Abstract
Type III interferon (IFN) is a novel member of the interferon family. Type III IFN utilizes a receptor complex different from that of type I IFN, but both types of IFN induce STAT1, STAT2, and STAT3 activation. Here we describe a detailed comparison of signal transduction initiated by type I and type III IFN. Gene expression array analysis showed that IFN types I and III induced a similar subset of genes. In particular, no genes were induced uniquely by type III IFN. Next, we used chromatin immunoprecipitation (ChIP) analysis to investigate the promoter activation by types I and III IFN. The ChIP assays demonstrated that stimulation of cells with both type I and type III IFN resulted in the recruitment of ISGF3 transcription factor components to the promoter region of responsive genes and in an increase of polymerase II loading and histone acetylation. Whereas IFN type I signaling was observed for a broad spectrum of cell lines, type III IFN signaling was more restricted. The lack of IFN type III signaling was correlated with a low expression of the IL28Ra component of the IFN type III receptor, and IL28Ra overexpression was sufficient to restore IFN type III signaling. We also tested the activation of mitogen-activated protein (MAP) kinases by type III IFN and found that type III IFN relies strongly upon both p38 and JNK MAP kinases for gene induction.
Type I interferons (IFNs) are expressed as a first line of defense against viral infections. The primary role of type I IFN is to limit viral spread during the first days of a viral infection, allowing sufficient time for the generation of a strong adaptive immune response against the infection. Type I IFNs combat viral infection both directly by inhibiting viral replication in infected cells and indirectly by stimulating the adaptive immune system (3, 9, 26, 29). The central role of type I interferon in innate immunity toward viral infections was demonstrated by deleting the type I IFN receptor (IFNAR1) in mice. The IFNAR1 knockout mice were extremely susceptible to infection with a broad panel of viruses (17). Type II gamma IFN (IFN-γ) displays some of the antiviral properties of type I IFN, but the dominating biological role of IFN-γ seems to be stimulation of the adaptive immune system, primarily activation of T cells (3, 17). Both type I and II IFNs are members of the class II cytokines. This family, which constitutes a functionally diverse but crucial set of cytokines (10, 20), also includes interleukin (IL)-10, IL-19, IL-20, IL-22, and several other related interleukins. The search for novel members of the class II cytokine or class II cytokine receptor family led to the identification of IFN-λ, also named IL-28/IL-29 by two independent groups. Kotenko et al. identified an orphan receptor which was a member of the cytokine class II family of receptors and IFN-λ as the ligand for this receptor (10). Sheppard et al. identified a novel set of human genes with weak similarities to type I IFN and IL-10 (27). By testing for binding to different class II receptors, they identified the same orphan receptor that Kotenko et al. found as the IFN-λ receptor. Humans carry three genes for IFN-λ, encoding IFN-λ1 (IL-29), IFN-λ2, and IFN-λ3 (IL-28A/B), and this group of cytokines is now collectively referred to as type III IFNs.
IFN-λ functionally resembles type I IFN, inducing antiviral protection in vitro (10, 23, 27) as well as in vivo (1). Activation of the IFN-λ receptor leads to the phosphorylation of STAT1, STAT2, and STAT3 and the formation of the interferon-stimulated gene factor 3 (ISGF3) transcription factor (10) and to the induction of typical IFN-induced genes like the OAS and MxA genes. IFN-λ can reduce cell growth in vitro and possesses antitumor activity in several rodent models (11, 25). However, a number of cytokines with very different biological effects activate STAT transcription factors, and pronounced functional differences between type I and type III IFNs exist. The in vivo antiviral activity of IFN-λ against herpes simplex virus 2 (HSV-2) has been shown to be comparable to that of IFN-α in a systemic model. However, a model for the clinically relevant vaginal HSV-2 infection revealed an antiviral activity of IFN-λ that surpassed that of IFN-α (1).
The biological effect of the cytokine-receptor system is determined primarily by three factors: the expression profile of the cytokine itself, the expression profile of the receptor, and the set of target genes for regulation. We decided to start our investigation of the function of the IFN-λ system by asking which genes are regulated by IFN-λ. A gene array experiment covering the whole human genome revealed that all IFN-λ-induced genes were also induced by type I IFN. Thus, no genes were identified as being uniquely regulated by IFN-λ. Using the gene chip data as the starting point, we selected a set of model genes for investigation of the signal transduction by the IFN-λ receptor complex. The data indicated that despite the use of different receptors for type I and type III IFNs, both types of interferon result in activation of the ISGF3 transcription factor. Furthermore, both IFNs lead to activation of the mitogen-activated protein (MAP) kinases p38 and JNK but not ERK, and IFN-λ depends on this activation for gene induction.
MATERIALS AND METHODS
Cell culture.
Cell lines HepG2, SW13, MCF-7, MDA-MB-231, HEK293, HT1080, and derivative cell lines were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (FCS) and penicillin and streptomycin. Raji cells were grown in RPMI medium (Invitrogen) supplemented with 10% FCS and penicillin and streptomycin. Cells were stimulated with human IFN-α2a (catalog no. 11100-1; Stratech Scientific Ltd.) or recombinant human IFN-λ1/IL-29 (catalog no. 1598-IL; R&D Systems) as indicated.
RNA extraction and real-time quantitative PCR.
RNA extraction was performed with TRI-reagent (Sigma) according to the manufacturer's instructions. Reverse transcription (RT) was performed with an iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR (qPCR) was performed using an iCycler with SYBR Green PCR master mix (FINNZYMES) with the PCR program as follows: 95°C for 15 min (94°C for 15 s, 58°C for 20 s, and 72°C for 20 s) for 40 cycles, then 72°C for 10 min, followed by melting curve analysis. Expression values were normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level. Information concerning primer sequences is available upon request.
Gene chip data.
RNA was extracted from Raji cells stimulated with 200 units of IFN-α or 10 ng/ml IFN-λ for 4 h. RNA quality was confirmed (by Agilent analysis) and processed for analysis on Affymetrix S130 high-density oligonucleotide arrays, according to Affymetrix standard procedures (Aros Biotechnology, Aarhus, Denmark). Three different induction experiments were analyzed for IFN-α and IFN-λ stimulation, along with a triplicate analysis of the unstimulated control cell RNA. Data were calculated using Affymetrix software, and the subsequent statistical analyses were performed using MeV software from the TM4 suite (24). To determine the genes that were differentially expressed, either analysis by Student's t test or a significance analysis of microarray (SAM) test was used.
