Abstract
Human cytomegalovirus (HCMV) establishes a lifelong infection with the potential for reinfection or viral transmission even in the presence of strong and diverse CD8 T-lymphocyte responses. This suggests that the CMVs skew the host T-cell response in order to favor viral persistence. In this study, we hypothesized that the essential, nonstructural proteins that are highly conserved among the CMVs may represent a novel class of T-cell targets for vaccine-mediated protection due to their requirements for expression and sequence stability, but that the observed subdominance of these antigens in the CMV-infected host results from the virus limiting the T-cell responses to otherwise-protective specificities. We found that DNA immunization of mice with the murine CMV (MCMV) homologs of HCMV DNA polymerase (M54) or helicase (M105) was protective against virus replication in the spleen following systemic challenge, with the protection level elicited by the M54 DNA being comparable to that of DNA expressing the immunodominant IE1 (pp89). Intracellular gamma interferon staining of CD8 T cells from mice immunized with either the M54 or M105 DNAs showed strong primary responses that recalled rapidly after viral challenge. M54- and M105-specific CD8 T cells were detected after the primary MCMV infection, but their levels were not consistently above the background level. The conserved, essential proteins of the CMVs thus represent a novel class of CD8 T-cell targets that may contribute to a successful HCMV vaccine strategy.
Human cytomegalovirus (HCMV) is an opportunistic betaherpesvirus that establishes a lifelong infection in 50 to 100% of the adult population worldwide (16). While primary infection of the immunocompetent host is usually asymptomatic, HCMV can cause severe disease and death in the immunocompromised or immunologically immature host. HCMV disease in solid organ or allogeneic stem-cell transplant recipients continues to be a significant clinical problem, and in the newborn, HCMV is the leading infectious cause of birth defects. Despite the morbidity and mortality associated with HCMV disease and the clear need for a vaccine (21), no experimental HCMV vaccine to date has shown significant protective efficacy in phase 3 clinical testing.
Our current understanding of the mechanisms of both host immune surveillance and viral immune evasion of HCMV is based upon results both from clinical studies and from animal CMV models (19). The murine CMV (MCMV) model has been particularly useful in characterizing some of the molecular mechanisms involved in the elicitation and subversion of host immune responses. While innate cell-mediated immunity (CMI), especially that provided by natural killer (NK) cells, is a major determinant in early disease progression, adaptive CMI—provided by the CD8 and CD4 T lymphocytes—plays the key role in resolving the primary infection and providing long-term viral immune surveillance and clearance of replicating virus. The inverse correlation between HCMV-specific T-lymphocyte responses and HCMV disease has been demonstrated by clinical data in transplant recipients with absent or impaired CMI, and CD8 T cells were found to be important mediators of protection in bone marrow/allogeneic stem-cell transplant patients.
The ability of the CMVs to persist in the immunocompetent host and to spread both horizontally and vertically in the face of a wide complement of antiviral CMI and antibody responses suggests that the viruses have successfully adapted to the immune pressures of their respective hosts. Additional evidence of the importance of T-cell-mediated viral control mechanisms is the discovery that the CMVs have each evolved specific mechanisms that interfere with the ability of the host cells to present viral-peptide-loaded major histocompatibility complex (MHC) class I and class II molecules to their cognate CD8 and CD4 T cells, respectively (for reviews, see references 18 and 20). However, CMV peptide-specific CD8 T cells are still generated in their respective hosts, and recent comprehensive studies of the CD8 T-cell specificities following infection have been used to identify the viral protein targets of HCMV or MCMV. In a landmark report by Sylwester et al., CD8 T-cell responses were detected against overlapping peptides from 107 of the 213 (50.2%) open reading frames (ORFs) when tested by cytokine flow cytometry of the peripheral blood mononuclear cells from 33 HCMV-seropositive subjects (22). The seropositive subjects directed their CD8 T-cell responses to as few as 1 HCMV ORF and as many as 31 (median, 8), indicating a highly individualized and often diverse response. Peptides derived from UL48, UL83 (pp65), and UL123 (IE1) were recognized by CD8 T cells in ≥50% of the subjects, and the latter two ORFs have been the most widely studied CD8 T-cell targets and have formed the basis of specifically targeted cytoimmunotherapies. A similar study of the CD8 T-cell specificities to MCMV during the acute phase of infection (day 7) was performed by stimulating the pooled splenocytes of C57BL/6 mice with H-2b stimulator cells that were transiently transfected with one of each of the 170 cloned ORFs (14). Twenty-seven ORFs (16% of those tested) were found to elicit a CD8 T-cell response that was >5-fold above the background level. Based on either experimental findings or homology with HCMV ORFs, the most antigenic peptides were encoded primarily by early (E) genes, with no immediate-early (IE) genes identified. Subsequently, two epitopes of the IE3 gene product were found to be recognized in chronically, but not acutely, infected C57BL/6 mice (13). These data together demonstrate the complexity of the dynamic CD8 T-cell response to acute and chronic CMV infection.
Our laboratory has used the MCMV model to study the basis of protective immunity, with the goal of developing a vaccine that provides sterilizing immunity against acute and latent CMV infection. Initially we found that intradermal (i.d.) immunization of BALB/c mice with plasmid DNA expressing the immunodominant MCMV IE1 gene product pp89 under the control of the HCMV major IE promoter/enhancer could elicit MCMV-specific CD8 T-cell responses and protection against the replication of virus in the spleen following sublethal, systemic challenge (6). Subsequently, we found that an E nonstructural MCMV homolog of HCMV UL83 (pp65), M84 (3, 9), was similarly protective against viral replication in the spleen and that coimmunization of mice with the two protective plasmids expressing IE1 and M84 elicited CD8 T-cell responses to both antigens and provided a synergistic level of protection (10). While plasmids expressing the putative tegument and capsid genes of MCMV (M32, M48, M56, M69, M82, M83, M85, M86, and M99) or the nonstructural M112-M113 (e1) were not protective when tested separately or in small groups, immunization with a pool of all 10 DNAs reduced the viral load in the spleen following low to intermediate challenge doses (12). Mice coimmunized with a pool of IE1, M84, and the matrix and capsid genes above had reductions in their splenic viral titers of 104 compared with the titers of controls immunized with vector alone: the highest levels of DNA-mediated protection we had elicited to date. Because the reduction in viral load in the salivary glands that was afforded by this vaccine was only 10-fold relative to the load in controls, we developed a prime-boost strategy in which DNA-primed mice were subsequently boosted with formalin-inactivated MCMV (FI-MCMV) (12). We found that only the mice that were immunized with a pool of 13 plasmid DNAs followed by FI-MCMV, and not those immunized with either component alone, had undetectable levels of virus in the spleen (<10 PFU per spleen) and salivary glands (<50 PFU in the salivary glands) after sublethal, systemic challenge (12). Finally, we demonstrated that priming with a trivalent vaccine containing IE1, M84, and M55 (glycoprotein B) DNAs and boosting with FI-MCMV provide long-term complete protection against viral replication in the spleen, lungs, liver, and salivary glands following systemic challenge (11). In addition, we found that this vaccination strategy was efficacious against mucosal challenge, with 1,000- to 2,000-fold reductions in viral titers in the lungs and undetectable levels in the salivary glands, spleen, and liver in the majority of immunized mice.
