucleotide level and appear to have evolved independently prior to appearing almost simultaneously in commercial swine herds (8, 11, 26). Both European and North American PRRSV cause reproductive failure in sows and gilts (late-term abortions, stillbirths, and mummies), high mortality rates among nursery pigs, and respiratory disease in swine of all ages. The disease has been the subject of recent reviews (17, 20, 25, 26).
Fully differentiated primary porcine alveolar macrophage (PAM) constitute the predominant cell target for viral replication (4, 5). A published example of an immortalized PAM cell line, however, is not permissive for PRRSV infection (33). In addition, monocytes from peripheral blood are largely refractory to PRRSV infection. A previous study demonstrated that heparin sulfate serves as an attachment factor in the binding and internalization of PRRSV but is not required for internalization (2). In PAM cells, initial binding of virions is mediated by interaction of the viral matrix protein with heparin sulfate proteoglycans (3). Virus particles are internalized in clathrin-coated vesicles and released following acidification (21). Internalization may be facilitated by sialoadhesin, a 210- or 220-kDa membrane glycoprotein in the siglec family of sialic acid binding immunoglobulin-like lectins (32), since incubation of PAM cells with monoclonal antibody (MAb) to this polypeptide blocks PRRSV infection (6, 34). Transfection of the nonpermissive PK-15 (porcine kidney) cell line with porcine sialoadhesin conferred the ability to internalize PRRSV particles, but there remained an apparent block at the uncoating stage, since virions entered into cellular vesicles but did not undergo nucleocapsid disintegration and vesicle membrane fusion (32).
Apart from primary PAM, the only other cell type known to be fully permissive for the growth of PRRSV in vitro is the immortalized monkey kidney cell line MA-104 and its derivatives, such as MARC-145 (12). In MARC-145 cells, the internalization of the virus by endocytosis and subsequent uncoating in low-pH vesicles seem to mimic similar events in PAM (14). However, MAbs that bind to porcine sialoadhesin fail to detect a homologous protein on the surfaces of MARC-145 cells (6, 34), suggesting that MARC-145 cells may use a divergent member of the same protein family or a different receptor altogether. Recently Kim et al. (13) reported that simian vimentin is a part of the PRRSV receptor complex. The following data support this hypothesis: (i) vimentin is expressed on the surface of MARC-145; (ii) MAb to vimentin blocked PRRSV infection; (iii) introduction of simian vimentin protein rendered BHK-21 and CRFK cells susceptible to PRRSV entry; and (iv) viral RNA was detected in association with vimentin-transfected cells following incubation with PRRSV. However, the authors did not provide any evidence that cells loaded with heterologous vimentin protein supported a productive infection (i.e., production of infectious progeny virus), nor was any evidence presented that endogenous expression from vimentin cDNA conferred susceptibility to nonpermissive cells.
In this report we demonstrate that CD163, a cellular protein in the scavenger receptor cysteine-rich (SRCR) superfamily, functions as a cellular receptor for PRRSV infection. Transfection with CD163 cDNA is necessary and sufficient to render a variety of nonpermissive cell lines fully permissive to PRRSV infection with production of progeny virus.
MATERIALS AND METHODS
Cells and viruses.
Primary PAM were harvested from 4- to 6-week-old PRRSV-negative pigs (Rural Technology Inc., Brookings, SD). Porcine kidney cells (PK032495), Norden Laboratories feline kidney cells (NLFK), Norden Laboratories swine testis (NLST) cells, Norden Laboratories dog kidney (NLDK) cells, Vero (African green monkey kidney) cells, and baby hamster kidney (BHK-21) cells were obtained from Biologicals Quality Control of Pfizer Animal Health (Lincoln, NE). RL (rabbit lung; ATCC CCl-193), DH82 (derived from canine histocytosis; ATCC CRL-10389), U937 (derived from human histiocytic lymphoma; ATCC CRL-1593.2), and PK-15 (porcine kidney; ATCC CCL-33) cell lines were purchased from ATCC (Manassas, VA). MARC-145 cells (African green monkey kidney) (12) were obtained from the National Veterinary Services Laboratories (NVSL) (Ames, IA). Cells were propagated and maintained in either OptiMEM I or Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 2 to 10% gamma-irradiated fetal bovine serum (JRH, Lenexa, KS), 1% antibiotic/antimycotic, and 1% GlutaMax (Invitrogen). For DMEM, 1% sodium pyruvate (Invitrogen) was also added. Geneticin (G418 sulfate, a neomycin sulfate analog; Invitrogen) at 500 to 1,000 μg/ml was used in growth medium for selection and maintenance of stable recombinant CD163-expressing cell lines. PRRSV isolates P129, IND5, P201, P3412, and P1151 were isolated and characterized by Gregory Stevenson, William Van Alstine, Charles Kanitz, and Ching Ching Wu at the Animal Disease Diagnostic Laboratory, Purdue University (West Lafayette, IN). PRRSV isolate NVSL 94-3 was obtained from NVSL, and PRRSV isolate EU98V226 was kindly provided by Hans J. Nauwynck (Ghent University, Ghent, Belgium). Prior to initiation of these studies, viruses were passaged once through pigs. Multiple aliquots of virus-containing sera were collected and stored at −80°C. This was done in order to amplify the viruses and to ensure that they retained the ability to infect pigs. Serum samples were used as inocula for further propagation in either PAM cells or stable cell lines expressing CD163. Preparation of recombinant P129-GFP plasmid and generation of P129-GFP virus have been described previously (35). Briefly, green fluorescent protein (GFP) was inserted between ORF1b and ORF2a/b and was expressed as an additional subgenomic RNA from the ORF2 transcriptional regulation sequence. A second copy of the ORF6 transcriptional regulation sequence was inserted downstream of GFP to allow expression of ORF2a and ORF2b. This virus retains the “green” phenotype for at least 37 cell culture passages.
Library construction and screening methods.
A cDNA library was constructed in the pCMV-Sport6.1 plasmid vector (Invitrogen) using RNA isolated from primary PAM. Plasmids containing library cDNA were cut with NotI and fractionated by size on agarose gels. Linear DNA was extracted from gel slices using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). The library was also digested with various restriction enzymes, including PmeI, SphI, AscI, NruI, and BamHI (Invitrogen).
BHK-21 cells were transiently transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The transfection mixture was removed after overnight incubation and replaced with fresh medium prior to infection of the cells with P129-GFP virus (35). Inocula were removed after adsorption and replaced with fresh medium. Infection was monitored via GFP expression using a fluorescence microscope.