Chromatin immunoprecipitation.
The two-step cross-linking method for chromatin immunoprecipitation (ChIP) was performed as described previously (19). The antibodies used for ChIP included H-205 (for IRF-1), C-19 (for IRF-2), IRF9 and H-143 (for ISGF-3g p48), E-23 (for STAT1 p84/p91), C-24 (for STAT1a p91), C-20 (for STAT2), K-15 (for STAT3), and H-224 (for Pol II; Santa Cruz Biotechnology). Histone H3 acetyl K9 (catalog no. ab4441) was obtained from Abcam. For quantitative detection of retained DNA, qPCR analysis was performed in triplicate. The primer sets used analysis of the promoter and 5′-transcribed region of the ISG56 gene, and the GAPDH control for normalization is available upon request. Also, the sequence of additional primers used in ChIP analysis is available upon request.
Inhibition of MAP kinases.
Raji cells were treated for 15 min with 5 μM of U0126 or with 50 μM of SB202190 or SP600125. Mock treatment was used as control. The cytokine was added to the media for 4 h, RNA was harvested, and expression levels of ISG56 and 9-27 were measured by real-time RT-PCR. The chemical inhibitors used were SB202190 (Sigma-Aldrich) (for p38), Sp600125 (Sigma-Aldrich) (for JNK), and U0126 (Sigma-Aldrich) (for MEK-1).
Detection of phosphoproteins.
For detection of the phosphorylation status of STAT2, p38, JNK, and ERK, we used Luminex technology. Briefly, the filter plate was washed with assay buffer, and 50 μl of freshly vortexed antibody-conjugated beads was added to each well. The plate was washed with assay buffer, and samples and standards were added. After a brief shake (30 s at 1,100 rpm), the plate was incubated at 4°C overnight in the dark with light shaking (300 rpm). After one wash step, 25 μl of the detection antibody was added to each well, and the plate was shaken and incubated as above. Subsequently, the plate was washed and incubated for 30 min with 50 μl of a streptavidin-phycoerythrin solution with shaking (30 s at 1,100 rpm and 10 min 300 rpm). Finally, the plate was washed, and 125 μl of assay buffer was added to each well, and the plate was shaken for 10 s at 1,100 rpm and read immediately on a Bio-Plex Luminex reader.
RESULTS
Expression array analysis of IFN-α- and IFN-λ-regulated genes.
We stimulated Raji cells with 200 units of IFN-α or 10 ng/ml of IFN-λ for 4 h, harvested total RNA 4 h later, and conducted an expression array experiment using Affymetrix S130 high-density oligonucleotide array chips. Each experiment was performed in triplicate. Data were calculated using Affymetrix software, and subsequent statistical analyses were performed using MeV software from the TM4 suite (24). First we used a Student t test for finding genes which displayed significantly different expression levels between the control and either the IFN-α or IFN-λ treatment. Furthermore, we omitted genes which displayed less than twofold up- or down-regulation relative to that of the control group. Results are presented in Table 1, and the full lists of IFN-α- and IFN-λ-regulated genes are shown in Table S1 and Table S2 in the supplemental material. Our initial analysis showed that 313 entries were significantly up-regulated at least twofold following IFN-α treatment and 38 entries were up-regulated following IFN-λ treatment. As several genes are represented multiple times at the gene chips, the 38 entries up-regulated by lambda interferon represent 27 unique genes.
TABLE 1.
Fold change of IFN-λ-regulated gene expression
| Gene title | Gene product | Fold induction by IFN-γ |
|---|---|---|
| Interferon-induced protein 44 | IFI44 | 13.6 |
| Chromosome 1 open reading frame 29 | C1orf29 | 9.0 |
| Chromosome 7 open reading frame 6 | C7orf6 | 8.1 |
| Interferon-induced protein with tetratricopeptide repeats 5 | IFIT5 | 7.0 |
| Epithelial stromal interaction 1 (breast) | EPSTI1 | 6.7 |
| Chromosome 7 open reading frame 6 | C7orf6 | 5.8 |
| Ubiquitin-specific protease 18 | USP18 | 5.4 |
| 2′-5′-Oligoadenylate synthetase 2, 69-71 kDa | OAS2 | 5.1 |
| 2′-5′-Oligoadenylate synthetase 1, 40-46 kDa | OAS1 | 4.6 |
| Interferon, alpha-inducible protein (clone IFI-15K) | G1P2 | 4.4 |
| Interferon regulatory factor 7 | IRF7 | 3.9 |
| Signal transducer and activator of transcription 1, 91 kDa | STAT1 | 3.9 |
| Tripartite motif-containing 22 | TRIM22 | 3.8 |
| Interferon-induced protein 35 | IFI35 | 3.6 |
| Peroxisomal proliferator-activated receptor A-interacting complex | PRIC285 | 3.2 |
| Tudor domain containing 7 | TDRD7 | 3.2 |
| Interferon regulatory factor 9 | IRF9 | 2.7 |
| Unknown | EST | 2.6 |
| B aggressive lymphoma gene | BAL | 2.6 |
| Zinc finger CCCH type domain containing 1 | ZC3HDC1 | 2.5 |
| Alu repetitive element | EST | 2.3 |
| Unknown | cDNA | 2.3 |
| Sterile alpha motif domain containing 9 | SAMD9 | 2.3 |
| Phospholipid scramblase 1 | PLSCR1 | 2.3 |
| Unknown | EST | 2.2 |
| Apolipoprotein L, 3 | APOL3 | 2.1 |
| SP110 nuclear body protein | SP110 | 2.0 |
Next, we determined whether any genes were regulated by IFN-λ but not by IFN-α. We did this by performing a SAM test with the IFN-λ data and comparing the results to those of the grouped control and IFN-α data. We also manually inspected the raw data for any gene significantly induced by IFN-λ but not IFN-α. In both cases, we failed to detect any genes which were uniquely regulated by IFN-λ. Thus, IFN-λ-induced genes are a subset of the IFN type I-induced genes. Since we detected 313 IFN-α-induced genes and only 38 IFN-λ-induced genes in our initial analysis, one could expect a larger number of IFN-α-specific genes. Nevertheless, the SAM test detected only 10 genes which were induced specifically by IFN-α in the expression array experiment. This is primarily because the IFN-λ signal is weaker, and many of the genes do display IFN-λ regulation but do not reach a twofold cutoff.