In this study, we sought to use the murine model to determine whether a novel class of CMV antigens that could elicit T-lymphocyte-mediated protection could be identified that could also have predictive value for protection against HCMV disease. While recent studies in healthy, seropositive volunteers have rapidly expanded the list of HCMV antigens capable of eliciting specific T-cell responses, far less is known regarding the antiviral activities of these cells, due to the logistical challenges and cost of clinical trials. The utility of the murine model for predicting the basis of protection against CMV would be greatly increased by the identification of a class of MCMV antigens that share a high degree of biological similarity with their HCMV counterparts. Based on our results that the protective MCMV antigens we had identified by DNA immunization were nonstructural IE, E, or E/L genes, we focused in this study on E genes. We also hypothesized that the gene products of the highly conserved, essential genes might be excellent targets for primed CD8 T cells. Because of the need for their expression during productive infection, the virus would not likely be able to abrogate or severely downregulate their expression without negatively affecting viral replication. In addition, the high degree of amino acid conservation required for the maintenance of the enzymatic activities of these gene products may allow a DNA vaccine to be effective against a wide range of HCMV strains. Of the essential gene products that are most highly conserved between HCMV and MCMV and that have been shown to have E gene expression kinetics, many are nonstructural proteins involved with DNA replication. We also noted that many of these proteins were mostly subdominant for memory CD8 T-cell responses to HCMV and acute-phase responses to MCMV, and speculated that the CMVs may have evolved mechanisms to diminish the priming of naïve T cells against these antigens or the maintenance of memory cells. However, these antigens might elicit a protective response with DNA immunization. Computer-based epitope analysis has revealed that these HCMV antigens contain high densities of putative human class I epitopes (Peter Barry, personal communication), and thus, the results with these highly conserved genes might be directly applicable to the development of novel strategies for a human vaccine.
MATERIALS AND METHODS
Mice, cells, and viruses and viral purification.
Three- to four-week-old specific-pathogen-free female BALB/c mice were purchased from Harlan Sprague Dawley, Inc., and housed in microisolator-covered cages in the Pacific Hall vivarium, University of California, San Diego, CA. The mice were allowed to acclimate for at least 1 week prior to immunization or MCMV infection.
NIH 3T3 (ATCC CRL 1658), COS-7 (ATCC CRL 1651), and BALB/c mouse embryonic cells were propagated as previously described (24). BALB SV40 cells, a simian virus 40 (SV40)-transformed H-2d cell line (1), were grown in COS-7 medium (24).
Salivary gland-derived MCMV strain K181 (SG-MCMV) was propagated in BALB/c mice, and tissue culture-derived MCMV strain K181 (TC-MCMV) was prepared in mouse embryonic cells as previously described (2, 5). The titers of these stocks were determined by plaque assay on NIH 3T3 cells (6). The 50% lethal dose (LD50) of the SG-MCMV stock in BALB/c mice in this study was 8 × 105 PFU (11).
Plasmid construction and expression.
The construction of pc3Δneo-pp89, expressing the cDNA of IE1 (pp89), was described previously (24). The cloning of the M54, M70, and M105 genes of MCMV MW97.01, a virus derived from a bacterial artificial chromosome of Smith strain MCMV, into pcDNA3.1/V5-His-TOPO (Invitrogen Life Technologies) has also been described previously (14). For the studies reported here, the complete sequences of the three cloned ORFs were determined (Eton Biosciences, Inc., San Diego, CA) and compared with the published sequence of MCMV Smith (GenBank accession number U68299) (17). The coding sequences of the M54 and M105 ORFs were identical to those in the published sequence (data not shown). The sequence of the M70 ORF was identical to that in the Smith strain sequence except for a single T insertion between the 3′ end of the MCMV-encoded ORF and the vector-encoded epitope tag sequences, a mutation that resulted in an immediate termination codon (data not shown). This mutation yielded a complete, wild-type, but untagged, coding sequence for the M70 protein. Confirmation of complete, continuous reading frames was provided by coupled in vitro transcription-translation (TNT T7 Quick coupled transcription/translation system; Promega Corporation) using [35S]methionine, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and autoradiography, following the manufacturer's recommendations. To facilitate subsequent Western blot analysis of the levels of expression in transiently transfected COS-7 cells, the extraneous T of the M70 clone was deleted by QuikChange site-directed mutagenesis (Stratagene, Inc.), following the manufacturer's recommendations, and the resulting clone was verified by DNA sequencing.
The levels of protein expression were confirmed by transient transfection of COS-7 cells using Effectene (QIAGEN), followed by Western blot analysis. At 48 h posttransfection, cell lysates were made in a reducing SDS sample buffer (3), solubilized at 42°C, and resolved by SDS-PAGE, and the proteins were transferred to nitrocellulose membranes. Plasmid-expressed MCMV proteins were detected with a mouse anti-V5 tag-specific monoclonal antibody (Invitrogen) and SuperSignal West Pico reagent (Pierce), according to the manufacturers' recommendations.
For i.d. immunization, plasmids were purified using QIAGEN EndoFree Plasmid Mega or Giga kits and resuspended to approximately 2 mg of DNA per ml of endotoxin-free 10 mM Tris-HCl (pH 8).
Immunization, virus challenge, and virus titration.
Plasmids were diluted immediately before injection with endotoxin-free 10 mM Tris-HCl (pH 8)-buffered saline. Mice were i.d. injected with 30 μl of diluted plasmid either into three sites (10 μl per site) in the shaved flank near the base of the tail or into one i.d. site in the tail, approximately 1.5 to 2 cm from the base (see Results for specific DNA doses). The mice were injected three times within 2 weeks and challenged 2 or 3 weeks after the last injection by intraperitoneal (i.p.) injection with 0.5 ml of Dulbecco's phosphate-buffered saline containing various sublethal doses of SG-MCMV (see Results).
On day 6 post-i.p. challenge, the mice were sacrificed and the spleens were aseptically removed and washed with Dulbecco's phosphate-buffered saline. The spleens were homogenized in Dulbecco's modified Eagle medium containing 10% heat-inactivated newborn calf serum (Invitrogen Life Technologies) and 10% dimethyl sulfoxide, using 7 ml Tenbröeck homogenizers, and the homogenates were separated into aliquots and stored at −80°C until titration. The titer of infectious MCMV was determined by a plaque assay of the clarified homogenates using NIH 3T3 cells in 24-well dishes as previously described (6). The limit of sensitivity for this initial assay was 100 PFU per spleen. If the viral titer of a spleen was less than or equal to five times the detection limit (≤500 PFU per spleen), 100 μl of clarified homogenate from another aliquot of the homogenate was used in a more-sensitive plaque assay on NIH 3T3 cells in 10-cm dishes, as previously described (11, 12), except that the concentration of heat-inactivated newborn calf serum in the medium was increased to 10%. The limit of sensitivity of this assay was empirically confirmed to be 10 PFU per spleen. The log10 values of the individual viral titers in each group were taken, and the mean of the log10 values was calculated.