Construction of pRSV-Script vector containing neomycin resistant gene.
The plasmid pRc/RSV (Invitrogen) was used as a template for PCR amplification of the Rous sarcoma virus (RSV) promoter. The RSV promoter sequence between nucleotides 209 and 604 of pRc/RSV was amplified using the forward primer (5′-ACACTCGACATGTCGATGTACGGGCCAGATATACGCGT-3′ with a built-in AflIII site) and a reverse primer (5′-TTCCTTACAGAGCTCGAGGTGCACACCAATGTGGTGAA-3′ with a built-in SacI site). The human cytomegalovirus immediate-early promoter (CMV promoter) in pCMV-Script (Stratagene, La Jolla, CA) was replaced with the RSV promoter-containing PCR fragment by digestion of the PCR product and pCMV-Script with AflIII and SacI, gel purification of appropriate fragments, ligation, and transformation of Escherichia coli DH5α. This construct was named pRSV-Script.
In vitro ligation to generate a linear CD163 expression construct.
The MARC CD163v2 cDNA proved difficult to clone into the pRSV-Script vector. To efficiently place the MARC CD163v2 gene downstream of the RSV promoter, a noncloning procedure was developed to produce microgram quantities of linear DNA suitable for use in generating stable cell lines. The process involved isolation and ligation of two gel-purified pieces of DNA, one containing the neomycin resistance gene cassette and the RSV promoter derived from pRSV-Script and the other containing the CD163 coding sequence. To accomplish this, pRSV-Script was linearized with DraIII upstream of the neomycin gene and then blunted with the Klenow fragment of E. coli DNA polymerase. This plasmid was then digested with NotI immediately downstream of the RSV promoter. The pCDNA3.1D MARC CD163v2 expression plasmid was digested upstream of the CD163 gene with NotI to generate compatible ends. The final construct is depicted in Fig. 1A. A large-scale ligation reaction was carried out as follows. Approximately 20 μg of each DNA fragment was incubated in a volume of 600 μl with 15 U of T4 DNA ligase. Following ligation, a linear piece of DNA containing the appropriate elements was purified by agarose gel electrophoresis and extraction using QIAquick kits (QIAGEN).
FIG. 1.
Constructs used for generating cell lines FK-D4 (feline kidney cells expressing simian CD163) and PK-9 (porcine kidney cells expressing porcine CD163). (A) The RSV promoter (pRSV) and neomycin resistance cassette were isolated from the pRSV-Script vector. Separately, the MARC-CD163v2 gene with a NotI 5′ restriction endonuclease recognition site was removed from pCDNA3.1D. The in vitro-ligated materials were separated on a preparative agarose gel. Linear DNA containing the CD163 gene under control of the RSV promoter was excised from the gel. The purified DNA was used for transfection of NLFK cells. (B) A circular pCMV-Script plasmid containing susCD163v1 and a neomycin resistance cassette was used for transfection of PK032495 cells. SV40pA, simian virus 40 polyadenylation signal; pSV40, simian virus 40 promoter; pCMV, human CMV immediate-early promoter.
Cloning and sequencing CD163 genes.
PrimerSelect (DNASTAR Inc., Madison, WI) was used to design several reverse transcription (RT)-PCR primer pairs based on the known CD163 sequences from pig (GenBank accession no. AJ311716), human (GenBank accession no. BC051281), and mouse (GenBank accession no. AF274883). Total cellular RNA was isolated from PAM, U937, MARC-145, Vero, dexamethasone-induced primary mouse peritoneal macrophage (29), and DH82 cells using the RNeasy minikit (QIAGEN, Santa Clarita, CA). U937 cells were treated with phorbol 12-myristate 13-acetate (100 ng/ml) for 3 days before harvesting of total cellular RNA in order to induce CD163 expression (10). CD163 transcripts were amplified in a single-tube RT-PCR using the SuperScript One-Step RT-PCR System for Long Templates kit (Invitrogen) according to the manufacturer's instructions. The primer pairs used for RT-PCR amplification were 5′-CGGAATGCGGCCGCTGTAATAATACAAGAAGATTTAAATGG-3′ and 5′-CGGTTGGTACCCAGCAATATTCTTTTTTATTTAATGCC-3′ for PAM, 5′-CACCGCGGCCGCGAAGTTATAAATCGCCACCATGAGCAAACTCAGAATGG-3′ and 5′-TGCTCCGGTACCTAGTCCAGGTCTTCATCAAGGTATCTTA-3′ for U937, 5′-CACCGCGGCCGCCACACGGAGCCATCAAAATCATCAA-3′ and 5′-GGTACCGCGAACAAGCAAACCAATAGCAATATTGTTTAATTCCCTC-3′ for mouse peritoneal macrophages, and consensus primers 5′-CACCGGAATGAGCAAACTCAGAATGG-3′ and 5′-TGCTCCGGTACCTAGTCCAGGTCTTCATCAAGGTATCTTA-3′ for MARC-145, Vero, and DH82 cells. RT-PCR parameters were as follows: 50°C for 30 min, 94°C for 2 min, 35 cycles (94°C for 30 s, 55°C for 30 s, and 68°C for 4 min), and 72°C for 10 min. RT-PCR products were cloned directly into pCMV-Script (Stratagene), pCR2.1-TOPO (Invitrogen), or pCDNA3.1 Directional/V5-His-TOPO vector (pCDNA3.1D; Invitrogen) according to the manufacturer's instructions. Based on restriction endonuclease enzyme analysis, E. coli colonies harboring plasmids with correct gene inserts were selected and gene sequences were determined using the Big Dye Terminator, version 1.0, sequence reaction kit (Applied BioSystems, Foster City, CA) and the Applied BioSystems 3730 DNA analyzer. Consensus sequences were assembled and edited using the EditSeq and SeqMan programs, and multiple sequence alignment analysis was done using MegAlign (DNASTAR Inc., Madison, WI) or Clustal W (v1.81) (31).
Establishment of stable cell lines expressing CD163.
