Abstract
The recent threat of an avian influenza pandemic has generated significant interest in enhancing our understanding of the events that dictate protective immunity to influenza and in generating vaccines that can induce heterosubtypic immunity. Although antigen-specific CD4 T cells are known to play a key role in protective immunity to influenza through the provision of help to B cells and CD8 T cells, little is known about the specificity and diversity of CD4 T cells elicited after infection, particularly those elicited in humans. In this study, we used HLA-DR transgenic mice to directly and comprehensively identify the specificities of hemagglutinin (HA)-specific CD4 T cells restricted to a human class II molecule that were elicited following intranasal infection with a strain of influenza virus that has been endemic in U.S. human populations for the last decade. Our results reveal a surprising degree of diversity among influenza virus-specific CD4 T cells. As many as 30 different peptides, spanning the entire HA protein, were recognized by CD4 T cells, including epitopes genetically conserved among H1, H2, and H5 influenza A viruses. We also compared three widely used major histocompatibility class II algorithms to predict HLA-DR binding peptides and found these as yet inadequate for identifying influenza virus-derived epitopes. The results of these studies offer key insights into the spectrum of peptides recognized by HLA-DR-restricted CD4 T cells that may be the focus of immune responses to infection or to experimental or clinical vaccines in humans.
Influenza virus is a major human pathogen. Despite the ready availability of vaccines, the infection of humans with influenza virus causes significant morbidity and mortality (reviewed in references 13, 37, 46, and 98). In industrialized societies, influenza is the leading virus-induced cause of death and is estimated to cause more than 40,000 deaths annually in the United States alone. In many more individuals, influenza virus infections cause significant debilitation that can be long lasting. In the United States, influenza virus infections are estimated to cause 100,000 hospitalizations annually. The factors that determine the impact of influenza virus infection on human health are diverse and include the immunological competence of the human host, the particular strain of infecting influenza virus, which determines overall pathogenicity (reviewed in references 50 and 69), and the genetic relatedness between the infecting virus and virus strains that were endemic previously or used in clinical vaccines (reviewed in references 37, 44, and 73). The recent threat of a new pandemic of avian influenza (44, 57, 73, 86, 90, 91, 106) has generated renewed interest in gaining better understanding of the factors that dictate protective immunity to influenza virus, including the enhancement of vaccines that induce heterosubtypic immunity.
It is now understood that protective immunity to influenza virus infection is due to the combined participation of many arms of the innate and adaptive immune responses. The components of the innate immune system, including interferons, macrophages, and natural killer cells, are triggered very early upon host infection with influenza virus and are typically effective at limiting early viral replication (reviewed in references 96 and 112). However, the ultimate clearance of influenza virus depends critically on the adaptive immune response and is mediated by antigen-specific T cells and B cells. Antigen-specific CD8 T cells are critical for the cytotoxic elimination of virus-infected cells in the lung, while antigen-specific B cells are an essential component of the protective immune response, producing neutralizing antibodies which provide the most significant source of protection from future infections (15, 96). Protective antibodies that inhibit viral entry and replication are typically of high affinity and are most commonly reactive with the virion envelope proteins hemagglutinin (HA) and, less commonly, neuraminidase, respectively (reviewed in references 29, 96, and 97).
Antigen-specific CD4 T cells play a central role in generating protective immunity to influenza virus through the provision of cognate help to B cells, a requisite event for immunoglobulin (Ig) switch and the affinity maturation of B cells (63, 64), and through their ability to help CD8 T cells, which appears to be essential for long-term CD8 memory responses (19, 41, 42, 48, 70, 93, 94). Finally, CD4 T cells may participate directly in viral clearance via cell-mediated cytotoxicity or the production of cytokines in the lung (reviewed in references 7, 95, and 99). Understanding the role of CD4 T-cell immunity to influenza virus and evaluating the efficacy of influenza vaccines in humans requires insight into the specificity and diversity of epitopes elicited upon infection and upon vaccination. Much of the data presently available on the specificity of CD4 T-cell responses to influenza virus are limited to a few peptide epitopes, typically those identified after long-term culture of T cells (3, 8, 22, 24-26, 53), which has the potential to significantly alter the representation of T-cell specificities and lead to oligoclonality in culture. Studies on the CD4 T-cell repertoire in humans is further complicated by the high level of representation of memory CD4 cells within peripheral blood, which may disproportionately contribute to the responses detected after acute infection or exposure to vaccines.