ICS assay.
The levels of specific CD8 T cells elicited by DNA immunization were measured by intracellular cytokine staining (ICS) assay using transfected stimulator cells as described previously (14). Three BALB/c mice per group were i.d. immunized in the tail three times in 2 weeks with 25 μg of either empty pc3Δneo vector DNA or DNA encoding IE1, M54, or M105. For the first ICS assay, a fourth immunization was given 1 week after the third injection and the assay was performed 2 weeks later. For the second ICS assay, a fourth immunization was given 3 weeks after the third injection and the assay was performed 3 weeks later. For comparative purposes, the CD8 T-cell levels resulting from MCMV infection were measured in 3 BALB/c mice per group that were i.p. infected with 1.2 × 105 PFU of TC-MCMV either 2 weeks (first ICS assay) or 4 weeks (second ICS assay) prior to the assay. For the assay, BALB SV40 (H-2d) cells were seeded into 96-well tissue culture plates and, 1 day later, the cells (ca. 60 to 75% confluent) were transfected with 0.5 μg of plasmid DNA and 1.25 μl of FuGene 6 (Roche) per well. To monitor the transfection efficiencies of the stimulator cells, additional wells were transfected with either IE1 DNA or DNA expressing enhanced green fluorescent protein (EGFP), pcDNA3-EGFP. Two days posttransfection, splenocytes from three mice per immunization or infection group were harvested, the erythrocytes were lysed (BD Pharm lyse; BD Biosciences), and 8 × 105 splenocytes from the immunized or infected mice were added to duplicate wells of transfected BALB SV40 cells in the presence of brefeldin A (GolgiPlug; BD Pharmingen). For peptide stimulation, duplicate wells containing 2 × 106 splenocytes each were stimulated with 1 μM of the Ld-restricted nonapeptide epitope of IE1 (168YPHFMPTNL176) in the presence of brefeldin A. Peptide-stimulated splenocytes served as gating controls for CD8 and gamma interferon (IFN-γ) staining. After 8 h of stimulation at 37°C and 7% CO2, duplicate wells of the splenocytes were combined into one well of a 96-well round-bottom plate for staining. The splenocytes were surface stained overnight with phycoerythrin-Cy5-conjugated anti-mouse CD8a (Ly-2) antibody clone 53-6.7 (eBioscience) and, following fixation and permeabilization (BD Cytofix/Cytoperm; BD Biosciences), the splenocytes were stained with fluorescein isothiocyanate-conjugated anti-mouse IFN-γ antibody clone XMG1.2 (eBioscience). The lymphocytes were gated and the dual-stained splenocytes were enumerated on a BD FACSCanto flow cytometer (BD Biosciences) with BD FACSDiva software at the Research Flow Cytometry Core Facility, VA Medical Center, La Jolla, California.
For the measurement of secondary CD8 T-cell responses, three BALB/c mice per group were i.d. immunized in the tail three times in 2 weeks with 25 μg of either pc3Δneo vector DNA or DNA encoding IE1, M54, or M105. For comparative purposes, three BALB/c mice were i.p. infected with 1.2 × 105 PFU of TC-MCMV as described above. Seven weeks after the last immunization or 6 weeks after the primary MCMV infection, the mice were i.p. infected with 1.2 × 105 PFU of SG-MCMV. On day 5 after infection with SG-MCMV, splenocytes were harvested and analyzed by ICS assay as described above.
RESULTS
Cloning, sequencing, and expression of MCMV DNA polymerase (M54), primase (M70), and helicase (M105).
We have used plasmid DNA immunization as a rapid means for identifying MCMV genes that are able to elicit protective CMI against challenge virus replication. With the exception of the glycoprotein B gene, the genes we have tested have encoded either nonstructural, tegument, or capsid genes. Based on our results that the MCMV antigens thus far identified as being protective by DNA immunization are nonstructural IE, E, or E/L genes, we sought to find additional protective MCMV antigens by focusing our attention on the E genes. In deciding which antigens to test, we hypothesized that essential genes that were highly conserved might be excellent targets for primed CD8 T cells but that during the infection there are mechanisms that prevent naïve T cells from being primed against these antigens or maintained as memory cells. Accordingly, these antigens might elicit a protective response with DNA immunization. Computer-based epitope analysis has revealed that these HCMV antigens contain high densities of putative human class I epitopes (Peter Barry, personal communication), and thus, the results with these highly conserved genes might be directly applicable to the development of novel strategies for a human vaccine. FASTA analyses of the deduced amino acid sequences of the DNA polymerase catalytic subunit (UL54), helicase (UL105), and primase (UL70) proteins of HCMV found these sequences to be particularly highly conserved with their respective homologs throughout the herpesviruses (data not shown), and these HCMV genes have been shown to be required for origin-dependent DNA replication (15). We therefore chose to test mammalian expression plasmids expressing the MCMV homologs of UL54, UL70, and UL105 (Table 1).
TABLE 1.
Conserved, essential genes used for DNA immunization
| MCMV ORF | HCMV homolog | FASTA identitya (%) | Overlap (aa)b |
|---|---|---|---|
| M54 | DNA polymerase catalytic subunit | 50 | 415 |
| M70 | Primase, helicase-primase subunit | 36 | 982 |
| M105 | Helicase, helicase-primase subunit | 43 | 861 |
FASTA identity for the overlap shown.
Overlap length of the aligned MCMV ORF and HCMV homolog sequences. aa, amino acids.
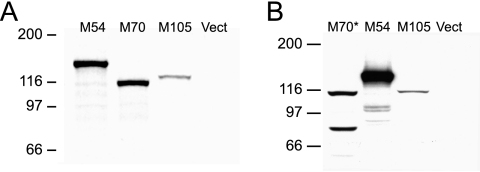
The M54, M70, and M105 genes were cloned from MCMV MW97.01, a Smith strain-derived bacterial artificial chromosome, into a vector that provides the carboxy-terminal V5 epitope and six-His tags (14). The complete sequences of the cloned M54 and M105 ORFs were identical to those of the published sequence of MCMV strain Smith (17) (data not shown). The sequence of the cloned M70 ORF was identical to that of MCMV Smith except for a single insertion mutation 3′ of the ORF that resulted in a complete, but untagged, coding sequence for the M70 protein (data not shown). Confirmation of complete, continuous reading frames for M54, M70, and M105 was provided by coupled in vitro transcription-translation reactions and [35S]methionine labeling (Fig. 1A). Each of the plasmids was found to express a single, labeled polypeptide with the predicted relative molecular mass: 128.8 kDa for M54, 109.6 kDa for the untagged M70, and 111.4 kDa for M105.
FIG. 1.