Real-time PCR analysis of IFN-α- and IFN-λ-induced genes.
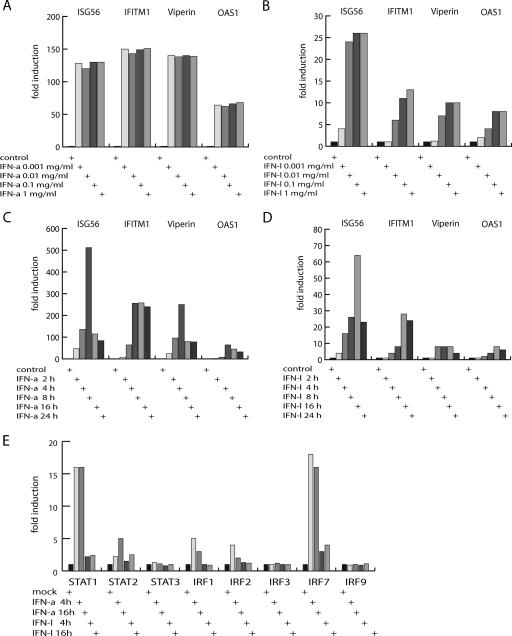
To evaluate the results from the expression array analysis, we selected a set of model genes and used a real-time PCR-based expression analysis to determine the kinetics of induction by IFN-α and IFN-λ with Raji cells. We tested several concentrations of IFNs as well as different time periods of stimulation. The genes examined by real-time PCR analysis were the ISG56/IFIT1, OAS1, IFITM1/9-27, and Viperin/cig5 genes, all of which were induced by IFN-α but of which only ISG56/IFIT1 and OAS1 were significantly induced by IFN-λ in the array analysis. Using different IFN-α concentrations, a robust and relatively concentration-independent response was detected for all four genes after a 4-h induction period (Fig. 1A). With IFN-λ stimulation, a concentration-dependent response was observed after a 4-h induction period (Fig. 1B). All four genes were induced by high IFN-λ concentrations, but none of them to the same extent as that observed for IFN-α stimulation. We also noted that IFITM1 and Viperin were only significantly induced starting at the minimal IFN-λ concentration of 0.01 μg/ml. By changing the stimulation time with a fixed concentration of IFN-α, maximum induction was observed after 8 h. For ISG56, a clear decrease in induction was observed at later time points, whereas Viperin, IFITM1, and OAS1 also showed a more constant induction level for later time points (Fig. 1C). With an IFN-λ concentration of 0.01 μg/ml, the maximal induction was observed after 16 h (Fig. 1D). Again, a clear decrease in ISG56 induction was observed after 24 h of induction. By performing real-time PCR analysis of IFN-α- and IFN-λ-stimulated HepG2 cells, it was similarly observed that genes induced by IFN-α also were induced by IFN-λ but to a lower level (see Fig. 3 and data not shown). In conclusion, the examined IFN-α-induced genes were all induced by IFN-λ, but the IFN-λ response was weaker and showed more pronounced sensitivity to concentration and induction time.
FIG. 1.
Response profiles of IFN-α- and IFN-λ-induced genes. (A) IFN-α concentration dependence for gene induction. The expression levels of ISG56/IFIT1, OAS1, IFITM1/9-27 h, and Viperin/cig5 were estimated by real-time PCR. The values represent combined results from two independent induction experiments. All values were normalized to the GAPDH expression level, which was invariable by IFN stimulation. The expression values obtained without IFN stimulation were normalized to 1. IFN-α was used in the concentrations 1 μg/ml, 0.1 μg/ml, 0.01 μg/ml, and 0.001 μg/ml to stimulate Raji cells for a 4-h period. (B) IFN-λ concentration dependence for gene induction. The experiments were performed as described in the legend for panel A, except IFN-λ was used at the following concentrations: 1 μg/ml, 0.1 μg/ml, 0.01 μg/ml, and 0.001 μg/ml. (C) IFN-α stimulation time dependence for gene induction. Experiments were performed as described in the legend to panel A, except IFN-α was used at the concentration of 0.01 μg/ml to stimulate Raji cells for 2 h, 4 h, 8 h, 16 h, or 24 h. (D) IFN-λ stimulation time dependence for gene induction. Experiments were performed as described in the legend to panel C, except IFN-λ was used at a concentration of 0.01 μg/ml to induce Raji cells for 2 h, 4 h, 8 h, 16 h, or 24 h. (E) IFN-α- and IFN-λ-stimulated induction of transcription factor genes. Raji cells were stimulated with 0.01 μg/ml IFN-α or 0.01 μg/ml IFN-λ for 4 h and 16 h. Using real-time PCR, the expression levels of STAT1, STAT2, STAT3, IRF1, IRF2, IRF3, IRF7, and IRF9 were estimated. Values were normalized to the expression level of GAPDH. The expression values obtained without IFN induction were normalized to 1.
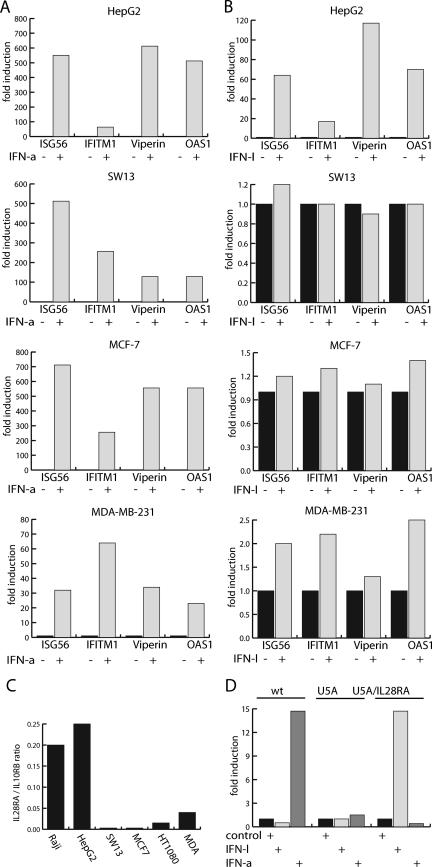
FIG. 3.