In this study, we have sought to directly and comprehensively identify the specificity of HA-specific CD4 T cells restricted to a human class II molecule that are elicited in naïve individuals after infection with an H1N1 strain of influenza A virus that has been endemic in North America within the last decade. We used DR1 transgenic mice for the identification of immunodominance patterns, an experimental strategy that allows analyses of de novo-generated immune responses without the complication of preexisting influenza virus-specific memory CD4 T cells and also the study of lymphocytes from lymphoid tissues that can be used immediately for the identification of peptide specificity. The HLA-DR1 molecule was chosen as a model human major histocompatibility complex (MHC) class II molecule because it is expressed in a large fraction of individuals in the United States (18, 62, 65) and because it shares peptide binding characteristics with a number of other DR molecules, including DR4 and DR7 (20, 40, 89), which are also expressed in a large proportion of individuals (14, 18, 65, 80, 89). We studied the specificity of primary CD4 T-cell responses to A/New Caledonia/20/99 virus, an H1N1 strain that has been circulating for almost a decade in the United States and has consequently been included in vaccines in North America for the past 7 years (27, 28, 49, 108, 111). For these studies, we used HLA-DR1 transgenic mice with MHC class II transgenes that encode peptide binding domains derived from the human class II molecule and membrane-proximal domains derived from the murine homolog of HLA-DR (I-E) to promote interactions with the murine CD4 protein expressed in these mice. The HLA-DR1 transgenic mice have been used extensively as a murine model of human arthritis (36, 54, 67, 68, 79), as well as an experimental tool to identify CD4 T-cell epitopes and study the MHC class II-restricted presentation of HLA-DR1 antigens (12, 72, 79).
To comprehensively evaluate the diversity of CD4 T cells that are specific for the HA protein, we obtained overlapping peptides representing the entire HA protein sequence and tested these peptides by using enzyme-linked immunospot (ELISPOT) assays (2, 23, 45, 100) to directly enumerate peptide-specific CD4 T cells isolated from the spleens of infected DR1 transgenic mice. The results of these experiments reveal a surprising degree of diversity among influenza virus-specific CD4 T cells and also suggest that presently available algorithms to predict CD4 epitopes are as yet inadequate for identifying influenza virus-derived epitopes. The results of these studies offer new insight into the diversity and specificity of influenza virus-specific, DR1-restricted CD4 T cells elicited during primary immune responses to influenza virus infection that correspond to likely candidate epitopes for human immune responses to infection or to experimental or clinical vaccines.
MATERIALS AND METHODS
Virus production.
Embryonated eggs, purchased from SPAFAS Inc. (North Franklin, CT), were incubated at 70°F and 100% humidity for 9 days, followed by the infection of the allantoic cavity with 100 μl of human influenza virus A/New Caledonia/20/99 (H1N1; generously provided by John Treanor at the University of Rochester) at a concentration of 103 50% egg infective doses (EID50), or 103 times the dose necessary to infect 50% of the embryonated eggs, per ml. The infected eggs were incubated for 48 h at 37°C and then for 24 h at 4°C. The allantoic fluid was harvested under sterile conditions and centrifuged at 2,000 × g for 20 min and 4°C. Small aliquots of the supernatant were frozen at −70°C, and the virus titer was determined by infecting new embryonated eggs with serial dilutions of the harvested allantoic fluid in triplicate. The resulting allantoic fluid was harvested from eggs infected with the serial dilutions and immediately titrated by an HA assay of chicken red blood cells according to the procedure recommended by the 2005-to-2006 World Health Organization bulletin for the identification of influenza virus isolates from subjects with influenza. The titer was reported as the number of EID50 per milliliter of allantoic fluid.
Cell lines and reagents.
Cell lines were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 5 mM HEPES, 2 mM l-glutamine, 1 mM nonessential amino acids, and 5 × 10−5 M 2-mercaptoethanol, all purchased from Invitrogen. DAP-3 cells transfected with the genes for DR1 were provided by E. Long, NIAID, NIH, and were screened periodically for class II surface expression by monoclonal antibody staining and fluorescence-activated cell sorter analysis. The DR1 transgenic mice (B10.M/J-TgN-DR1) (110) were obtained from D. Zaller (Merck) through Taconic laboratories and were maintained in the pathogen-free facility at the University of Rochester Medical Center according to institutional guidelines. As originally described (110), these mice express a chimeric HLA-DR1 class II molecule with the peptide binding membrane-distal domain derived from the human HLA-DR1 protein and the membrane-proximal domains derived from the murine I-E protein in order to promote interactions with the murine CD4 protein. Monoclonal antibody-producing cell lines used to purify CD4 T cells were originally obtained from the American Type Culture Collection and included 3.155 (anti-CD8), RA3/3A1/6.1 (anti-B220), L243 (anti-DR), and Y3P (anti-I-Af). Low-Tox M rabbit complement and Lympholyte-M used for the purification of CD4 T cells were both purchased from Cedarlane Laboratories. Antibodies used to assess the purity of the CD4 T cells included CD4-fluorescein isothiocyanate (CD4-FITC; clone RM4-4) from BD Biosciences, CD8a-FITC (Ly-2 clone 53-6.7) from eBiosciences, anti-HLA-DR-FITC (clone HK14) from Sigma, and anti-mouse Ig from BD Biosciences. The antibody used to detect I-Af was FITC-labeled I-Ak (Aβκ; 10-3.6) from BD Biosciences, which is known to cross-react with I-Af. Purified anti-mouse CD16/CD32 (FcγIII/II receptor) clone 2.4G2, purified rat anti-mouse interleukin-2 (IL-2; JES6-1A12), biotinylated rat anti-mouse IL-2 (JES6-5H4), rat anti-mouse gamma interferon (IFN-γ; clone R4-6A2), and biotinylated rat anti-mouse IFN-γ (clone XMG1.2) for ELISPOT assays were all purchased from BD Biosciences.