In vitro and in vivo expression of the MCMV homologs of HCMV DNA polymerase (M54), primase (M70), and helicase (M105). (A) The MCMV ORFs, cloned into pcDNA3.1/V5-His-TOPO, were expressed using the T7 promoter by coupled in vitro transcription/translation reactions (TNT T7 Quick) with [35S]methionine. A portion of each reaction mixture was subjected to reducing SDS-PAGE on a 7% polyacrylamide gel, and the labeled proteins were detected by autoradiography. The numbers and lines on the left indicate the positions and relative molecular masses (in kDa) of the proteins in the marker (not shown). Vect indicates the TNT reaction performed with the DNA vector alone. The predicted molecular masses for the encoded proteins are M54, 128.8 kDa; M70 (untagged), 109.6 kDa; and M105, 111.4 kDa. (B) After the M70 clone was mutated to yield the V5- and six-His-tagged M70*, plasmids were transiently transfected into COS-7 cells and the whole cells were lysed and solubilized in reducing SDS-PAGE sample buffer 48 h posttransfection. The lysate proteins were resolved by SDS-PAGE as described above and electroblotted to a nitrocellulose membrane that was subsequently probed with a mouse anti-V5 monoclonal antibody. The numbers on the left are as described for panel A, and Vect indicates the lysate from cells transfected with empty plasmid vector. The predicted molecular mass for the M70* protein is 114.7 kDa.
To facilitate subsequent Western blot analysis following in vivo expression, the extraneous T at the 3′ end of the M70 ORF was deleted by site-directed mutagenesis. The DNA sequence of the resulting M70 clone (designated M70*) was found to be identical to that of the parent except for the T deletion (data not shown). The M54, M70*, and M105 plasmids were transiently transfected into COS-7 cells for Western blot analysis using a V5-tag-specific monoclonal antibody. We found that the expression of M70* in COS-7 cells yielded a band corresponding to the predicted 114.7 kDa, as well as a faster-migrating band of equal intensity that may represent a proteolytic degradation product (Fig. 1B). No anti-V5 reactive band was observed following Western blot analysis of COS-7 cells transfected with the original mutant M70 clone (data not shown). In addition, the predominant bands seen in the M54 and M105 lanes were of the expected sizes observed following their expression in vitro (Fig. 1B). Duplicate blots were probed with a mouse anti-MCMV hyperimmune serum, but no seroreactive bands were detectable (data not shown). Taken together, these results demonstrate that the plasmids express the full-length, tagged ORFs but that the encoded MCMV antigens do not elicit detectable antibody responses following repeated infection of BALB/c mice.
DNA immunization with the M54 and M105, but not M70, genes elicits protective responses in BALB/c mice.
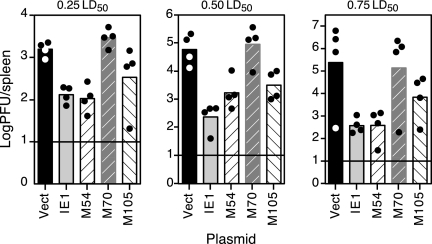
We next asked whether protective prophylactic responses could be generated using any of the three conserved, essential genes in BALB/c mice. Because the gene products of M54, M70, and M105 and their respective HCMV homologs are not likely to be part of the viral envelope, any protective responses elicited following DNA immunization using these genes would likely be from cell-mediated, not neutralizing antibody, responses. To test for the protective efficacies of these plasmid DNAs, four BALB/c female mice per group were i.d. immunized in the shaved flank three times over the course of 2 weeks with (i) either pc3Δneo vector alone (20 μg) or 10 μg of pc3Δneo and 10 μg of (ii) IE1 (pp89), (iii) M54, (iv) M70, or (v) M105. Two weeks after the last immunization, the mice were i.p. challenged with one of three sublethal doses of SG-MCMV: 0.25 × LD50 (2 × 105 PFU), 0.50 × LD50 (4 × 105 PFU), or 0.75 × LD50 (6 × 105 PFU). The spleens were harvested on day 6 postchallenge for determination of their MCMV titers.
We found that, in addition to the positive-control IE1 DNA, the DNAs encoding M54 and M105 were protective against viral replication in the spleen following all of the i.p. challenge doses (Fig. 2). Following challenge with the low dose of MCMV, the mean reductions in the titers provided by immunization with the M54 and M105 DNAs were comparable to that in the IE1 group (12- to 15-fold relative to that in mice immunized with vector alone). While the virus titer reductions in the spleens of the M54- and M105-immunized mice were not as high as those in the IE1-immunized mice following the intermediate challenge dose, both IE1 and M54 immunization resulted in >600-fold reductions in viral titers, and M105 resulted in a 60-fold reduction following the high challenge dose. Finally, we found that the M70 DNA, which, interestingly, has the lowest percent amino acid identity to its HCMV homolog, was not protective at any challenge dose level. These results identify M54 and M105 as protective members of a new class of protective antigens: the highly conserved, essential genes.
FIG. 2.
DNA immunization with M54 (DNA polymerase) or M105 (helicase) is consistently protective against challenge virus replication in the spleen. Four BALB/c mice per group were i.d. immunized in the shaved flank three times in 2 weeks with either empty pc3Δneo vector DNA (Vect) or DNA encoding IE1, M54, M70, or M105, as described in Materials and Methods. Two weeks after the last immunization, the mice were i.p. challenged with one of the three doses of SG-MCMV shown. On day 6 postchallenge, the spleens were removed and homogenized for determination of their MCMV titers. The bars and circles represent the means of the log10 values of the virus titers for each group and the log10 values of the individual virus titers for each of the spleens, respectively. The horizontal lines indicate the limits of detection for the plaque assay.
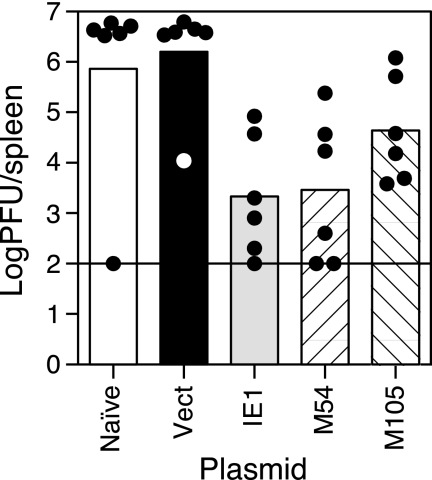
We performed an independent experiment to confirm the protective efficacies of the M54 and M105 DNAs. Groups of BALB/c mice either were left untreated (naïve) or i.d. immunized in the tail three times in 2 weeks with 50 μg of vector DNA or IE1, M54, or M105 DNA. Three weeks after the last immunization, the mice were i.p. challenged with 0.50 × LD50 (4 × 105 PFU) of SG-MCMV, and their spleens were harvested on day 6 postchallenge for determination of the viral titers. As shown in Fig. 2, we found that immunization with the IE1 or M54 DNAs resulted in mean viral titer reductions in the spleens of >700- and >500-fold, respectively, relative to the viral titers in the spleens of the controls immunized with vector DNA alone, with more variability in the levels of protection in individual mice in this experiment (Fig. 3) than in the previous experiment (Fig. 2). In this experiment, immunization with the M105 DNA resulted in more modest reductions in viral titers, with a mean level approximately 40-fold lower than that of the vector DNA group. It is unclear why we observe these outlier titers in the naïve and vector DNA groups, which should have no specific immunity to MCMV. Taken together, immunization with the M54 DNA provided a high level of protection, similar to that elicited by the immunodominant IE1 DNA, while immunization with the M105 DNA elicited more moderate reductions of virus titers.