Cell culture wells (35 mm) containing approximately 1 × 106 cells each were transfected with 2 to 4 μg of CD163 expression vector or a negative control plasmid (pPAMB) in serum-free and antibiotic-free medium (OptiMEM I or DMEM) and 10 μl of Lipofectamine 2000 according to the manufacturer's instructions. Cells were washed with phosphate-buffered saline (PBS), removed from the substrate using Accutase (Sigma), diluted in growth medium containing 500 to 1,000 μg/ml of Geneticin (G418 sulfate, a neomycin sulfate analog; Invitrogen), and seeded into 96-well plates at various densities to ensure recovery of single-cell clones after Geneticin selection. Throughout Geneticin selection, the growth medium was changed approximately every 3 to 5 days. After Geneticin selection, wells containing single cell-clones were split and expanded in duplicate 96-well plates. One plate was screened for susceptibility to virus by infecting it with P129-GFP, while the other plate was retained as a cell stock. All engineered cell lines were routinely maintained in G418 to ensure stability of the inserted genes.
Fluorescent-antibody (FA) assays.
Cell monolayers were washed once with PBS, fixed in 80% acetone for 5 to 10 min, and air dried. For detection of the PRRSV nucleocapsid (N) protein, MAb SDOW-17 (22) conjugated with fluorescein isothiocyanate (FITC) was added to each well. After 30 to 60 min of incubation, the antibody was aspirated and monolayers were washed three times with PBS prior to viewing under a fluorescence microscope.
Antibody blocking assay.
MARC-145 and FK-A6 cells (NLFK cells stably expressing human CD163v2) were incubated with dilutions of goat anti-human CD163 polyclonal antibody (R&D Systems, Minneapolis, MN) in a volume of 100 μl. As a control, equivalent amounts of normal goat immunoglobulin G (IgG) (R&D Systems) were used in replicate wells. Following a 1-h incubation at 37°C, monolayers were infected with a P129-GFP virus at a multiplicity of approximately 5. After a 1-h adsorption period, inocula were removed and cells were washed with PBS. After an additional incubation for 24 h at 37°C, cell monolayers were trypsinized, resuspended in 500 μl of PBS, and analyzed by flow cytometry. Uninfected cells were used to set the baseline for fluorescence detection, and approximately 100,000 events were recorded from each sample.
Western blot analysis.
Cell monolayers were washed once with PBS, lysed in 0.5% Triton X-100, and boiled for 5 min. Lysates were loaded immediately into wells of precast 4 to 12% Novex bis-Tris gels with morpholinepropanesulfonic acid-sodium dodecyl sulfate running buffer (NuPAGE electrophoresis system) according to the manufacturer's instructions (Invitrogen). After electrophoresis under reducing conditions, protein bands were transferred to polyvinylidene difluoride membranes (Invitrogen). Membranes were blocked with 1% bovine serum albumin in PBS and reacted with goat anti-human CD163 polyclonal antibody (R&D Systems) followed by rabbit anti-goat alkaline phosphatase conjugate (Kirkegaard & Perry Laboratories, Gaithersburg, MD). Color was developed using the Western Blue alkaline phosphatase substrate (Promega, Madison, WI).
Detection of porcine sialoadhesin transcripts.
A primer pair for the RT-PCR amplification of the porcine sialoadhesin mRNA (GenBank accession no. AF509585) was chosen using the PrimerSelect program (DNASTAR) and had the sequences 5′-GACGCCCACCATGACTGTTTTTGT-3′ and 5′-CTGCGTGGTTTCCTTCCGAGATAC-3′. Total cellular RNA was extracted from PK-15, PK032495, or primary PAM cells using the RNeasy minikit (QIAGEN). RT-PCRs were performed using the SuperScript One-Step RT-PCR System for Long Templates kit (Invitrogen).
Nucleotide sequence accession numbers.
The novel cDNA sequences described here have been deposited in the GenBank database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Accession numbers are as follows: porcine CD163 from primary alveolar macrophage, DQ067278 and DQ067279; African green monkey CD163 from MARC-145 cells, DQ067277; African green monkey CD163 from Vero cells, DQ060838, DQ060839, DQ060840, DQ060841, DQ060842, and DQ060843; human CD163 from U937 cells, DQ058615; canine CD163 from DH82 cells, DQ060836 and DQ060837; and murine CD163 from primary peritoneal macrophage, DQ058616 and DQ058617.
RESULTS
Porcine CD163 is a PRRSV receptor.
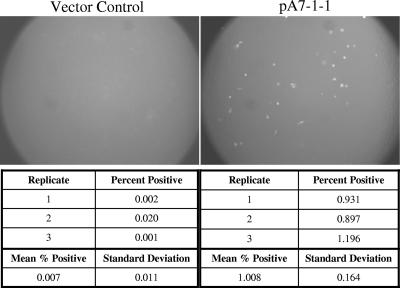
A cDNA library was constructed in the pCMV-Sport6.1 plasmid vector using RNA isolated from primary PAM. Transfection of the entire library into BHK-21 cells resulted in a few permissive cells following infection with P129-GFP virus (mock-transfected wells were negative). Fractionation of this cDNA library by size was achieved by gel purification of NotI-linearized cDNA. The resulting gel smear was cut into eight sections and used to generate sublibraries by retransformation of E. coli. Testing of these sublibraries by transfection into BHK-21 cells and infection with P129-GFP virus showed that the plasmids containing the largest inserts (>3 kb) contained the receptor activity. The sublibrary with the highest molecular weight (sublibrary no. 8) was chosen for use to generate libraries of decreasing complexity. To achieve this, sublibrary no. 8, which contained approximately 30,000 original colonies, was replated at multiple clone densities, from approximately 600 clones per library to approximately 9,000 clones per library. Testing DNA isolated from these sublibraries in the BHK-21 transfection/infection assay identified a sublibrary (sublibrary C1), containing approximately 1,200 clones, that contained the receptor activity. Using DNA from sublibrary C1, we were able to show that the restriction enzymes PmeI, SphI, AscI, NruI, and BamHI did not destroy the receptor activity. To generate sublibraries of further-decreasing complexity, DNA was isolated from sublibrary C1 and digested with the above enzymes before retransformation of E. coli. The resultant transformants were plated in libraries of approximately 75 to 200 clones. One of these sublibraries, containing 88 clones, was shown to contain receptor activity in the BHK-21 transfection/infection assay. Subsequent testing of individual colonies derived from this library identified a single clone (pA7-1-1) that possessed the receptor activity (Fig. 2). Throughout this process of enriching the library for the gene of interest, we observed a progressively higher frequency of positive cells in the transfection/infection assay.
FIG. 2.