Cell specificity of the IFN-λ response is receptor mediated and independent of a functional IFN-α receptor. (A and B) Different cell lines were stimulated with 0.02 μg/ml IFN-α (A) or 0.05 μg/ml IFN-λ (B), and the expression levels of ISG56/IFIT1, OAS1, IFITM1/9-27 h, and Viperin/cig5 were estimated by real-time PCR. Cell lines used were HepG2, MCF-7, MDA-MB-231, and SW13. All values were normalized to the GAPDH expression level, and for cells not stimulated by IFN, the expression level was normalized to 1. (C) Expression ratio of IL28Rα and IL10Rβ. The expression levels of the IFN-λ receptor subunits IL28Rα and IL10Rβ were estimated by real-time PCR in Raji, HepG2, MCF-7, MDA-MB-231, SW13, and HT1080 cells. Values were normalized to the corresponding GAPDH values. Data are presented as the expression ratio between IL28Rα and IL10Rβ. Note that IL10Rβ was expressed in a relative constant ratio to GAPDH in the examined cell lines. (D) Expression of the IFN-λ receptor IL28Rα subunit restores IFN-λ signaling, and IFN-λ signaling functions independent of the IFN-α receptor. HT1080 cells and HT1080-U5A cells were used in the analysis with the latter lacking a functional IFNAR2 receptor subunit. Cells were stimulated with 0.1 μg/ml IFN-λ or 1 kU/ml IFN-α. U5A/IL28Rα indicates that cells were transiently transfected with an expression vector for IL28Rα. Using real-time PCR, the expression level of ISG56 was determined and normalized to the GAPDH expression level and given the value 1 for the expression level in cells not stimulated by IFN.
One primary consequence of cellular stimulation with IFNs is the transcriptional and posttranscriptional activation of IFN response factors that mediates transcription. First, we compared the expression of STAT and IRF members by using the expression array test and found that only IRF1 and IRF2 differed significantly between IFN-α and IFN-λ treatment, with only IFN-α stimulation resulting in significant induction of IRF1 and IRF2. We investigated this further by RT-PCR. Raji cells were stimulated for 4 h or 16 h to compare the transcriptional responsiveness of IFN-signaling transcription factors for IFN-α and IFN-λ stimulation. Under these experimental conditions, we had observed a clear induction of the ISG56/IFIT1, OAS1, IFITM1/9-27, and Viperin/cig5 genes by both IFN-α and IFN-λ stimulation (Fig. 1D). The real-time PCR analyses showed robust induction of STAT1 and IRF7 by both IFN-α and IFN-λ, whereas no significant induction of IRF3, IRF9, or STAT3 was observed for either of the cytokines (Fig. 1E). STAT2, IRF1, and IRF2 were modestly induced by IFN-α, and of these, only STAT2 was also induced by IFN-λ stimulation to a measurable level (Fig. 1E). Thus, the RT-PCR experiments confirmed the findings from the gene array experiments. We note that for HepG2 cells, induction of IRF1 was observed after both IFN-α and IFN-λ stimulation, whereas no induction of IRF7 was observed with either type I or type III IFN (data not shown).
IFN-α and IFN-λ stimulate promoter recruitment of a common set of transcription factors.
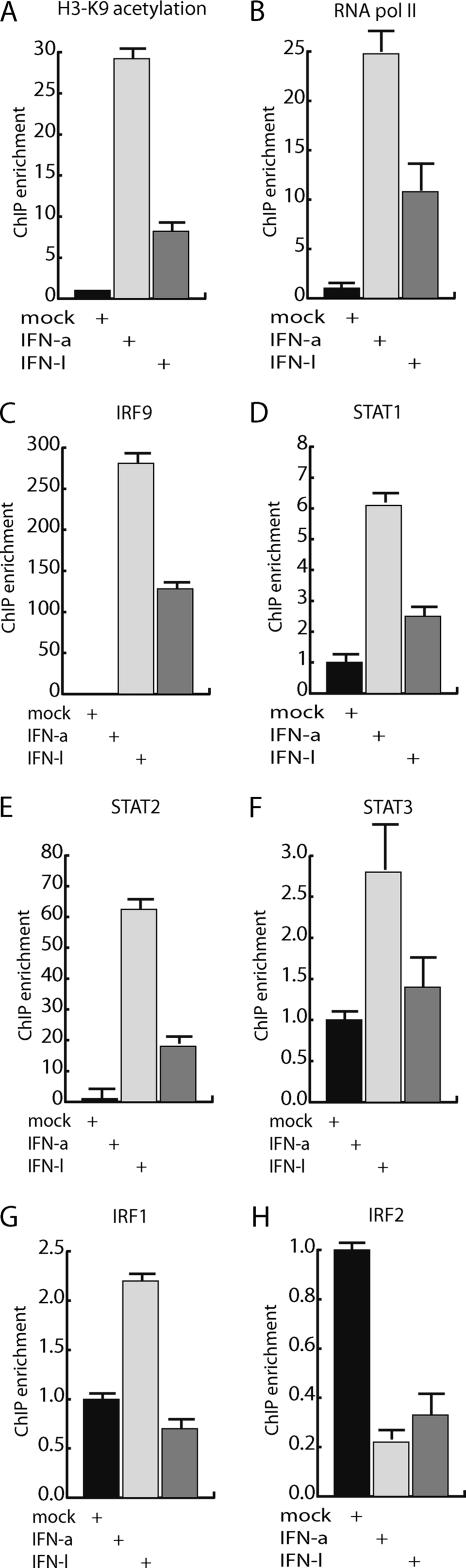
To monitor the recruitment of transcriptional regulatory proteins to promoters of responsive genes as a consequence of stimulation with IFN-α or IFN-λ, we utilized the ChIP assay. Chromatin extracts were generated from control cells and cells stimulated with either IFN-α or IFN-λ for 4 h. As shown by real-time PCR analysis, the ISG56 gene was robustly activated by IFN-λ; therefore, we focused the analysis on this promoter. Histone acetylation constitutes a mark for active genes. ChIP analysis with a histone H3 acetylation antibody showed that stimulation with IFN-α or IFN-λ resulted in an increased histone acetylation within the ISG56 promoter region (Fig. 2A). Also, an increase in the amount of RNA polymerase II situated on the ISG56 gene was identified as a response to IFN-α and IFN-λ stimulation (Fig. 2B).
FIG. 2.