FIG. 3.
Protection against virus replication in the spleen elicited by M54 and M105 in an independent experiment. Six BALB/c mice per group were either left untreated (Naïve) or i.d. immunized in the tail with 50 μg of empty pc3Δneo vector DNA (Vect) or DNA encoding IE1, M54, or M105. Three weeks after the last immunization, the mice were i.p. challenged with 0.5 × LD50 of SG-MCMV, and on day 6 postchallenge, the spleens were harvested and titered as described in the Fig. 2 legend. The bars, circles, and horizontal line are as described in the Fig. 2 legend.
Immunization with M54 or M105 elicits antigen-specific CD8 T-cell responses that increase rapidly following viral challenge.
Protective responses elicited by DNA immunization with the conserved, essential genes encoding proteins involved in viral DNA replication would likely be due to specific adaptive-cell-mediated immune responses, i.e., CD8 or CD4 T-lymphocyte responses, rather than neutralizing antibody responses. We have shown previously that DNA immunization with IE1- or M84-expressing plasmids, injected either alone or in combination, elicits strong CD8 T-cell responses and protection against viral replication in the spleen after sublethal i.p. challenge (10, 24). We subsequently found that the enumeration of M84-specific CD8 T cells in DNA-immunized or MCMV-infected mice by ICS assay showed consistently higher numbers of IFN-γ+ CD8+ T cells following the stimulation of splenocytes with cells expressing the full-length M84 protein than following stimulation with the epitope peptide defined by Holtappels et al. (8, 23). These results prompted us to measure cell-mediated responses against the full-length protective M54 and M105 gene products, rather than attempt to map all of the H-2d-restricted epitopes of these gene products and possibly overlook some of the protective specificities. We therefore used an ICS assay in which splenocyte stimulation was mediated by BALB SV40 cells, a highly transfectable SV40-transformed H-2d cell line (1), that were previously transfected with DNAs expressing full-length MCMV ORFs. This technique was recently used to characterize the specificities of the CD8 T-cell repertoire of MCMV-infected C57BL/6 mice, using plasmids encoding all 170 of the known MCMV ORFs (14).
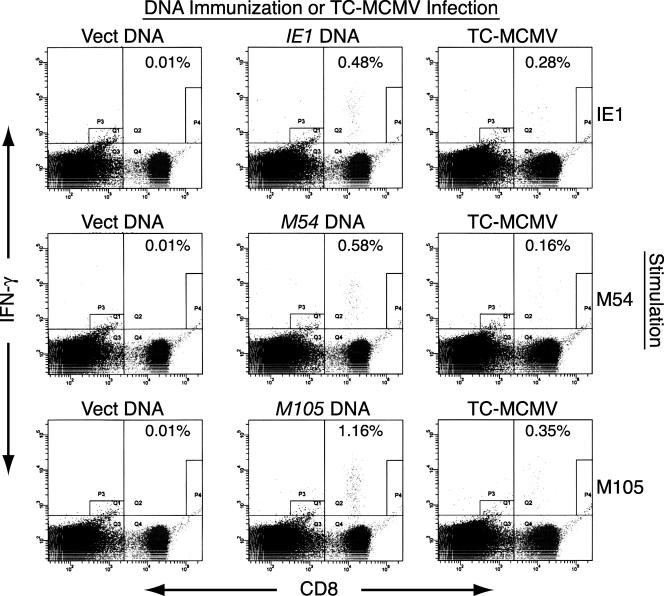
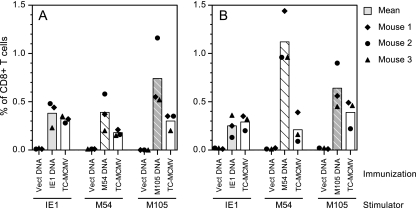
For the measurement of specific CD8 T-cell responses in DNA-immunized mice, BALB/c mice were i.d. immunized in the tail with 25 μg of either empty plasmid vector or plasmid DNA expressing IE1, M54, or M105 (Fig. 4 and 5). The mice were immunized three times in 2 weeks, and an additional immunization was given either 1 week after the third immunization and 2 weeks prior to the ICS assay (Fig. 4 and 5A) or 3 weeks after the third immunization and 3 weeks prior to the ICS assay (Fig. 5B). Additional groups of mice were i.p. infected with TC-MCMV either 2 weeks (Fig. 4 and 5A) or 4 weeks (Fig. 5B) prior to the ICS assay in order to compare the specific CD8 T-cell levels of DNA-immunized mice with those of MCMV-infected mice. For the ICS assay, SV40 BALB stimulator cells were transfected with either IE1, M54, or M105 DNA 48 h prior to harvesting the splenocytes from DNA-immunized or MCMV-infected mice. The splenocytes from three mice per group were incubated with stimulator cells in the presence of brefeldin A for 8 h, prior to staining with anti-CD8 and anti-IFN-γ antibodies and analysis by flow cytometry. Figure 4 shows the flow cytometric results for one representative mouse from each group (mouse 2). We found that the level of background staining of splenocytes from the vector-immunized mice was very low; between 0.01% and 0.02% of the CD8+ T cells stained IFN-γ positive, regardless of the DNA used to transfect the stimulator cells.
FIG. 4.
Flow cytometric analyses of the CD8 T-cell responses in BALB/c mice immunized with DNA expressing IE1, M54, or M105 by using transiently transfected stimulator cells. The mice were i.d. immunized four times in 3 weeks with either empty plasmid vector (Vect DNA) or plasmid DNA expressing IE1, M54, or M105. To measure the CD8 T-cell responses elicited by MCMV infection, another group of mice was i.p. infected with 1.2 × 105 PFU of TC-MCMV. Two weeks after the last DNA immunization or MCMV infection, the mice were sacrificed for ICS assay. As described in Materials and Methods, the splenocytes were incubated in the presence of brefeldin A with BALB SV40 (H-2d) stimulator cells that were transfected 48 h earlier with plasmid DNA expressing either IE1, M54, or M105, as shown (Stimulation). After stimulation, the splenocytes were surface stained with a phycoerythrin-Cy5-conjugated anti-mouse-CD8a, fixed, stained intracellularly with fluorescein isothiocyanate-conjugated anti-mouse-IFN-γ antibodies, and analyzed by flow cytometry. The scatter plot for one mouse per immunization/infection and stimulation group is shown, arbitrarily chosen as mouse 2. The percentages shown are the percentage of CD8+ T cells that were IFN-γ positive after stimulation, with the cell number for CD8 and IFN-γ double-positive cells calculated as the cell number in quadrant Q2 minus the background staining in gate P4.
FIG. 5.