PAM library clone A7-1-1 contains a putative PRRSV receptor. BHK-21 cells were transiently transfected (four replicates) with either the original PAM library-derived CD163-containing clone (A7-1-1) or a vector control plasmid (pPAMB) containing a small irrelevant PAM library insert. Following overnight incubation, the transfection mixture was removed and replaced with fresh medium. After several hours at 37°C, this medium was removed and cells were infected with PRRSV P129-GFP. At 1 day postinfection, three of the replicate wells were harvested for flow cytometry (cell counts, below) and the remaining well was analyzed by fluorescence microscopy (photomicrographs, above).
Sequencing revealed that the nucleotide sequence of this cDNA is 99.4% identical to a known porcine CD163 sequence (GenBank accession no. AJ311716) but contains additional 5′ and 3′ untranslated regions. In addition, the open reading frame of the identified clone from our PAM library, susCD163v1 (variant no. 1), differs from the published AJ311716 sequence in three ways: (i) a 738-bp internal deletion near the 5′ end, which represents an exact deletion of the first two SRCR repeats, (ii) a 15-bp extension of the 5′ end to an upstream ATG codon, and (iii) 16 nucleotide changes predicted to cause 10 amino acid differences. We were unable to amplify a corresponding CD163 transcript from PAM mRNA, suggesting that this deletion variant may have been generated during construction of the cDNA library or expansion in E. coli rather than representing a naturally occurring splice variant (data not shown).
Full-length CD163 transcripts were amplified by RT-PCR from PAM mRNA and cloned into either pCR2.1-TOPO or pCDNA3.1D. Sequence analysis revealed that the full-length porcine CD163 cDNA (susCD163v2) encodes 1,115 amino acids (GenBank accession no. DQ067278) and is 98.9% identical at the amino acid level to the AJ311716 sequence. SusCD163v2 also has an additional five amino acid residues at the extreme 5′ end, extending the open reading frame to an upstream ATG initiation codon as observed with susCD163v1. SusCD163v1 and susCD163v2 encode proteins with SRCR domain structures similar to that reported for human CD163 (16) (Fig. 3).
FIG. 3.
Structural domain organization of CD163 proteins. The deletion of two SRCR domains in variant susCD163v1 (middle) is compared to full-length susCD163v2 (top). A variant lacking the transmembrane domain is represented by VeroCD163 (TM −) (bottom).
Transient transfection of various cell lines with susCD163v1 confers permissivity to PRRSV.
Because the nonpermissive BHK-21 (18), PK032495, NLFK, NLST, and RL cell lines (unpublished data) have the ability to replicate PRRSV and produce progeny virus after transfection with infectious cDNA clones, these cell lines are assumed to have an early block in the viral replication cycle. After transient transfection with susCD163v1, all five of these cell lines became permissive to PRRSV infection as determined by FA staining for the PRRSV N protein. In most cases the level of N-protein expression appeared similar to that seen in PAM or MARC-145 cells, but individual positive cells were scattered throughout the monolayer (as one would expect following transient transfection with a receptor). Transient transfection with a negative control plasmid from the same library resulted in no virus infection in any of the cell lines as determined by FA staining. Three of the cell lines transfected with susCD163v1 (BHK-21, PK032495, and NLFK) produced detectable levels of progeny virus as determined by seeding of culture supernatants on MARC-145 cells. MARC-145 cells were used for detection of progeny virus because the P129 virus used in this study had been previously adapted to growth on MARC-145 cells. Progeny virus was not detected from the NLST and RL cell lines, most likely due to a low transfection efficiency, as demonstrated in parallel using a GFP-expressing control plasmid (pGreenLantern, Invitrogen).
Transient transfection of BHK-21 cells with diverse CD163 cDNAs confers permissivity to PRRSV.
After identification of porcine CD163 as a PRRSV receptor, we wanted to assess the ability of nonporcine CD163 cDNAs to confer permissivity. CD163 cDNAs were amplified by RT-PCR from total RNA extracted from five cell types, using primers and conditions as described in Materials and Methods. The MARC-145 cell line, derived from an African green monkey kidney, is known to support productive PRRSV infection. However, the Vero cell line, which was also derived from an African green monkey kidney, shows only very low levels of PRRSV susceptibility. Therefore, the CD163 cDNAs from both African green monkey cell lines were evaluated. The canine CD163 cDNA was also assessed, because the canine histocytosis cell line DH82 supports an abortive PRRSV infection (data not shown). Because macrophages are the natural target cells of arteriviruses, we also studied the CD163 cDNAs from the U937 human macrophage cell line and primary mouse peritoneal macrophages.
The CD163 cDNA we amplified and cloned from the MARC-145 cell line was 1,116 amino acids in length and was designated MARC CD163v2. The splice pattern of MARC CD163v2 is similar to that found in susCD163v2, and the two sequences share 84.8% amino acid identity. Relative to susCD163v2, MARC CD163v2 has single amino acid insertions between residues 138 and 139 and between residues 1041 and 1042 and a deletion of residue 1065. From Vero mRNA, eight cDNA clones with six discreet splicing patterns were identified, two of which lack the exon coding the transmembrane domain (Fig. 3). One CD163 splice variant was observed in cDNAs cloned from human U937 cells, whereas two splice variants were cloned from canine DH82 cells and two from mouse primary peritoneal macrophages. All of the above CD163 cDNAs, except the two Vero-derived cDNAs that lack the transmembrane domain, conferred PRRSV permissivity when transiently transfected into BHK-21 cells. This is consistent with a critical role of the transmembrane domain in PRRSV receptor function.
Stable cell lines expressing CD163 are permissive to PRRSV infection and produce progeny virus.
Recombinant cell lines were established from BHK-21, PK032495, and NLFK cell lines, using CD163 cDNA derived from porcine, human, or monkey cells and driven by either the CMV or RSV promoter (Table 1). At least one stable cell line that supported some degree of PRRSV replication was established from each combination of parental cell, CD163 cDNA, and promoter listed in Table 1. Susceptibility was determined by the observation of foci containing nucleocapsid-expressing cells following infection with PRRSV isolate P129, using FA staining with monoclonal antibody SDOW17-FITC. The three parental cell lines were completely negative for PRRSV infection. Differences in susceptibility between individual cell clones tended to be greater than differences due to the parental cell type, CD163 variant, or promoter. From these many cell clones, we chose a small subset that displayed good growth kinetics and high PRRSV susceptibly for further characterization. Two of these cell lines, FK-D4 (feline kidney cells transfected with MARC CD163v2 driven by the RSV promoter) and PK-9 (porcine kidney cells transfected with susCD163v1 driven by the CMV promoter), were used to passage two North American PRRSV isolates (P129 and P3412) for at least 40 passages. The growth of PRRSV on these stable cell lines was determined by observing the enlargement of infected foci through time (Fig. 4).