Influenza infections.
Unless otherwise indicated, HLA-DR1 transgenic mice were infected intranasally with A/New Caledonia/20/99 at 100,000 EID50 in 30 μl of phosphate-buffered saline (PBS). Groups of mice were 2 to 4 months old at the time of infection and were anesthetized by intraperitoneal injection with tribromoethanol (Avertin; 250 to 300 μl per mouse) prior to intranasal infection to ensure inhalation of the virus solution. Seven to eight days postinfection, the mice were sacrificed and spleens were excised and used as a source of CD4 T cells for ELISPOT analyses. Spleens from six to seven mice were pooled unless otherwise stated.
Cell purification.
A single suspension of cells from the spleens of infected mice was depleted of red cells by treatment for 5 min at room temperature with ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, and 0.1 mM Na2-EDTA in H2O, pH 7.2 to 7.4; GIBCO). The resultant lymphoid cells were depleted of non-CD4 T cells by incubation at 2 × 107 cells/ml with monoclonal antibody supernatants from cell lines 3.155 for CD8 cells, RA3/3A1/6.1 for B cells, L243 for DR cells, and Y3P for I-Af cells at a final dilution of 1:4. Cells were then washed and resuspended at 2 × 107 cells/ml in complement (Low Tox M; Cedarlane)-containing buffer at 37°C for 30 min. Dead cells were removed by density gradient centrifugation with Lympholyte-M according to the manufacturer's instructions. Viable cells were recovered and washed twice to remove residual Lympholyte-M. In some experiments, a second step of purification was taken to isolate T cells based on the expression of CD4. CD4 T cells were purified from antigen-presenting cells (APC) and CD8 cells by incubation with a limiting dilution (1:6,500) of allophycocyanin-conjugated anti-CD4 (RM4-5 [BD Biosciences, CA]), which had been previously titrated to determine the lowest dose that provided adequate separation from other cell types, and sorted by preparative flow cytometry. Sorting of the CD4-positive cells was performed using a BD FACSAria cell sorting system. The purity of the cell populations was assessed by collecting samples before and after complement-mediated lysis and after sorting by staining for CD4-FITC, CD8a-FITC, DR1-FITC, I-Af-FITC, and B cells by using an Ig-FITC antibody.
ELISPOT assays.
ELISPOT assays were performed as previously described (105), with some modifications. Briefly, 96-well filter plates (Millipore) were coated with 2 μg of purified rat anti-mouse IL-2/ml in PBS at room temperature for at least 2 h and then washed with cell culture medium to remove any unbound antibody. CD4 T cells (350,000) were cocultured with 30,000 DAP-3 fibroblasts that either did or did not express the DR1 MHC class II protein and with the indicated peptides at a final concentration of 10 μM in a total volume of 200 μl for 16 to 18 h at 37°C and 5% CO2. Cells were removed from the plates, and the plates were washed with wash buffer (PBS plus 0.1% Tween 20), after which 50 μl of a solution of 2 μg of biotin rat anti-mouse IL-2/ml prepared in wash buffer with 10% fetal bovine serum was added and the plates were incubated for a further 30 min at room temperature. The plates were washed again, blotted, and probed with a 1:1,000 dilution of alkaline phosphatase-strepavidin (Jackson ImmunoResearch, PA). The plates were then incubated at room temperature for 30 min, after which they were washed, blotted dry, and developed using Vector blue substrate kit III (Vector Laboratories, CA) prepared in 100 mM Tris, pH 8.2. After drying, the quantification of spots was performed with an Immunospot reader series 2A using Immunospot software, version 2.
Peptides.