Transcription factor binding analysis of the ISG56 promoter after IFN-λ and IFN-α stimulation. The occupancy of transcription factors and histone modifications on the promoter region of ISG56 was determined by ChIP analysis. HepG2 cells were stimulated with either 0.05 μg/ml IFN-λ or 0.01 μg/ml IFN-α and used for the ChIP assay. The ChIP analysis was done with both specific and preimmunized antibodies to determine the unspecific level of precipitation of DNA. Real-time PCR analysis of the ChIP material was done with primers corresponding to the ISG56 5′-transcribed region and the GAPDH 5′-transcribed region (A and B, respectively) or with primers corresponding to the ISG56 promoter and the GAPDH promoter (C to H). All ChIP data were normalized to the GAPDH values and used as baseline values to determine experimental variation. All experiments were performed in triplicate. For cells not stimulated with IFN, the values obtained from the ChIP experiment were normalized to 1, and promoter fragment retention as a response to IFN was visualized. The ChIP analysis was performed with antibodies against (A) histone H3 lysine 9 (K9) acetylation, (B) RNA polymerase II, (C) IRF9, (D) STAT1, (E) STAT2, (F) STAT3, (G) IRF1, and (H) IRF2.
A common theme in IFN type I-mediated gene induction is the induced binding of the ISGF3 transcription factor complex to responsive promoters. ISGF3 is composed of the STAT1, STAT2, and IRF9 transcription factors (22). ChIP analysis using antibodies against IRF9, STAT1, and STAT2 showed the recruitment of the ISGF3 components to the ISG56 promoter (Fig. 2C, D, and E). This was observed after stimulation with either IFN-α or IFN-λ. Recruitment of STAT3 and IRF1 was weak and only significant in response to IFN-α stimulation (Fig. 2F). The amounts of IRF2 present at the ISG56 promoter were decreased in response to IFN-α and IFN-λ stimulation (Fig. 2H). The overall picture from the ChIP analysis showed less efficient remodeling of transcription factor occupancy at the ISG56 promoter in response to IFN-λ stimulation compared to that of IFN-α stimulation (Fig. 2), in agreement with the lower observed transcriptional induction of the ISG56 gene by IFN-λ (Fig. 1). Examination of promoter regions for IFITM1, Viperin, and OAS1 showed comparable ChIP assay results for ISGF3 recruitment and, again, with a less efficient transcription factor recruitment and histone acetylation effect through IFN-λ stimulation compared to that of IFN-α stimulation (data not shown). In conclusion, cellular stimulation with either IFN-α or IFN-λ results in similar remodeling of transcription factor occupancy at an IFN-responsive promoter, and the observed weaker gene induction potential of IFN-λ is reflected in a less efficient transcription factor remodeling.
Specificity of the IFN-λ response is regulated through limited receptor expression.
Our experiments suggest that the signal transduction pathways of type I and type III IFN converge at the nuclear response via activation of ISGF3. However, in the cell lines we used, IFN-α induces a stronger signal than IFN-λ. Clearly, the signal strength will depend upon receptor expression in the given cell line. IFN-α-stimulated gene induction is observed with a large number of cell types, in accordance with a broad expression pattern of the receptor subunits IFNAR-1 and IFNAR2, and also with the effecting transcription factors (16). Very little is currently known about the IFN-λ receptor (IL-28Rα) expression. We examined IFN-λ signaling in a set of cell lines by estimating the induction of the ISG56, IFITM1, Viperin, and OAS1 genes. In contrast to Raji and HepG2 cells, which both responded well to IFN-λ, no significant response was observed for Sw13 adrenal carcinoma cells and MCF-7 breast cancer cells. In MDA-MB-231 breast cancer cells, a very modest induction was observed (Fig. 3B). In all of the cell lines tested, the ISG56, IFITM1, Viperin, and OAS1 genes were induced strongly by IFN-α (Fig. 3A). The IFN-λ receptor complex consists of IL28Rα and IL10Rβ, with the latter involved in a variety of cellular functions and with a ubiquitous expression profile (6). By using real-time PCR, we estimated the ratios of expression levels for IL28Rα and IL10Rβ (Fig. 3C). The ratio between the receptor components was decreased in the cell lines not responding to IFN-λ compared to those of the Raji and HepG2 cell lines (Fig. 3C). Thus, the limitation of the IL28Rα receptor component could account for the lack of IFN-λ response.
To further explore the receptor dependencies for IFN-λ signaling, we utilized HT1080 cells and the HT1080 derivative cell line HT1080-U5A, which lacks the functional IFNAR2 subunit (12, 14). By examining the expression level of ISG56 resulting from IFN-α stimulation in HT1080 and HT1080-U5A cells, we found, as expected, a strong induction in HT1080 cells but no induction in HT1080-U5A cells (Fig. 3D). Neither HT1080 nor HT1080-U5A cells responded to IFN-λ stimulation (Fig. 3D). This is in accordance with an IL28Rα/IL10Rβ ratio within HT1080 cells, as in the other cell lines tested, that is unresponsive to IFN-λ stimulation (Fig. 3C). To determine if overexpression of IL28Rα could result in a gain in IFN-λ signaling, we transiently transfected HT1080-U5A cells with an expression vector encoding IL28Rα. In the transfected HT1080-U5A cells, the ISG56 gene was induced pronouncedly in response to IFN-λ stimulation (Fig. 3D). Transfection of HT1080 cells, regardless of the vector used, resulted in activation of the ISG56 gene independently of IFN stimulation (data not shown). This is most likely caused by the activation of IFN-α/β production by the transfection with foreign DNA and was not observed for HT1080-U5A cells due to the lack of the type I IFN receptor (30). The introduction of IL28Rα into MCF-7 cells also resulted in de novo IFN-λ-mediated induction of the ISG56 gene (data not shown). In conclusion, IFN-λ signaling can be restored through increasing the IL28Rα receptor expression, and IFN-λ signaling functions independently of a functional IFN-α receptor complex.
Role of MAP kinases in IFN-λ signaling.