CD8 T-cell responses in BALB/c mice immunized with DNA expressing IE1, M54, or M105. (A) Mice were i.d. immunized in the tail four times in 3 weeks or i.p. infected with 1.2 × 105 PFU of TC-MCMV as described in the Fig. 4 legend, and 2 weeks after the last DNA immunization or MCMV infection, three mice per group were sacrificed for ICS assay. The complete data for the experiment are shown in Fig. 4, with bars representing the mean percentages of CD8+ T cells that were IFN-γ positive for each vaccine group and symbols representing the individual value for each mouse in the group. Note that splenocytes from the same three TC-MCMV-infected mice were tested with those from each of the three stimulator groups. (B) Mice were i.d. immunized three times in 2 weeks, a fourth immunization was given 3 weeks later, and the ICS assay was performed 3 weeks after the last DNA immunization or 4 weeks after infection with TC-MCMV as described for panel A. Vect DNA, DNA vector alone.
Figure 5A shows the resulting CD8 T-cell levels in the mice at 2 weeks after the last DNA immunization or 2 weeks after MCMV infection. The levels of IE1-specific CD8 T cells in IE1 DNA-immunized or MCMV-infected mice were comparable, with mean levels of 0.38% and 0.32%, respectively. Overall, these levels are lower than those obtained when splenocytes were stimulated with the dominant IE1 epitope peptide, most likely as a result of differences in the assays. At the time of stimulation, transfected stimulator cells were routinely 85 to 100% confluent and 60 to 70% of cells were transfected, as measured either by direct fluorescence of EGFP-transfected cells or by immunofluorescent staining of IE1-transfected cells (data not shown). All mice immunized with M54 DNA or infected with MCMV had M54-specific CD8 T cells detectable after stimulation with M54-transfected stimulator cells, with the DNA-immunized mice having slightly higher, but more variable, levels (mean, 0.39%; range, 0.20% to 0.58%) relative to those in the MCMV-infected mice (mean, 0.18%; range, 0.15 to 0.21%) (Fig. 5A). Similarly, immunization with M105 DNA or MCMV infection elicited M105-specific CD8 T cells, with DNA immunization again eliciting higher but more variable levels (mean, 0.74%; range, 0.52 to 1.16%) compared with the levels elicited by MCMV infection (mean, 0.30%; range, 0.20 to 0.35%).
The CD8 T-cell levels were subsequently tested in mice that were rested an additional week after the last DNA immunization (3 weeks total) or 2 weeks after MCMV infection (4 weeks total). Overall, these results (Fig. 5B) were very similar to those described above. The most notable exception is that the M54-specific CD8 T-cell responses in the M54 DNA-immunized mice (mean, 1.1%; range, 0.96 to 1.44%) were further increased relative to those in the MCMV-infected mice. The mean M54-specific CD8 T-cell responses in the MCMV-infected mice were comparable between the two experiments (0.18% and 0.21%), suggesting that the increases observed in the M54 DNA-immunized mice may have resulted from the slower kinetics of the CD8 T-cell response to M54 DNA compared with those of the IE1 or M105 DNA. Taken together, the results of these two experiments demonstrate that the M54 and M105 DNAs consistently elicited significant levels of antigen-specific CD8 T-cell responses, with the response to the M54 DNA perhaps having slower kinetics. M54- and M105-specific CD8 T-cell responses were also detectable following acute i.p. infection with MCMV. However, there was a strong trend toward higher levels of M54- and M105-specific CD8 T cells following DNA immunization than following MCMV infection.
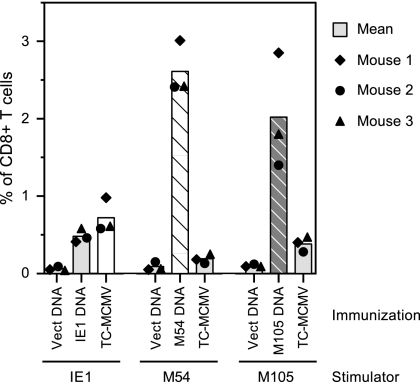
We were also interested in measuring the secondary response to the M54 and M105 gene products, to determine how the CD8 T cells primed by DNA immunization responded to viral challenge. We previously showed that MCMV infection does not prime significant levels of IE1- or M84-specific CD8 T cells by day 5 postchallenge in mice immunized with vector alone (24). In contrast, CD8 T cells in IE1 or M84 DNA-immunized mice were able to respond vigorously by this early time point, indicating that a secondary response had occurred. We were also interested in determining whether the low M54- or M105-specific CD8 T-cell levels primed by MCMV infection would increase following subsequent viral challenge or whether these antigens' subdominance during infection was also observed following reinfection. To this end, three mice per group were DNA immunized three times in 2 weeks and then i.p. challenged with 1.2 × 105 PFU of SG-MCMV 7 weeks after the last immunization, while three of the MCMV-infected mice were similarly reinfected with SG-MCMV 6 weeks after the primary MCMV infection. On day 5 postchallenge or reinfection, splenocytes were isolated and analyzed by ICS assay as described above.
We found that the mean levels of specific CD8 T cells on day 5 postchallenge in the mice immunized with vector alone were 0.06%, 0.09%, and 0.10% when the stimulation was with cells expressing the IE1, M54, or M105 DNAs, respectively (Fig. 6). These results indicate that the primary responses to these antigens were low overall at this time postinfection and show the magnitude of their possible contribution to the levels in the DNA-immunized or MCMV-infected mice. The mean percentage of IE1-specific CD8 T cells in the IE1 DNA-immunized mice was 0.48% (Fig. 6). This value was slightly higher than the peak level observed prechallenge (0.38%; Fig. 5A). By comparison, the mean in the MCMV-infected mice after reinfection was 0.73%, which was at least twofold higher than the peak prechallenge level of 0.32% (Fig. 5A).
FIG. 6.
Levels of CD8 T-cell responses in DNA-immunized or TC-MCMV-infected mice on day 5 after challenge with SG-MCMV. Three BALB/c mice were immunized three times in 2 weeks with either empty vector or plasmid DNA expressing either IE1, M54, or M105. Another group of mice was i.p. infected with TC-MCMV as described in the Fig. 4 legend. Seven weeks after the last DNA immunization or 6 weeks after MCMV infection, the mice were i.p. challenged with 1.2 × 105 PFU of SG-MCMV, and an ICS assay was performed on the splenocytes of these mice on day 5 postchallenge, as described in Materials and Methods. The bars and symbols are as described in the Fig. 5 legend.
The CD8 T-cell levels in the mice that were MCMV challenged at week 7 after the last immunization with M54 or M105 DNA were also two- to threefold higher than their respective peak levels prechallenge. In contrast, in the mice previously infected with MCMV, the postchallenge levels of CD8 T cells specific for M54 and M105 were almost identical to their respective peak prechallenge levels. Although it is not known what the levels of CD8 T cells specific to M54 and M105 in this group of DNA-immunized mice would have been without restimulation with challenge virus, we previously found that IE1 or M84 DNA-immunized mice had CD8 T-cell levels that either decreased or remained constant 2 weeks after the last immunization (24). In these previous experiments, the level of IE1 peptide-specific CD8 T cells was 5.2% at week 2 after the last immunization and then declined to 1.7% at week 4, while the level of M84 peptide-specific CD8 T cells was 1.1% at week 2 and 1.0% at week 4. Taken together, our data showed that the M54- and M105-specific CD8 T cells in the DNA-immunized mice were able to respond rapidly to viral challenge with increasing levels of IFN-γ+ cells, while the low levels of M54- and M105-specific cells in MCMV-infected mice were not increased by subsequent reinfection.