18-mer peptides overlapping by 11 amino acids to encompass the entire sequence of the HA protein from the H1N1 strain of influenza virus A/New Caledonia/20/99 were synthesized by Mimotopes (Clayton, Victoria, Australia). The peptides were reconstituted at 10 mM in PBS, with or without added dimethyl sulfoxide for hydrophobic peptides and 1 mM dithiothreitol for cysteine-containing peptides. The stocks were stored at −20°C. Working stocks (100 μM) in Dulbecco's modified Eagle's medium (Invitrogen Corp., Carlsbad, CA) were made, sterilized by filtration, and stored at −20°C. Individual peptides were synthesized by Biopeptides (San Diego, CA) and dissolved and stored as described above.
Binding predictions.
The entire sequence of the HA protein from the human influenza virus A/New Caledonia/20/99 (H1N1; accession number, AAP34324) was input into the websites for Rankpep (http://bio.dfci.harvard.edu/Tools/rankpep.html), SYFPEITHI (http://www.syfpeithi.de/home.htm), and ProPred (http://www.imtech.res.in/raghava/propred/). Different threshold values were used to perform the query for each algorithm. With Rankpep, 11 peptides scored above the recommended binding threshold of 8.01. With SYFPEITHI, the top 11 peptides were selected, while for ProPred, the threshold was fixed at 1%, which is considered to provide a high level of stringency with a low rate of false positives. Under these conditions, nine of the top peptides were identified.
RESULTS
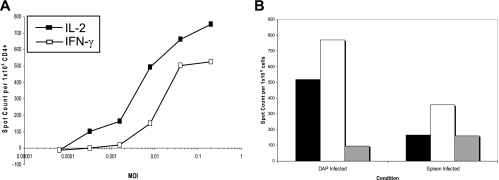
To evaluate the specificity of HLA-DR1-restricted CD4 T cells elicited in response to the A/New Caledonia/20/99 influenza virus strain in DR transgenic mice, we first established conditions for T-cell priming in response to increasing doses of infectious virus introduced intranasally in PBS, an infection route meant to mimic infection in humans. Lymphoid cells were isolated from spleens and lymph nodes of infected mice at 7 to 8 days after the introduction of the virus, and CD4 T cells were purified from the cell population by the depletion of CD8 and MHC class II protein-positive cells. To evaluate the antigen specificity of influenza virus-primed CD4 T cells, we used ELISPOT assays, which allow the direct ex vivo quantification of antigen- or peptide-reactive lymphocytes without any extended in vitro culture (21, 45, 100, 101). Because early experiments indicated that, in our hands, IL-2 production was more sensitive than IFN-γ production for the detection of influenza virus-specific CD4 T cells (Fig. 1A), IL-2 ELISPOT assays were used to enumerate antigen-reactive CD4 T cells for the remainder of the study.
FIG. 1.
Comparison of results from IL-2 and IFN-γ ELISPOT assays for the enumeration of influenza virus-specific CD4 T cells and the titration of the virus in mice. (A) Mice were infected intranasally with 100,000 EID50 of A/New Caledonia/20/99 virus. Seven days later, mice were sacrificed and CD4 T cells were purified from splenocytes and cocultured with an influenza virus-infected DR1 homozygous human B-cell line, LG2. The number of cytokine-producing cells per 300,000 input CD4 T cells was quantified by IL-2 or IFN-γ ELISPOT assays. Shown are the average numbers of spots from triplicate wells of cytokine-secreting cells as calculated by subtracting the numbers of spots detected with uninfected cells (85 and 8 for IL-2 and IFN-γ, respectively). Similar results were obtained with infected DR1 gene-transfected DAP-3 cells and with selected antigenic peptides as stimulators on DR1-positive uninfected cells (data not shown). Spleen cells from three mice were pooled prior to analyses. (B) Mice were infected intranasally with 200,000 (black bars), 20,000 (white bars), 2,000 (gray bars), or 200 EID50 of A/New Caledonia/20/99 influenza virus. The number of IL-2-producing, influenza virus-specific CD4 T cells from spleens was determined 7 days later by 18-h stimulation with infected DAP.3-5.3.1 DR1-positive transfectants or syngeneic spleen cells infected with virus at a multiplicity of infection (MOI) of 5. IL-2 ELISPOT results were quantified with an automated plate reader as the averages for triplicate wells by subtracting the background values obtained with uninfected APC (28 and 65 for spleen and DAP-3 cells, respectively). The response of the mice infected at 200 EID50 was below the background level and is not shown. Spleen cells from two mice per group were pooled prior to analyses.