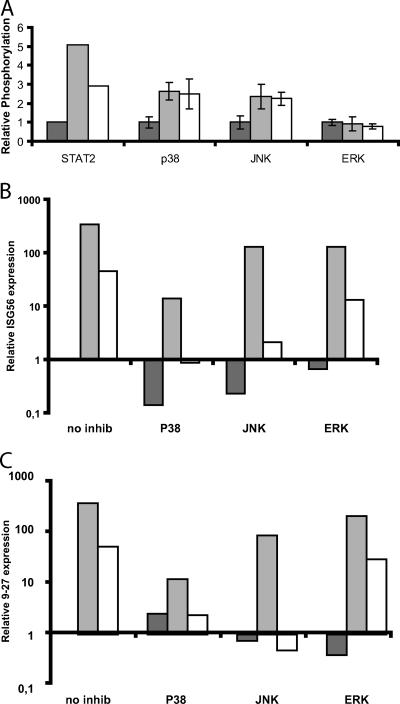
In addition to activating the Jak-STAT pathway, type I IFNs activate kinases of the MAP kinase pathway (18, 21, 28). In order to determine whether this also occurred in the cellular system used in this study and to compare type I and III IFNs, we stimulated cells with IFN-α or IFN-λ and harvested whole-cell extracts at 20 and 60 min posttreatment. The lysates were examined for the phosphorylation status of the MAP kinases p38, JNK, and ERK, using Luminex. Figure 4A shows the relative phosphor ylation after 20 min of treatment with interferon. Similar data were obtained after 60 min of stimulation (data not shown). Stimulation with either IFN-α or IFN-λ elevated the levels of phosphorylation of p38 and JNK. By contrast, the level of ERK phosphorylation was not affected by treatment with either IFN. The two types of interferon resulted in similar levels of p38 and JNK phosphorylation. This is in contrast to STAT2 phosphor ylation, where IFN-α produces a stronger response than IFN-λ.
FIG. 4.
Role of the MAP kinase pathways in IFN-α or IFN-λ signaling. (A) Relative phosphorylation of the indicated proteins after 20 min of stimulation of Raji cells with either IFN-α or IFN-λ. The phosphorylation in untreated cells was set to 1. Data are the means of four experiments, except for STAT2, where only two experiments were conducted. (B) Relative expression of the ISG56 mRNA after IFN stimulation with or without inhibitors of the indicated MAP kinases as measured by real-time RT-PCR. The expression level in untreated cells is set to 1, and the remaining data are normalized to this value. The y axis shows the induction/reduction change (n-fold) after treatment with IFN and/or the relevant inhibitor. Note the logarithmic scale. Data are the mean values of two experiments. Dark gray bars represent samples not treated with interferon; light gray bars are samples treated with IFN-α, and open bars are samples treated with IFN-λ. (C) Data are shown as in panel B but with the relative expression of the IFITM1/9-27 gene.
The ability of type I and III IFNs to activate p38 and JNK prompted us to investigate how these signaling pathways affected the gene expression induced by these cytokines. Therefore, we treated the Raji cells with inhibitors against p38 (SB202190), JNK (SP600125), or the ERK upstream kinase MEK1 (U0126) and subsequently stimulated cells with IFNs. RNA was harvested 4 h later for measurement of specific mRNA species. Figure 4B shows that induction of ISG56 by IFN-α was strongly reduced by the p38 inhibitor but was only marginally affected by the JNK inhibitor. The IFN-λ induction of ISG56 was almost completely abolished by the p38 inhibitor and strongly reduced by the JNK inhibitor. Highly similar results were found when we tested the effect upon the IFITM1/9-27 gene (Fig. 4C). It should be noted that both the p38 and JNK inhibitors led to a significant reduction of the basal level of ISG56; this effect was less prominent for the IFITM1/9-27 gene. The ERK inhibitor had no significant effect on either interferon type.
DISCUSSION
We used gene array analysis to determine the set of genes regulated by IFN-α and IFN-λ. The statistical cutoff level determined for significant genes is always a point of debate when dealing with gene array data. We decided to use two different strategies, both of which applied very strict statistical criteria. First, we identified genes induced by IFN-α and IFN-λ. We did this by first performing a t test and then by selecting the genes which were significantly regulated in the t test and that showed at least a twofold change. We selected this approach from among several possibilities because it was highly robust. By this analysis, we identified 313 entries regulated by IFN-α and 38 entries regulated by IFN-λ. All of the 38 entries regulated by IFN-λ stimulation were included in the group of genes regulated by IFN-α simulation. It should be noted that there appeared to be several duplicates of identical genes on the gene array; thus, for interferon lambda, the 38 entries regulated represent 27 unique genes. Manual inspection of the IFN-α-induced genes revealed that the majority of these genes were also induced upon IFN-λ treatment but failed to meet the stringent statistical criteria (see the supplemental material). Most obvious is the ISG56 gene, which failed the t test because one sample is an outlier. However, as our subsequent RT-PCR data show, this gene is clearly IFN-λ regulated. Others have also reported the gene array data for IFN-λ (7, 15). Doyle et al. reported 35 entries upregulated in HEP2G cells, with 10 of those overlapping genes found by us. However, it should be noted that Doyle and coworkers used five times the concentration of IFN-λ (50 ng/ml) and 6 h of induction, in addition to using a different cell line; therefore, a straightforward comparison can be difficult. As mentioned above, the low dose and short time of treatment used by us mean that a number of IFN-λ-induced genes fail to meet the stringent statistical criteria used. We encourage the reader to use the full data supplied in Table S1 and Table S2 in the supplemental material for comparison.
In an alternative approach to determine if any genes were regulated selectively by either IFN-α or IFN-λ, we performed a SAM test. The SAM test was performed with the IFN-α or the IFN-λ data relative to the grouped data of the control and the reciprocal treatment. Thus, it tests if any genes are selectively regulated by one of the two cytokines. This analysis showed that no gene was regulated by IFN-λ alone and that a small group of genes (10) appeared to be regulated IFN-α alone. However, when we tested several of these genes by RT-PCT, we found that they could be induced by higher concentrations of IFN-λ (see below).