TABLE 1.
Stable cell lines expressing CD163
| Parental cell line | Promoter | CD163 gene |
|---|---|---|
| NLFK | CMVa | susCD163v1 |
| RSV | susCD163v1 | |
| CMV | humCD163v2b | |
| RSV | MARC CD163v2c | |
| PK032495 | CMV | susCD163v1d |
| CMV | susCD163v2 | |
| RSV | susCD163v1 | |
| CMV | humCD163v2 | |
| BHK-21 | CMV | susCD163v1 |
| RSV | susCD163v1 | |
| CMV | susCD163v2 | |
| CMV | humCD163v2 |
Human CMV immediate-early promoter.
Includes the FK-A6 cell line and a subclone, FK-A6.A2.
Includes the FK-D4 cell line.
Includes the PK-A10 cell line and a subclone, PK-9.
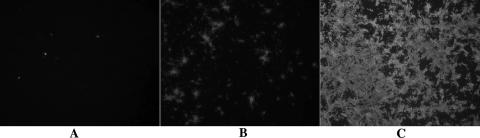
FIG. 4.
Growth of PRRSV on stably transfected cells expressing CD163. Replicate wells of PK-9 cells were infected with P129/p16 (passaged 16 times in PK-9 cells). Cells were fixed daily with 80% acetone and stained with FITC-conjugated MAb SDOW-17. The PRRSV N protein was visualized by fluorescence microscopy. Development of infection started with a few infected cells on day 1 (A), followed by enlargement of foci on day 2 (B). By day 3 postinfection, approximately 80% of the cells were infected.
Initially, PRRSV primary isolates grew slowly and to low titers in CD163-expressing cell lines. Ten or more carefully monitored serial passages were required to gradually adapt primary field isolates to consistent and reliable growth on FK-D4 and PK-9 cell lines. Adaptation involved infection of replicate subconfluent monolayers with several dilutions of supernatant fluids from the previous virus passage and harvesting of progeny virus fluids at days 2, 3, and 4 postinfection. Monolayers were immediately fixed and subsequently stained with FITC-labeled MAb SDOW17 and observed by FA assay. Selection of the most appropriate fluids to initiate the next passage was based on the degree of infection of the corresponding monolayer. A similar (but typically shorter) period of adaptation is required when primary PRRSV field isolates are grown on MARC-145 cells, and this corresponds to genetic changes in the virus (30). Some field isolates (e.g., P3412) could not be adapted to MARC-145 cells even after several attempts but could be adapted to growth on both FK-D4 and PK-9 cells. After adaptation to stable CD-163-expressing cell lines, PRRSV grew rapidly and reached titers similar to those on MARC-145 cells. For viruses passaged 15 to 40 times, typical peak titers are between 5 and 6 log10 50% tissue culture infective doses (TCID50)/ml when titrated on the same cell line (PK-9-adapted virus on PK-9 cells, FK-D4-adapted virus on FK-D4 cells, MARC-145-adapted virus on MARC-145 cells). Poor growth was seen when MARC-145-adapted virus was used to infect PK-9 cells or vice versa. Interestingly, all viruses adapted to grow on kidney cells of various types (MARC-145, PK-9, and FK-D4) retained the ability to replicate well on PAM cells (the natural host cell of PRRSV) for many passages. In fact, these viruses typically titrate 0.5 to 1.5 log10 TCID50/ml higher on PAM cells than on the appropriate kidney cell line. For these reasons, PAM cells were used for many (but not all) of the titrations described in this study. Figure 5A shows growth curves of PRRSV isolate P129, adapted for 19 passages on PK-9 cells, grown on PK-9 cells (at two multiplicities of infection), and titrated on PK-9 cells. At a multiplicity of 0.1, progeny virus in the culture fluid increases by more than 3 logs in the first 24 h. A peak titer of 4.9 log10 TCID50/ml was reached by 36 h postinfection. At a much lower multiplicity (0.001), there was a lag before progeny virus was first detected in the culture fluids. Eventually, titers reached 4.7 log10 TCID50/ml at 72 h postinfection.
FIG. 5.
Time courses of PRRSV replication on PK-9 and FK-D4 cells. (A) Growth kinetics were determined for PRRSV isolate P129 (passage 19 on PK-9 cells) using a multiplicity of 0.1 (⧫) and a multiplicity of 0.001 (▪) and titrated on PK-9 cells. After adsorption, inocula were removed and cells were washed with fresh medium. Culture fluids were harvested at 12-h intervals and virus titers were determined. (B) Growth curves for PRRSV P129/p38 (passage 38 on FK-D4 cells) were determined on FK-D4 cells at three passage levels to show stability of the PRRSV-permissive phenotype. Cells were evaluated at passage 19 (⧫), passage 29 (▪), and passage 46 (▴). Culture fluids were titrated on PAM cells. (C) Growth curves for P3412/p17 (passage 17 on FK-D4 cells) were evaluated on FK-D4 cells at passage 19 (⧫), passage 29 (▪), and passage 46 (▴). Inocula were removed after adsorption, and cells were washed with fresh medium. Culture fluids were titrated on PAM cells.
The growth curves in Fig. 5B and C show the phenotypic stability of the FK-D4 cell line. Isolate P129 adapted to FK-D4 for 38 passages (Fig. 5B) or isolate P3412 adapted for 17 passages (Fig. 5C) was used to initiate infection of FK-D4 cells at three different passage levels (cell passages 19, 29, and 46). Culture supernatants were collected periodically and titrated for progeny PRRSV on PAMs. PAMs were used due to their sensitivity and because they proved to be the best choice for titrating viruses adapted to diverse cell lines. Regardless of the cell passage level, P129 progeny virus increased more than 4 logs to reach a titer of 5.5 log10 TCID50/ml at 40 h postinfection. Similarly, isolate P3412, which grows slowly on all cell lines tested, grew to a titer of at least 4 log10 TCID50/ml on all three passage levels of FK-D4 cells. Thus, the FK-D4 cell line, when maintained in medium containing G418, appears to stably express MARC CD163v2 for at least 46 cell passages. Similarly, the PK-9 cell line has maintained a PRRSV-permissive phenotype for more than 80 cell passages (data not shown).