Shown in Fig. 1B are the results of ELISPOT assays following the titration of the virus in vivo in the DR1 transgenic mice, with the infecting dose introduced intranasally in PBS as EID50 of virus grown in fertilized chicken eggs and recovered in allantoic fluid. Priming in the infected mice was assessed by quantifying IL-2 immunospots produced by CD4 T cells isolated from DR1 transgenic mice 7 days postinfection in response to virus-infected syngenic splenocytes (Fig. 1B, left) or fibroblasts transfected with the gene encoding HLA-DR1 (Fig. 1B, right). These experiments indicated that the A/New Caledonia/20/99 H1N1 strain of influenza virus, which has never been adapted to grow in mice, shows good infectivity in mice and primes T cells when introduced at relatively low doses (<2,000 EID50/mouse). The highest dose tested, 200,000 EID50, was not lethal to the animals, and all mice appeared healthy at 7 to 10 days postinfection, independent of the dose of virus used for infection. For the remainder of the assays, 100,000 EID50 per animal was used to infect mice and prime influenza virus-specific CD4 T cells.
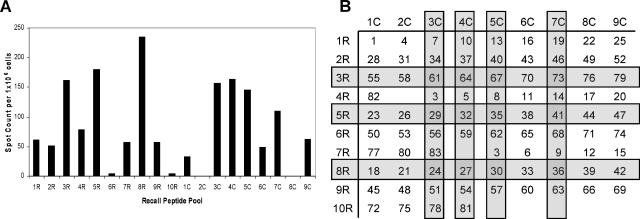
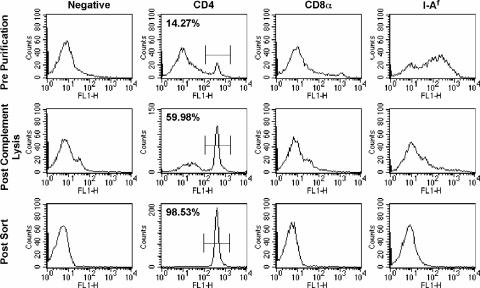
To assess the peptide specificity of the influenza virus-specific CD4 T cells, synthetic overlapping 18-mer peptides, offset by 7 amino acids, representing the entire translated sequence of HA expressed in the A/New Caledonia/20/99 virus were used. The HA0 precursor protein of HA is more than 550 amino acids in length, and therefore, more than 80 peptides were synthesized in order to span its entire sequence. Because of the high number of peptides in the peptide scan, we used a peptide-pooling strategy (100, 101) to initially identify individual HA peptides to which CD4 T cells respond. Shown in Fig. 2B is the layout of the matrix with the number of the individual peptide shown. Figure 2A shows the results of an HA-specific ELISPOT analysis of the peptide pools, with the results presented as the number of IL-2 spots detected per 106 CD4 T cells after subtracting the number of background spots (<20). Pools displaying positive reactivity (Fig. 2, 1R, 2R, 3R, 4R, 5R, 7R, 8R, 9R, 1C, 3C, 4C, 5C, 6C, 7C, and 9C) each contained candidate peptides, while negative pools (Fig. 2, 6R, 10R, 2C, and 8C) were presumed to have no or very weak CD4 T-cell epitopes. Peptides within these negative pools were eliminated from further consideration. Peptides belonging to more than one strongly positive pool were considered to be candidates for the most dominant T-cell epitopes and were tested as single peptides in IL-2 ELISPOT assays. For these studies, we used an additional step of purification to be certain that the CD4 T cells detected were indeed restricted to DR1. The repertoire of DR1 transgenic mice is thought to be largely restricted to the human class II molecule expressed by a transgene, but the mice do express low levels of an endogenous class II protein (I-Af) that is thought to poorly select and activate CD4 T cells (110). Although the T-cell population was completely depleted of CD8 T cells prior to the addition of T cells to the ELISPOT assay, the depletion of class II protein-positive cells was only about 90% complete (Fig. 3). It was therefore conceivable that some of the peptide epitopes detected in the pool arrays were restricted to the endogenous I-Af class II molecule expressed in the B10.M host. Therefore, in all of the subsequent analyses, we adopted an even more rigorous purification strategy to eliminate any endogenous APC present among the spleen cells from the infected donors. Spleen cells were depleted of APC and CD8 T cells by treatment, as before, with a cocktail of specific anti-CD8, anti-class II molecule, and anti-B-cell antibodies and complement. CD4 T cells were further purified from the residual viable cells by using a positive selection strategy involving preparative flow cytometry with limiting concentrations of an anti-CD4 antibody that was found not to inhibit T-cell responses (data not shown). Figure 3 shows that this strategy led to the isolation of CD4 T cells with more than 98% purity. We found in separate experiments that the use of higher concentrations of the anti-CD4 antibody GK1.5 blocked the antigen-dependent IL-2 production, attesting to the functionality of the CD4 T-cell-class II molecule interactions in the HLA-DR1 transgenic mice.