DISCUSSION
In this study, we focused our search for protective CD8 T-cell targets of MCMV on the E genes whose homologs in HCMV have been found to be essential for viral replication in cultured cells. HCMV UL54 and UL105 were demonstrated to be essential for oriLyt-dependent DNA replication in a transient transfection system (15), and their requirement for viral replication in vivo has been confirmed by the independent mutagenesis studies by Dunn et al. and Yu et al. (4, 25). UL54 encodes the DNA polymerase catalytic subunit, and based on amino acid homology with the HSV-1 helicase protein encoded by UL5, HCMV UL105 is believed to contain the helicase activity of the proposed helicase-primase complex (UL70-UL102-UL105). Thus, it is unlikely that MCMV would be able to severely downregulate or abrogate the expression of M54 and M105 in vivo in a productive infection in the face of cognate effector CD8 T cells. However, it remains a possibility that MCMV could downregulate the expression of M54 or M105 in infected tissues to a level that promotes a lower rate of replication and reduced pathogenicity but avoids the surface presentation of sufficient M54- or M105-derived peptides to be recognized by their cognate CD8 T cells. The similar protective abilities of the IE1 and M54 DNAs shown by our protection assay of the levels of acute-phase splenic viral replication following relatively high-dose systemic challenge with virulent virus do not illustrate a qualitative difference between the net protective abilities of IE1- and M54-specific CD8 T cells, but work is in progress to determine if there are any differential antiviral effects of these cells during chronic or latent infection or in tissues of clinical relevance for HCMV disease such as the lungs.
We found, by ICS assay, that DNA immunization with M54 or M105 consistently elicited CD8 T-cell responses capable of IFN-γ secretion upon short-term incubation with transfected H-2d stimulator cells. Thus, there exists at least one H-2d-restricted antigenic peptide within each polypeptide sequence. It also appeared that the peak response to M54 may be delayed relative to the peak response to M105. This is consistent with our previous finding that the kinetics of IE1- and M84-specific CD8 T-cell responses also differed slightly (24). It is also possible that immunization with the M54 and/or M105 DNAs elicited specific CD4 T-cell responses that either mediated or contributed to the observed protection, either by direct lytic or helper functions, and additional studies would be required to determine the precise immune correlates of protection.
When we examined whether MCMV infection generated CD8 T-cell responses to either M54 or M105, we found that the acute infection primed specific responses to these antigens, but that the responses were slightly lower than those in the DNA-immunized mice. The M54-specific responses to acute MCMV infection in individual mice were between 0.09% and 0.39% in two experiments, while the M105-specific responses in these experiments ranged between 0.20% and 0.49%. We conclude that MCMV infection does elicit specific CD8 T-cell responses to antigenic peptide(s) of both M54 and M105. Although the mean and individual CD8 T-cell responses that were specific for M105 and IE1 were similar for the MCMV-infected mice within each of the two experiments shown (Fig. 5A and B), we must refrain from making quantitative comparisons between the transfection groups because the assay has not been rigorously optimized for each antigen. However, because the M54 and M105 DNAs were each able to elicit strong peak responses to their encoded antigens in each experiment relative to those elicited by MCMV infection (unlike the IE1 responses), we suspect that further analyses would classify M54 and M105 among the subdominant antigens of MCMV in BALB/c mice. While acute MCMV infection may not prime high levels of CD8 T cells that are specific to M54 and M105, the efficacy of the CD8 T cells primed by DNA immunization demonstrates that M54- and M105-derived antigenic peptides are indeed presented by infected splenocytes in vivo.
Sylwester et al. and Munks et al. have comprehensively documented the repertoires of CD8 T-cell immunity to HCMV and MCMV infection, respectively (14, 22). In the case of HCMV, of the 33 seropositive subjects, only 3 (9%) had detectable UL54 peptide-specific CD8 T-cell responses, and the levels were less than 1% (22). UL105 was recognized by CD8 T cells in these subjects at a slightly higher frequency, as 6 of 33 subjects (18%) had detectable UL105-specific CD8 T-cell responses, but only 1 subject had a level that was greater than 1%. By comparison, for UL83, the second-most-frequently detected ORF in this study, 18 of 33 subjects (55%) had detectable UL83-specific CD8 T-cell responses, with 7 subjects having levels greater than 1%. Although these CD8 T-cell response frequencies and magnitudes suggest that UL54 is among the subdominant antigens, these data describe the HCMV specificities of the memory pool, and it is not known whether the responses to UL54 and UL105 were higher during the acute infection.
In a pool of T cells from three acutely infected C57BL/6 mice, the CD8 T-cell repertoire included a low, but >6-fold above the background, response to M54 (0.13% of CD8 T cells). However, there was no measurable response to M105 (14). In our experiments with acutely infected BALB/c mice, using the same methodology as used in the C57BL/6 study, we found similar mean responses to M54, of 0.2% of CD8 T cells, but higher levels of M105-specific CD8 T cells, of 0.3 to 0.4%. While these responses primed to M54 and M105 during acute MCMV were low, we were interested in determining whether reinfection with MCMV would increase these CD8 T-cell levels or whether the mechanism(s) that governs the apparent subdominance of these antigens during infection would prevent an increased secondary response. We found, upon reinfection of mice, that the levels of M54- and M105-specific CD8 T cells (Fig. 6) were not appreciably higher than their respective levels in the acutely infected mice at 4 weeks postinfection (Fig. 5B). However, it is possible that there were further declines in CD8 T-cell levels between week 4 (the last time point measured before reinfection) and week 6 (when mice were reinfected) after the primary infection and that the subsequent reinfection resulted in increases of CD8 T-cell responses back to their week-4 levels. In any event, the responses to M54 and M105 in the reinfected mice contrasted with the IE1-specific CD8 T-cell levels, which had increased at least twofold upon reinfection with MCMV at the time points measured. Thus, if preexisting immunity to MCMV in the infected mice had prevented the subsequent infecting virus (or viral antigen) from entering the splenic compartment to restimulate secondary responses to M54 or M105, the secondary response to IE1 was not similarly abrogated. The generation of a secondary response to IE1 likely has a temporal advantage over the response to the M54 and M105 E proteins, and thus, increases in the IE1-specific CD8 T cells, which are highly protective, may have precluded sufficient E gene expression in infected splenocytes to restimulate the CD8 T cells primed against M54 and M105 epitopes. It is not known whether an increased dose of challenge virus would overcome the lack of secondary responses against M54 and M105, as secondary responses to these antigens in the DNA-immunized mice were clearly observed. The secondary responses in the IE1 DNA-immunized mice were not as high as those we have observed previously in ICS assays using the immunodominant IE1 Ld-restricted nonapeptide epitope (24), possibly due to differences in the ICS assays used.