The induction profile for a subset of genes identified from the array was also analyzed by quantitative real-time PCR. In the gene array experiment, the ISG56 and OAS1 genes were induced by both IFN-α and IFN-λ, whereas the Viperin and IFITM1 genes represented the group of 10 genes induced only by IFN-α. Real-time PCR analysis showed that high doses of IFN-λ did have the capacity to induce all four of these genes. Similar results were obtained with other genes which in the array experiment were induced significantly only by IFN-α (Fig. 1D and data not shown). Note that we used concentrations of 10 ng/ml of IFN-λ1 (IL-29) in the gene array experiment, which was shown in the literature to result in optimal induction (27). However, under our experimental conditions, 100 ng/ml appears to be optimal. In conclusion, the gene array data combined with real-time PCR data show the absence of genes robustly induced by IFN-λ but not those induced by IFN-α. The data also clearly indicate that genes induced by IFN-α also can be induced by IFN-λ but to a lower level. One possible implication of this effect could be that genes only modestly induced by IFN-α, as exemplified by the IRF1 and IRF2 genes analyzed here, will respond to IFN-λ stimulation to a degree that is hardly detectable under standard experimental settings. Induction of the IRF1 gene by both IFN-α and IFN-λ was shown with Hep2G cells, whereas IRF2 induction was also at the borderline of detection with this cell line (data not shown). It should be noted that the kinetics of IRF2 induction by lipopolysaccharides in macrophages was previously shown to be slower than that for IRF1 induction (2, 8). It remains to be clarified if the induction of IRF1 by IFN-α results in functional differences.
We used ChIP assays to investigate the activation of the ISG56 promoter and loading of transcription factors in response to either IFN-α or IFN-λ. IFN stimulation resulted in increased histone acetylation and RNA polymerase II recruitment. We also tested which transcription factors associated with the promoter after IFN treatment. A clear increase in STAT1, STAT2, and IRF9 recruitment was seen after stimulation with both IFNs, suggesting that recruitment of activated ISGF3 to the ISG56 promoter is the result of both IFNs. Similar results were obtained by examining the promoter regions for other IFN-induced genes (data not shown). We also observed a weak IFN-α-dependent STAT3 and IRF1 recruitment to the ISG56 promoter. This was not observed for IFN-λ; however, this could be due to the detection limit of the assay. IFN-α and IFN-λ stimulation resulted in a decreased ISG56 promoter presence of IRF2. It should be noted that the different IRF factors have overlapping recognition sequences on the ISG56 promoter, according to the possibility of competition for binding. Taken together, the mRNA expression and ChIP results suggest that signal transduction by IFN-α and IFN-λ grossly converges at the nuclear response and that activation of ISGF3 appears to be a primary force in gene induction by both IFN-α and IFN-λ. Moreover, under the experimental conditions, the lower efficiency of IFN-λ to transcriptionally induce genes is reflected in a lower transcription factor occupancy at responsive promoter regions.
If activation of the type I or type III IFN receptor complex leads to induction of approximately the same subset of genes, one might ask why nature maintains two apparently redundant systems. One obvious answer is that the distribution of type I and type III receptor differs and, thus, the biological response will differ, since different cellular subsets are effected by either IFN type I or IFN type III production. We tested a number of cell lines for their responses to IFN-λ and for expression of the IL28Rα receptor chain. Raji and HepG2 cells had robust responses to IFN-λ and showed the highest expression of the IL28Rα receptor. MDA cells had a weak response and a correspondingly low receptor expression level. SW13, MCF-7, and HT1080 cells did not respond and showed very low levels of the receptor. Thus, a good correlation exists between IL28Rα receptor expression and the response to IFN-λ. Expression of the IL28Rα receptor appears much more restrictive than that of the type I IFN receptor complex. This could suggest that the biological role for the type III IFN system lies within the ability to activate a more defined subset of cells within the body and thereby avoid some of the toxicity associated with type I IFN. Thus, IFN-λ stimulation could result in an IFN response which differs substantially, both spatially and temporally, from that of type I IFN but which utilizes the same basic set of IFN-regulated genes. Accordingly, an important future task is the establishment of the tools to determine the distribution of the functional IFN type III receptor complex in vivo.
It is now well-established that IFNs require the coordination and cooperation of multiple distinct signaling pathways for a full interferon response (21). To address this issue in relation to type III IFN, we examined whether the MAP kinase pathways were activated by type I and type III IFN stimulation and how activation contributed to the IFN-stimulated response. In agreement with the literature, we found significant levels of activation of both p38 and JNK kinases in response to IFN-α, whereas ERK activation was marginal in the Raji cells in our studies (5, 32). IFN-λ leads to levels of p38 and JNK phosphorylation similar to that of IFN-α but lower than STAT2 phosphorylation, in agreement with the lower level of STAT2 recruitment to the ISG56 promoter in response to IFN-λ, as seen with the ChIP assays.
The ability of IFN-λ to activate members of the MAP kinase family has previously been examined by Brand et al., who showed that IFN-λ activates JNK and also ERK1/2 in intestinal epithelial cells (4). However, the authors did not investigate the functional consequences of the ERK activation in relation to interferon treatment of cells. In our studies, we used Raji cells and found that IFN-λ activated p38 and JNK but not ERK1/2. At present, we cannot explain the discrepancy between these findings, but it is possible that the panel of MAP kinases activated by IFN-λ displays some degree of cell type specificity. For type I IFN, it has been reported that, depending on the cellular system, different MAP kinases are activated and contribute to the biological activity (5, 32).
Subsequently, we tested the effect of chemical inhibitors of the different MAP kinases upon the induction of ISG56 in response to either IFN-α or IFN-λ stimulation. IFN-λ displayed a stronger dependency upon p38 and JNK than IFN-α. A detailed understanding of the mechanistic role played by MAP kinases in the gene induction by interferons is not yet available; however, it is generally believed that MAP kinase activation leads to the recruitment of auxiliary transcription factors that cooperate with the ISGF3 complex in the activation of transcription (13, 31). A possible interpretation of our data is that the weaker activation of ISGF3 seen by IFN-λ increases the dependency upon these auxiliary factors. However, clearly, more experiments are required to determine the role of MAP kinases in the IFN-λ signaling.
Supplementary Material
Acknowledgments
We thank George Stark for the gift of U5A cells and for helpful discussions.
The project was supported by the Novo Nordisk Foundation, the Lundbeck Foundation, the Danish Medical Research Council (grant numbers 22-03-0183 and 22-04-0704), the Augustinus Foundation, Fonden til Lægevidenskabens Fremme, and the Danish Cancer Association. Z.Z. and N.A. were supported by fellowships from the Faculty of Health Sciences, Aarhus University.