Anti-human CD163 antibody blocks PRRSV infection.
FK-A6 cells, which express the human CD163v2 gene, were treated with various concentrations of goat anti-human CD163 antibody or control goat IgG (from normal goat serum) for 1 h followed by infection with recombinant P129-GFP (preadapted to MARC-145 cells). At 24 h postinfection, flow cytometric analysis of the cells indicated that anti-CD163 antibody was able to block virus infection in a dose-dependent fashion (Fig. 6A). Anti-CD163 antibody at 0.3125 μg was able to reduce the frequency of infected FK-A6 cells by 70%, while 10 μg resulted in a greater than 90% reduction. Control goat IgG did not reduce the frequency of infected cells. In similar experiments with MARC-145 cells, a dose-dependent inhibition of infection was observed. The percentage of infected cells decreased from 67% without antibody to less than 2% when cells were pretreated with anti-CD163 antibody (Fig. 6B). This clearly demonstrates that expression of CD163 (a protein normally restricted to macrophages) is responsible for the unanticipated susceptibility of MARC-145 cells to PRRSV infection. The overall lower percentage of infected FK-A6 cells than of MARC-145 cells was most likely due to the use of MARC-145-adapted P129-GFP virus, which had not yet been adapted to grow on feline kidney cells expressing human CD163.
FIG. 6.
Blocking PRRSV infection with anti-human CD163 antibody. Infected cells were trypsinized, washed with PBS, and analyzed by flow cytometry, with at least 100,000 cells analyzed per sample. (A) Recombinant FK-A6 cells stably expressing humCD163v2 were incubated with either goat anti-human CD163-specific antibody (▴) or normal goat IgG (▪) and infected with P129-GFP virus. At 24 h postinfection, the percentage of GFP-expressing cells was determined by flow cytometry. (B) MARC-145 cells were incubated with either goat anti-human CD163-specific antibody (▴) or normal goat IgG (▪) and infected with P129-GFP virus. At 24 h postinfection, the percentage of GFP-expressing cells was determined by flow cytometry. Error bars represent the standard deviation of three replicate counts.
Recombinant cell lines express CD163.
To demonstrate the expression of CD163 in our recombinant cell lines, FA staining and Western blot analysis were employed. By FA staining, both FK-A6 (humCD163v2) and FK-D4 (MARC CD163v2) cells exhibited weak staining, while the parental NLFK cell line was negative for CD163 (not shown). As shown in Fig. 7, CD163 proteins had a molecular mass of approximately 130 kDa. A higher-molecular-mass band of unknown identity was also detected in the FK-A6.A2 cell lysates (FK-A6.A2 is a subclone of FK.A6). The level of CD163 expression was high in FK-A6.A2 cells. It appears that FK-D4 (MARC CD163v2) expresses smaller amounts of CD163 than FK-A6.A2 (human CD163v2), but this may be due in part to a lower specificity of the antibody, which was raised against human CD163. Comparing the two cell lines that express MARC CD163 and have similar susceptibilities to PRRSV infection (MARC-145 and FK-D4), we observed that the level of CD163 expression is much lower in MARC-145 cells. This suggests that even low-level expression of CD163 is sufficient to confer PRRSV permissiveness.
FIG. 7.
Western blot analysis of CD163 proteins. Lysates of MARC-145, FK-A6.A2, FK-D4, or parental NLFK were separated under reducing conditions. Proteins were transferred to a polyvinylidene difluoride membrane and incubated with goat anti-human CD163 polyclonal antibody followed by rabbit anti-goat antibody labeled with alkaline phosphatase. Membranes were developed in a Western Blue alkaline phosphatase substrate. Arrows indicate the CD163 protein at approximately 130 kDa. MW, prestained molecular mass markers (Invitrogen).
CD163 is a receptor for both genotype I and genotype II PRRSV.
Three recombinant CD163 cell lines were assessed for permissivity to PRRSV isolates (Table 2). FK-D4 (NLFK expressing MARC CD163v2), PK-A10 (PK032495 expressing susCD163v1), and PK-9 (a subclone of PK-A10) were infected with 0.1 ml of PAM-amplified PRRSV isolates. As controls, MARC-145, parental FK, and parental PK032495 cells were infected in parallel. PRRSV infectivity was determined by staining fixed monolayers with FITC-conjugated MAb SDOW-17. All three recombinant cell lines were susceptible to infection with both genotype I and genotype II PRRSV isolates. FK-D4 cells performed as well as or better than MARC-145 cells for all isolates except NVSL 94-3, which replicated better in MARC-145 cells. This may reflect the passage history of NVSL 94-3, which had previously been adapted to growth on MARC-145 cells at the National Veterinary Services Laboratory. Both nontransfected parental cell lines were completely negative for infection with all virus isolates.
TABLE 2.
Susceptibility of cell lines to infection with PRRSV genotypes I and IIa
| Cell line | Susceptibility to PRRSV isolate
|
|||||
|---|---|---|---|---|---|---|
| Genotype I, EU98V226 | Genotype II
|
|||||
| P129 | P201 | P1151 | NVSL 94-3 | IND5 | ||
| FK-D4 | ++ | + | +++ | +++ | ++ | ++++ |
| PK-9 | + | + | ++ | + | + | ++ |
| PK-A10 | + | + | ++ | ++ | ++ | ++ |
| MARC-145 | ++ | + | +++ | + | ++++ | +++ |
| Parental PK032495 | − | − | − | − | − | − |
| Parental NLFK | − | − | − | − | − | − |
Cells were infected with various PRRSV isolates. At 3 days postinfection, cells were fixed and stained with MAb SDOW-17-FITC and examined under a fluorescence microscope for nucleocapsid expression. ++++, many positive cells; +++, moderate number of positive cells; ++, some positive cells; +, very few positive cells; −, negative.
Sialoadhesin mRNA is present in PAM but not detectable in PK cells.
To determine the presence of sialoadhesin mRNA, cellular RNA from PK-15 (ATCC CCL-33), PK032495, or PAM cells was used as a template in RT-PCRs. A 469-bp PCR product was readily amplified from PAM but was not detected in either PK-15 or PK032495 cells (Fig. 8). Sequencing of the gel-purified PCR product revealed that this 469-bp fragment was identical to the published porcine sialoadhesin sequence (GenBank accession no. AF509585; nucleotide positions 4607 to 5073).