FIG. 2.
Peptide screening by the peptide-pooling matrix method. Mice were infected intranasally with 100,000 EID50 of A/New Caledonia/20/99 virus. The number of IL-2-producing CD4+ splenocytes was determined 8 days later by 18-h coculture with DAP-3 DR1-positive transfectants cultured with the pools of peptides shown in panel B. The averages of results for duplicate wells as calculated by subtracting the background value obtained with no peptide (48.5) are shown in panel A. The highlighted rows and columns in panel B identify those pools with high levels of IL-2 ELISPOT responses, and the intersections of the rows and columns correspond to predicted epitopes that elicit high numbers of responding CD4 T cells. Peptides belonging to more than one of the most stimulatory pools are shaded and were considered to be candidates for the most dominant T-cell epitopes. Spleen cells from six mice were pooled for this experiment.
FIG. 3.
Purification of CD4 T cells for ELISPOT analyses. Splenocytes were harvested from infected mice, and a portion was reserved for staining (top row, prepurification). The remainder of the cell set was depleted of class II protein-positive and CD8 cells by using antibodies and complement. A portion of the remaining cells was reserved for staining (middle row) and exemplifies the typical level of purity of cell groups used in the experiments described in the legends to Fig. 1 to 3. The remaining portion was incubated with a subsaturating concentration of anti-CD4 antibody, purified by preparative flow cytometry as described in Materials and Methods, and reanalyzed by analytical flow cytometry (bottom row). All cell populations were stained with FITC-labeled anti-CD4 (second column), anti-CD8a (CD8α; third column), or anti-Aβk (10-3.6) that cross-reacts with I-Af (fourth column). Results for unstained cells are shown in the first column. The percentage of CD4-positive cells in each population is shown in the top left corner of each CD4 graph. The results shown for the population of CD4 T cells obtained after preparative flow cytometry are typical of results from the experiments described in the legends to Fig. 4 and 5.
FIG. 4.
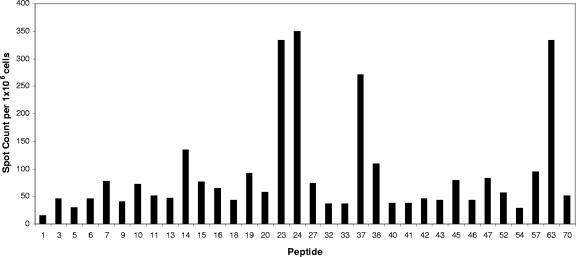
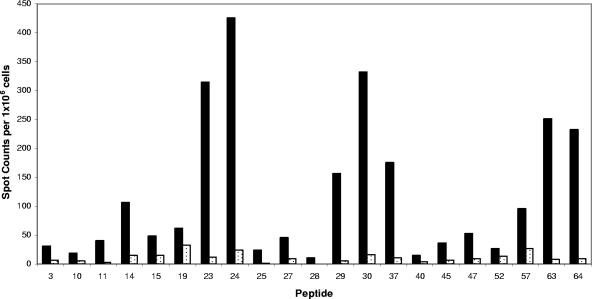
Representative results from a single-peptide screen demonstrate the diversity of the primary CD4 response to influenza virus HA. Mice were infected intranasally with A/New Caledonia/20/99 influenza virus, and the number of IL-2-producing CD4 splenocytes was determined 8 days later by 18-h in vitro stimulation with DAP.3-5.3.1 DR1-positive transfectants cultured with the single 18-mer peptides of interest at a 10 μM final concentration. IL-2 ELISPOT results quantified as the averages for duplicate wells with the background value (11 spots) subtracted are given. The results shown are representative of those of two to three independent experiments for each peptide. Spleen cells from six to seven mice were pooled for each experiment.
FIG. 5.
Confirmation of DR1 restriction of CD4 T cells. Mice were infected intranasally with A/New Caledonia/20/99 virus, and the number of IL-2-producing CD4 T cells from spleens was determined 8 days later by 18-h in vitro stimulation with DAP.3-5.3.1 DR1-positive (black bars) or DAP.3 DR1-negative (stippled bars) fibroblasts cultured with single peptides at a 10 μM final concentration. Results of the IL-2 ELISPOT analyses were quantified as the averages for duplicate wells after the background value (10 spots) was subtracted. Spleen cells from seven mice were pooled prior to analyses.
FIG. 7.