Having found that the responses to two of the three conserved, essential E genes of MCMV tested were protective against viral replication prompts us to speculate whether the CD8 T-cell response to MCMV infection is purposely skewed by the virus to limit the responses to these antigens. The skewing in favor of an immunodominant, but ultimately ineffective, CD8 T-cell response has been demonstrated with the lack of viral control in immunoablated C57BL/6 mice reconstituted with M45-specific CD8 T cells (7). It was shown that the Db-restricted M45 peptide is very effective at priming a specific CD8 T-cell response, presumably through cross-presentation of the M45 protein in uninfected dendritic cells, but that the MHC class I presentation of this peptide in the infected tissues is sufficiently blocked by the immunoevasin m152 to limit the effector mechanisms of viral control. While these results show how the viral immunoevasins may negatively regulate the antiviral effects of primed virus-specific CD8 T cells, a mechanism by which negative skewing of the primary CD8 T-cell response to viral antigens may occur is more difficult to explain, given the existing state of knowledge of T-cell priming through direct and cross-presentation. However, the CMVs have successfully coevolved with and spread among their respective hosts over the last ∼70 million years, since the time of the mammalian radiation. Undoubtedly, the selective immune pressures of the host have helped to shape the immunodominance hierarchy and the virus-host balance to establish a T-cell repertoire that favors lifelong, persistent infection that is normally not pathogenic. A benefit of using DNA immunization to elicit virus-specific CD8 T cells is that responses can be generated in the absence of this immunodominance hierarchy or any undiscovered viral mechanisms that could limit the priming of otherwise highly effective CD8 T-cell subsets. However, the primed CD8 T cells would still require that their cognate peptides are presented in infected tissues in the face of the immunoevasins in order for the DNA vaccine to be protective. Our results using the BALB/c mouse model do not predict whether human vaccination with DNAs expressing HCMV UL54, UL105, or any of the other conserved, essential genes would elicit a CD8 T-cell response or protection. In the case of UL54 and UL105, while these antigens have been found to elicit CD8 T-cell responses in humans (22), the ability of the corresponding DNA vaccines to provide CD8 T-cell responses across multiple HLA types is not known, and a multivalent vaccine may be required to give broad coverage. Our hope is that a CMV vaccine that can prime virus-specific CD8 T-cell specificities that are normally negatively skewed during infection may be able to generate a CD8 T-cell repertoire that is qualitatively different than that normally elicited by the natural infection. The vaccine may be able to succeed where natural immunity fails, in the generation of sterilizing immunity or immune control of vertical or horizontal virus transmission.
Acknowledgments
This work was supported by research grants 1-FY2002-4360 from the March of Dimes Birth Defects Foundation and AI 051557 from NIH.
We thank Neil Sekiya for the flow cytometry analyses and Paul Erlich and Christian Welch for technical assistance.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Arase, H., E. S. Mocarski, A. E. Campbell, A. B. Hill, and L. L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323-1326. [DOI] [PubMed] [Google Scholar]
- 2.Brune, W., H. Hengel, and U. H. Koszinowski. 1999. A mouse model for cytomegalovirus infection, p. 19.17.11-19.17.13. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology, vol. 4. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed] [Google Scholar]
- 3.Cranmer, L. D., C. L. Clark, C. S. Morello, H. E. Farrell, W. D. Rawlinson, and D. H. Spector. 1996. Identification, analysis, and evolutionary relationships of the putative murine cytomegalovirus homologs of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J. Virol. 70:7929-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott, R., C. Clark, D. Jaquish, and D. H. Spector. 1991. Transcription analysis and sequence of the putative murine cytomegalovirus DNA polymerase gene. Virology 185:169-186. [DOI] [PubMed] [Google Scholar]
- 6.González Armas, J. C., C. S. Morello, L. D. Cranmer, and D. H. Spector. 1996. DNA immunization confers protection against murine cytomegalovirus infection. J. Virol. 70:7921-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holtappels, R., J. Podlech, M. F. Pahl-Seibert, M. Julch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holtappels, R., D. Thomas, and M. J. Reddehase. 2000. Identification of a K(d)-restricted antigenic peptide encoded by murine cytomegalovirus early gene M84. J. Gen. Virol. 81:3037-3042. [DOI] [PubMed] [Google Scholar]
- 9.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morello, C. S., L. D. Cranmer, and D. H. Spector. 2000. Suppression of murine cytomegalovirus (MCMV) replication with a DNA vaccine encoding MCMV M84 (a homolog of human cytomegalovirus pp65). J. Virol. 74:3696-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morello, C. S., M. Ye, S. Hung, L. A. Kelley, and D. H. Spector. 2005. Systemic priming-boosting immunization with a trivalent plasmid DNA and inactivated murine cytomegalovirus (MCMV) vaccine provides long-term protection against viral replication following systemic or mucosal MCMV challenge. J. Virol. 79:159-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morello, C. S., M. Ye, and D. H. Spector. 2002. Development of a vaccine against murine cytomegalovirus (MCMV), consisting of plasmid DNA and formalin-inactivated MCMV, that provides long-term, complete protection against viral replication. J. Virol. 76:4822-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munks, M. W., K. S. Cho, A. K. Pinto, S. Sierro, P. Klenerman, and A. B. Hill. 2006. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J. Immunol. 177:450-458. [DOI] [PubMed] [Google Scholar]
- 14.Munks, M. W., M. C. Gold, A. L. Zajac, C. M. Doom, C. S. Morello, D. H. Spector, and A. B. Hill. 2006. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J. Immunol. 176:3760-3766. [DOI] [PubMed] [Google Scholar]
- 15.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pass, R. F. 2001. Cytomegaloviruses, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 17.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 19.Reddehase, M. J. 2000. The immunogenicity of human and murine cytomegaloviruses. Curr. Opin. Immunol. 12:390-396. [DOI] [PubMed] [Google Scholar]
- 20.Reddehase, M. J., C. O. Simon, J. Podlech, and R. Holtappels. 2004. Stalemating a clever opportunist: lessons from murine cytomegalovirus. Hum. Immunol. 65:446-455. [DOI] [PubMed] [Google Scholar]
- 21.Stratton, K., J. Kurch, and R. Lawrence. 2000. Vaccines for the 21st century: a tool for decision making. National Academy Press, Washington, DC. [PubMed]
- 22.Sylwester, A. W., B. L. Mitchell, J. B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P. R. Sleath, K. H. Grabstein, N. A. Hosken, F. Kern, J. A. Nelson, and L. J. Picker. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202:673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye, M., C. S. Morello, and D. H. Spector. 2004. Multiple epitopes in the murine cytomegalovirus early gene product M84 are efficiently presented in infected primary macrophages and contribute to strong CD8+-T-lymphocyte responses and protection following DNA immunization. J. Virol. 78:11233-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye, M., C. S. Morello, and D. H. Spector. 2002. Strong CD8 T-cell responses following coimmunization with plasmids expressing the dominant pp89 and subdominant M84 antigens of murine cytomegalovirus correlate with long-term protection against subsequent viral challenge. J. Virol. 76:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]