FIG. 8.
Detection of sialoadhesin mRNA. A primer pair located at the 3′ end of the sialoadhesin gene was used in RT-PCRs to amplify sialoadhesin mRNA from PAM, PK-15, and PK032495 cells. RT-PCR products were separated on a preformed 0.8% E-gel (Invitrogen) and visualized under the UV light. A 469-bp RT-PCR fragment was amplified from PAM cells (arrow) but not from either PK cell line. M, Ready Load 1-kb ladder (Invitrogen).
DISCUSSION
In the current study, we provide evidence that CD163 is a putative PRRSV receptor, closely associated with infection. Transient transfections of CD163 cDNAs from six cell types (encompassing five animal species) are capable of rendering nonpermissive cell lines permissive to PRRSV infection. Stable transfection with a CD163 cDNA was necessary and sufficient to generate fully permissive recombinant cell lines from established porcine kidney, feline kidney, and hamster kidney cell lines. Some of these cell lines retained the PRRSV-permissive phenotype for at least 80 cell passages, supported at least 60 serial virus passages, and produced in excess of 105 TCID50/ml of progeny virus.
CD163 is a scavenger receptor (SR) protein. The SR proteins comprise a large number of cell surface and soluble glycoproteins involved in the recognition of various ligands, including proteins, polyribonucleotides, polysaccharides, and lipids (28). These proteins are therefore capable of binding a wide range of host molecules and pathogens. Sequencing revealed that SR genes could be further divided into different types based on the commonality of motifs or domains. The SRCR domain has been found in more than 25 different secreted and/or membrane-anchored proteins. Many SRCR proteins are expressed by leukocytes and are involved in the development of the immune system and in regulation of the immune response (28).
The domain that defines the SRCR family of proteins consists of 100 to 110 amino acid residues. Molecules with SRCR domains are further divided into two groups based on the location and number of cysteine residues. Members of group A have six cysteine residues, and those of group B have eight cysteine residues. CD163, which was originally identified as a specific differentiation protein of macrophages and monocytes, is now known to be a group B SRCR protein containing eight cysteine residues per domain (7).
CD163 is a type 1 membrane protein. The extracellular domain of CD163 consists of nine SRCR tandem repeats, followed by a transmembrane segment and an intracellular cytoplasmic tail. Several variants of CD163 with different cytoplasmic domains have been described and are the result of alternative splicing of the CD163 primary transcript. Expression of CD163 is low in undifferentiated cells (10) and generally increases following stimulation and activation of macrophages. Human CD163 lacking the transmembrane domain sheds into the bloodstream and exhibits cytokine-like functions (9, 19). One well-characterized function of CD163 involves scavenging of hemoglobin, which is mediated by endocytosis of haptoglobin-hemoglobin complexes (15).
SusCD163v1 is missing the entire first and second SRCR repeats, yet it still confers permissivity to PRRSV infection, indicating that these two domains are not required for binding of PRRSV. Of six Vero cell splice variants isolated, only the two that lack the hydrophobic transmembrane domain (Vero CD163v4 and v5) failed to function as PRRSV receptors when transfected into BHK-21 cells. This suggests that CD163 in the type 1 membrane protein configuration (the form that scavenges haptoglobin-hemoglobin complexes) is preferred over soluble forms as the active PRRSV receptor. All other CD163 cDNAs tested in this study, from human, pig, mouse, dog, or African green monkey (MARC-145 and Vero) cells, contained the transmembrane domain and functioned as PRRSV receptors. Among these are alternative splice patterns that encode cytoplasmic tails in different reading frames, as has been reported for human CD163 transcripts (24). Therefore, sequence variations within the cytoplasmic domain do not appear to determine PRRSV receptor function.
Sánchez-Torres et al. (27) reported that CD163 is involved in the uptake of another porcine virus, African swine fever virus (ASFV), and that expression of CD163 on porcine macrophages and/or monocytes correlates with susceptibility to ASFV. When CD163+ and CD163− cells were separated, susceptibility to ASFV was associated with the CD163+ cell population. The authors demonstrated an association between CD163 expression and ASFV infection but did not provide evidence that CD163 is capable of converting nonsusceptible cells to ASFV susceptibility. Our study is the first to firmly establish a role for CD163 in viral entry.
Given the strict tropism of PRRSV for pigs, it is somewhat unexpected that CD163 homologs from divergent mammalian species (human, monkey, dog, and mouse) can functionally replace porcine CD163 in several cell lines. It would appear that the species specificity of PRRSV infection might not be based on receptor binding alone but also on downstream cellular processes in the replication cycle. For example, canine DH82 cells support PRRSV internalization and gene expression but do not yield progeny virus, indicating a block in one or more late stages of viral replication (data not shown). Consistent with a role in the early stages of viral infection, CD163 cDNA from DH82 cells is sufficient to convert nonsusceptible BHK-21 cells to PRRSV susceptibility.
Our results do not exclude the possibility of an unidentified cofactor, which is present in some established CD163− cell lines (such as PK032495, NLFK, and BHK-21) but absent in some CD163+ cells (such as primary mouse peritoneal macrophages and differentiated human U937 cells). Hence, the presence of both CD163 and cofactor(s) may be required for efficient attachment, entry, and uncoating.
A previous study suggested that sialoadhesin is a binding factor that is capable of mediating attachment and internalization of PRRSV (32). It is possible that binding to sialoadhesin is a necessary first step in a pathway that also includes CD163, leading to uncoating and release of viral RNA into the cytoplasm. However, the PK-15 cell line used in that study did not express detectable levels of sialoadhesin (32). Our RT-PCR results further confirmed that sialoadhesin mRNA was not detectable in PK-15 or PK032495 cells under conditions that readily amplified sialoadhesin mRNA from PAMs. In spite of this, introduction of a CD163 gene into PK032495 cells was sufficient to render them fully permissive to PRRSV infection, arguing against a requirement for sialoadhesin in CD163-mediated initiation of infection. Additional studies are needed to identify possible cofactor(s) and to further dissect the interactions between CD163 and PRRSV.