Sequence alignment of immunodominant peptides in HA. Shown is the ClustalW alignment of HA protein sequences from the New Caledonia/20/99 (H1N1), Canada/720/05 (H2N2), and Hong Kong/156/97 (H5N1) strains of influenza A virus. The regions conserved among these strains are highlighted in green. Other sequences are highlighted as noted at the bottom of the figure. Peptides were grouped according to the criteria indicated in the legend to Fig. 6. Because the responses corresponded to a number of adjacent peptides, in many cases the stimulatory peptides overlap in this figure.
FIG. 6.
Summary of empirically observed immunodominant HA peptides and comparison with peptides predicted to be presented by HLA-DR1 by algorithms available on the Web. The HA sequences of 18-mer overlapping peptides are shown as a ladder. The underlined peptide sequences are those eliciting a response as determined by the number of cytokine-producing CD4 T cells generated compared to the number generated in response to one of the major immunodominant peptides (peptide 63). Any peptide that elicited less than 5 percent of the response elicited by peptide 63 was considered negative for reactivity, while those with a score of 6 to 24% were considered weak but positive for epitopes and are indicated by a single underline. Those peptides eliciting between 25 and 60% of the number of IL-2-producing CD4 T cells generated in response to peptide 63 were considered subdominant and are underlined twice, while those peptides that elicited at least 61% of the CD4 T-cell response elicited by peptide 63 were considered to be immunodominant epitopes and are indicated by triple underlining. Highlighted regions of the peptide sequences predicted by the algorithms tested to bind to HLA-DR1 are coded as noted in the upper right corner of the figure. Epitopes that were predicted by more than one algorithm are highlighted in green or hatched colors. cds, coding sequence.
HA-derived peptides were tested individually in several subsequent IL-2 ELISPOT assays using purified CD4 T cells isolated from mice infected 8 days previously. The results of a sample IL-2 ELISPOT assay with 21 single peptides are presented in Fig. 4. These results show the typical range in reactivity observed in independent assays, with several peptides, such as peptide 23 and peptide 63, eliciting the greatest number of DR1-restricted T cells while other peptides elicited intermediate (e.g., peptide 14), low (e.g., peptide 27), or undetectable (e.g., peptide 1) responses. We then sought to formally test the MHC class II restriction of the candidate peptides identified through the course of experiments using individual peptides. CD4 T-cell ELISPOT assays with the individual candidate HA peptides were performed in the presence of DR1-positive fibroblasts that expressed DR1 from transfected genes as their only MHC class II molecule, with endogenous MHC class I molecules being derived from the strain of origin of the fibroblasts (C3H [H-2k]). Untransfected fibroblasts were used as negative-control APC. The results of this experiment, presented in Fig. 5, show that all of the provisionally identified HA-derived CD4 T cells were indeed restricted to the HLA-DR1 class II molecule. Even the relatively minor specificities detected, exemplified by peptides 11, 27, and 45, were restricted to HLA-DR1. None of the influenza virus-specific CD4 specificities identified in our studies were detectable in uninfected mice (data not shown).
After comprehensively identifying the HA-derived peptides recognized by CD4 T cells, we asked how well these empirically defined epitopes compared to those selected by algorithms presently used to predict class II epitopes. Three programs available on the Web (reviewed in references 9, 75, and 84) to predict the binding of peptides to MHC molecules, ProPred (1, 66, 87), SYFPEITHI (31, 35, 74), and Rankpep (16, 32), which have been used previously to predict T-cell epitopes, were used to scan the entire HA sequence for peptides predicted to bind to HLA-DR1. These programs implement different algorithms to predict peptide binding to HLA-DR1. Each algorithm identified approximately 12 to 15% of the peptides (e.g., 9 to 11 out of a total of more than 75 peptides) as candidate immunodominant peptides. These predicted peptides are highlighted in Fig. 6, which shows a ladder representation of each peptide contained in the HA peptide set. The empirically defined peptide epitopes are indicated in this figure as well, with the most immunodominant, the intermediate, and the minor peptides denoted. The criteria used for grouping the peptides are indicated in the legend to Fig. 6. Table 1 shows the results of multiple independent experiments with each of the identified peptides, allowing for significant confidence that the relative immunodominance depicted in Fig. 6 is reproducible.
TABLE 1.