Footnotes
Published ahead of print on 16 May 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ank, N., H. West, C. Bartholdy, K. Eriksson, A. R. Thomsen, and S. R. Paludan. 2006. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80:4501-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, S. A., M. J. Fultz, C. A. Salkowski, and S. N. Vogel. 1995. Differential expression of interferon regulatory factor 1 (IRF-1), IRF-2, and interferon consensus sequence binding protein genes in lipopolysaccharide (LPS)-responsive and LPS-hyporesponsive macrophages. Infect. Immun. 63:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron, C. A. 1994. Cytokines in the generation of immune responses to, and resolution of, virus infection. Curr. Opin. Immunol. 6:530-538. [DOI] [PubMed] [Google Scholar]
- 4.Brand, S., F. Beigel, T. Olszak, K. Zitzmann, S. T. Eichhorst, J. M. Otte, J. Diebold, H. Diepolder, B. Adler, C. J. Auernhammer, B. Goke, and J. Dambacher. 2005. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am. J. Physiol. Gastrointest. Liver Physiol. 289:G960-G968. [DOI] [PubMed] [Google Scholar]
- 5.Caraglia, M., A. Abbruzzese, A. Leardi, S. Pepe, A. Budillon, G. Baldassare, C. Selleri, S. D. Lorenzo, A. Fabbrocini, G. Giuberti, G. Vitale, G. Lupoli, A. R. Bianco, and P. Tagliaferri. 1999. Interferon-alpha induces apoptosis in human KB cells through a stress-dependent mitogen activated protein kinase pathway that is antagonized by epidermal growth factor. Cell Death Differ. 6:773-780. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly, R. P., F. Sheikh, S. V. Kotenko, and H. Dickensheets. 2004. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 76:314-321. [DOI] [PubMed] [Google Scholar]
- 7.Doyle, S. E., H. Schreckhise, K. Khuu-Duong, K. Henderson, R. Rosler, H. Storey, L. Yao, H. Liu, F. Barahmand-pour, P. Sivakumar, C. Chan, C. Birks, D. Foster, C. H. Clegg, P. Wietzke-Braun, S. Mihm, and K. M. Klucher. 2006. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44:896-906. [DOI] [PubMed] [Google Scholar]
- 8.Harada, H., T. Fujita, M. Miyamoto, Y. Kimura, M. Maruyama, A. Furia, T. Miyata, and T. Taniguchi. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729-739. [DOI] [PubMed] [Google Scholar]
- 9.Ida-Hosonuma, M., T. Iwasaki, T. Yoshikawa, N. Nagata, Y. Sato, T. Sata, M. Yoneyama, T. Fujita, C. Taya, H. Yonekawa, and S. Koike. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 79:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 11.Lasfar, A., A. Lewis-Antes, S. V. Smirnov, S. Anantha, W. Abushahba, B. Tian, K. Reuhl, H. Dickensheets, F. Sheikh, R. P. Donnelly, E. Raveche, and S. V. Kotenko. 2006. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 66:4468-4477. [DOI] [PubMed] [Google Scholar]
- 12.Lewerenz, M., K. E. Mogensen, and G. Uze. 1998. Shared receptor components but distinct complexes for alpha and beta interferons. J. Mol. Biol. 282:585-599. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y., A. Sassano, B. Majchrzak, D. K. Deb, D. E. Levy, M. Gaestel, A. R. Nebreda, E. N. Fish, and L. C. Platanias. 2004. Role of p38alpha Map kinase in type I interferon signaling. J. Biol. Chem. 279:970-979. [DOI] [PubMed] [Google Scholar]
- 14.Lutfalla, G., S. J. Holland, E. Cinato, D. Monneron, J. Reboul, N. C. Rogers, J. M. Smith, G. R. Stark, K. Gardiner, K. E. Mogensen, et al. 1995. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 14:5100-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcello, T., A. Grakoui, G. Barba-Spaeth, E. S. Machlin, S. V. Kotenko, M. R. MacDonald, and C. M. Rice. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131:1887-1898. [DOI] [PubMed] [Google Scholar]
- 16.Meager, A., K. Visvalingam, P. Dilger, D. Bryan, and M. Wadhwa. 2005. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine 31:109-118. [DOI] [PubMed] [Google Scholar]
- 17.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen, V. A., J. Chen, F. Hong, E. J. Ishac, and B. Gao. 2000. Interferons activate the p42/44 mitogen-activated protein kinase and JAK-STAT (Janus kinase-signal transducer and activator transcription factor) signaling pathways in hepatocytes: differential regulation by acute ethanol via a protein kinase C-dependent mechanism. Biochem. J. 349:427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. BioTechniques 39:715-725. [DOI] [PubMed] [Google Scholar]
- 20.Pestka, S., C. D. Krause, and M. R. Walter. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8-32. [DOI] [PubMed] [Google Scholar]
- 21.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat. Rev. Immunol 5:375-386. [DOI] [PubMed] [Google Scholar]
- 22.Qureshi, S. A., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1995. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. USA 92:3829-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robek, M. D., B. S. Boyd, and F. V. Chisari. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 25.Sato, A., M. Ohtsuki, M. Hata, E. Kobayashi, and T. Murakami. 2006. Antitumor activity of IFN-lambda in murine tumor models. J. Immunol. 176:7686-7694. [DOI] [PubMed] [Google Scholar]
- 26.Sen, G. C., and P. Lengyel. 1992. The interferon system. A bird's eye view of its biochemistry. J. Biol. Chem. 267:5017-5020. [PubMed] [Google Scholar]
- 27.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 28.Stancato, L. F., M. Sakatsume, M. David, P. Dent, F. Dong, E. F. Petricoin, J. J. Krolewski, O. Silvennoinen, P. Saharinen, J. Pierce, C. J. Marshall, T. Sturgill, D. S. Finbloom, and A. C. Larner. 1997. Beta interferon and oncostatin M activate Raf-1 and mitogen-activated protein kinase through a JAK1-dependent pathway. Mol. Cell. Biol. 17:3833-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 30.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 31.Uddin, S., B. Majchrzak, J. Woodson, P. Arunkumar, Y. Alsayed, R. Pine, P. R. Young, E. N. Fish, and L. C. Platanias. 1999. Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem. 274:30127-30131. [DOI] [PubMed] [Google Scholar]
- 32.Verma, A., D. K. Deb, A. Sassano, S. Uddin, J. Varga, A. Wickrema, and L. C. Platanias. 2002. Activation of the p38 mitogen-activated protein kinase mediates the suppressive effects of type I interferons and transforming growth factor-beta on normal hematopoiesis. J. Biol. Chem. 277:7726-7735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.