Acknowledgments
The studies described here were funded entirely by the Animal Health Division of Pfizer Inc.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 2.Delputte, P. L., S. Costers, and H. J. Nauwynck. 2005. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparin sulfate and sialoadhesin. J. Gen. Virol. 86:1441-1445. [DOI] [PubMed] [Google Scholar]
- 3.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparin like receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan, X., H. J. Nauwynck, and M. B. Pensaert. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus. Arch. Virol. 142:2483-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan, X., H. J. Nauwynck, and M. B. Pensaert. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 56:9-19. [DOI] [PubMed] [Google Scholar]
- 6.Duan, X. B., H. J. Nauwynck, H. W. Favoreel, and M. B. Pensaert. 1998. Identification of a putative receptor for porcine reproductive and respiratory syndrome virus on porcine alveolar macrophages. J. Virol. 72:4520-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabriek, B. O., C. D. Dijkstra, and T. K. van den Berg. 2005. The macrophage scavenger receptor CD163. Immunobiology 210:153-160. [DOI] [PubMed] [Google Scholar]
- 8.Forsberg, R. 2005. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 22:2131-2134. [DOI] [PubMed] [Google Scholar]
- 9.Frings, W., J. Dreier, and C. Sorg. 2002. Only the soluble form of the scavenger receptor CD163 acts inhibitory on phorbol ester-activated T-lymphocytes, whereas membrane-bound protein has no effect. FEBS Lett. 526:93-96. [DOI] [PubMed] [Google Scholar]
- 10.Gronlund, J., L. Vitved, M. Lausen, K. Skjodt, and U. Holmskov. 2000. Cloning of a novel scavenger receptor cysteine-rich type I transmembrane molecule (M160) expressed by human macrophages. J. Immunol. 165:6406-6415. [DOI] [PubMed] [Google Scholar]
- 11.Hanada, K., Y. Suzuki, T. Nakane, O. Hirose, and T. Gojobori. 2005. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol. Biol. Evol. 22:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogenous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. K., A. M. Fahad, K. Shanmukhappa, and S. Kapil. 2006. Defining the cellular targets of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 80:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreutz, L. C., and M. R. Ackermann. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res. 42:137-147. [DOI] [PubMed] [Google Scholar]
- 15.Kristiansen, M., J. H. Graversen, C. Jacobsen, O. Sonne, H. J. Hoffman, S. K. Law, and S. K. Moestrup. 2001. Identification of the haemoglobin scavenger receptor. Nature 409:198-201. [DOI] [PubMed] [Google Scholar]
- 16.Law, S. K., K. J. Micklem, J. M. Shaw, X. P. Zhang, Y. Dong, A. C. Willis, and D. Y. Mason. 1993. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur. J. Immunol. 23:2320-2325. [DOI] [PubMed] [Google Scholar]
- 17.Mengeling, W. L., and K. M. Lager. 2000. A brief review of procedures and potential problems associated with the diagnosis of porcine reproductive and respiratory syndrome. Vet. Res. 31:61-69. [DOI] [PubMed] [Google Scholar]
- 18.Meulenberg, J. J. M., J. N. A. Bosderuijter, R. Vandegraaf, G. Wensvoort, and R. J. M. Moormann. 1998. Infectious transcripts from cloned genome-length cDNA of porcine reproductive and respiratory syndrome virus. J. Virol. 72:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller, H. J., N. A. Peterslund, J. H. Graversen, and S. K. Moestrup. 2002. Identification of the hemoglobin scavenger receptor/CD163 as a natural soluble protein in plasma. Blood 99:378-380. [DOI] [PubMed] [Google Scholar]
- 20.Murtaugh, M. P., Z. Xiao, and F. Zuckermann. 2002. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 15:533-547. [DOI] [PubMed] [Google Scholar]
- 21.Nauwynck, H. J., X. Duan, H. W. Favoreel, P. Van Oostveldt, and M. B. Pensaert. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J. Gen. Virol. 80:297-305. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385-392. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, M. J., M. Madsen, H. J. Moller, and S. K. Moestrup. 2006. The macrophage scavenger receptor CD163: endocytic properties of cytoplasmic tail variants. J. Leukoc. Biol. 79:837-845. [DOI] [PubMed] [Google Scholar]
- 25.Nodelijk, G. 2002. Porcine reproductive and respiratory syndrome (PRRS) with special reference to clinical aspects and diagnosis. A review. Vet. Q. 24:95-100. [DOI] [PubMed] [Google Scholar]
- 26.Plagemann, P. G. 2003. Porcine reproductive and respiratory syndrome virus: origin hypothesis. Emerg. Infect. Dis. 9:903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Torres, C., P. Gómez-Puertas, M. Gómez-del-Moral, F. Alonso, J. M. Escribano, A. Ezquerra, and J. Dominguez. 2003. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 148:2307-2323. [DOI] [PubMed] [Google Scholar]
- 28.Sarrias, M. R., J. Gronlund, O. Padilla, J. Madsen, U. Holmskov, and F. Lozano. 2004. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24:1-37. [DOI] [PubMed] [Google Scholar]
- 29.Schaer, D. J., F. S. Boretti, A. Hongegger, D. Poehler, P. Linnscheid, H. Staege, C. Muller, G. Schoedon, and A. Schaffner. 2001. Molecular cloning and characterization of the mouse CD163 homologue, a highly glucocorticoid-inducible member of the scavenger receptor cysteine-rich family. Immunogenetics 53:170-177. [DOI] [PubMed] [Google Scholar]
- 30.Tan, C., L. Chang, S. Shen, D. X. Liu, and J. Kwang. 2001. Comparison of the 5′ leader sequences of North American isolates of reference and field strains of porcine reproductive and respiratory syndrome virus (PRRSV). Virus Genes 22:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderheijden, N., P. L. Delputte, H. W. Favoreel, J. Vandekerckhove, J. Van Damme, P. A. van Woensel, and H. J. Nauwynck. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77:8207-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weingartl, H. M., M. Sabara, J. Pasick, E. van Moorlehem, and L. Babiuk. 2002. Continuous porcine cell lines developed from alveolar macrophages: partial characterization and virus susceptibility. J. Virol. Methods 104:203-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wissink, E. H., H. A. van Wijk, J. M. Pol, G. J. Godeke, P. A. van Rijn, P. J. Rottier, and J. J. Meulenberg. 2003. Identification of porcine alveolar macrophage glycoproteins involved in infection of porcine respiratory and reproductive syndrome virus. Arch. Virol. 148:177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo, D., S. K. Welch, C. Lee, and J. G. Calvert. 2004. Infectious cDNA clones of porcine reproductive and respiratory syndrome virus and their potential as vaccine vectors. Vet. Immunol. Immunopathol. 102:143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]