Reproducibility of responses to individual peptides tested by ELISPOT analysesa
| Peptide no. | No. of indicated responses/no. of tests
|
|||
|---|---|---|---|---|
| Negative | Weak | Moderate | Strong | |
| 1 | 2/2 | |||
| 2 | 2/2 | |||
| 3 | 3/3 | |||
| 4 | Mtx | |||
| 5 | 2/2 | |||
| 6 | 1/2 | 1/2 | ||
| 7 | 3/3 | |||
| 8 | 2/2 | |||
| 9 | 2/2 | |||
| 10 | 4/4 | |||
| 11 | 3/3 | |||
| 12 | Mtx | |||
| 13 | 1/2 | 1/2 | ||
| 14 | 2/5 | 3/5 | ||
| 15 | 3/3 | |||
| 16 | 3/3 | |||
| 17 | Mtx | 1/1 | ||
| 18 | 3/3 | |||
| 19 | 1/3 | 2/3 | ||
| 20 | 2/2 | |||
| 21 | Mtx | |||
| 22 | Mtx | |||
| 23 | 4/4 | |||
| 24 | 7/7 | |||
| 25 | 1/2 | 1/2 | ||
| 26 | Mtx | |||
| 27 | 3/3 | |||
| 28 | Mtx, 1/1 | |||
| 29 | 4/5 | 1/5 | ||
| 30 | 2/2 | |||
| 31 | Mtx | |||
| 32 | 1/2 | 1/2 | ||
| 33 | 1/2 | 1/2 | ||
| 34 | 1/1 | |||
| 35 | Mtx | |||
| 36 | 1/2 | 1/2 | ||
| 37 | 4/6 | 2/6 | ||
| 38 | 2/2 | |||
| 39 | Mtx | |||
| 40 | 1/3 | 2/3 | ||
| 41 | 2/3 | 1/2 | ||
| 42 | 1/2 | 1/2 | ||
| 43 | 3/3 | |||
| 44 | Mtx | 1/1 | ||
| 45 | 3/3 | |||
| 46 | 2/3 | 1/3 | ||
| 47 | 3/3 | |||
| 48 | Mtx | |||
| 49 | Mtx | |||
| 50 | Mtx | 1/1 | ||
| 51 | Mtx | 1/1 | ||
| 52 | 3/3 | |||
| 53 | Mtx | |||
| 54 | 1/2 | 1/2 | ||
| 55 | 2/2 | |||
| 56 | Mtx | |||
| 57 | 3/3 | |||
| 58 | Mtx | |||
| 59 | Mtx | |||
| 60 | Mtx, 1/1 | |||
| 61 | Mtx, 1/1 | |||
| 62 | Mtx | |||
| 63 | 10/10 | |||
| 64 | 2/2 | |||
| 65 | Mtx, 1/1 | |||
| 66 | Mtx | |||
| 67 | Mtx, 1/1 | |||
| 68 | Mtx | |||
| 69 | 2/2 | |||
| 70 | 1/3 | 2/3 | ||
| 71 | Mtx | 1/1 | ||
| 72 | Mtx | |||
| 73 | 1/2 | 1/2 | ||
| 74 | Mtx | |||
| 75 | Mtx | |||
| 76 | Mtx | 1/1 | ||
| 77 | Mtx, 1/2 | 1/2 | ||
| 78 | 1/4 | 2/4 | 1/4 | |
| 79 | Mtx | |||
| 80 | Mtx, 1/1 | |||
| 81 | Mtx, 1/1 | |||
| 82 | Mtx | |||
Shown are the results of independent ELISPOT analyses of individual peptides. Responses were scored based on the number of IL-2 spots relative to the number produced in response to peptide 63, as described in the legend to Fig. 6. Also shown is the number of times each peptide was tested. Mtx indicates that the peptide was excluded by the matrix screening.
Several interesting observations emerge from the analyses shown in Fig. 6. First, different peptides were predicted by each of the three different algorithms, with only two 9-mer peptides (in peptide 1 and peptide 48) selected by all three. Four peptides were selected by at least two algorithms, but the algorithms selected different peptides within the 18-mer peptides. More than 10 9-mer peptides were selected by only a single algorithm, and of the algorithms that were tested, Rankpep appeared to give results that were the most discordant from those from the other two. The other remarkable observation made from this type of analysis is that many of the empirically defined epitopes were not predicted by any of the three algorithms. Two of the four most immunodominant peptides (contained within peptide 37 and peptides 63 and 64) and four of the six subdominant peptides were not predicted to be immunodominant by any of the algorithms that were used to scan the HA sequence. This result suggests that presently available algorithms are as yet inadequate for predicting peptides that will be the focus of the CD4 T-cell response, at least to influenza virus.
DISCUSSION
In the studies reported here, we have comprehensively identified the major HLA-DR1-restricted peptide epitopes from influenza A virus HA that are the focus of the primary CD4 T-cell response that occurs after intranasal infection with influenza virus. We conclude from this series of experiments that the repertoire of HA-specific CD4 T cells elicited in a primary response to influenza virus infection is quite diverse and encompasses more than 30 different peptide specificities, including at least four dominant and five to six subdominant epitopes and more than 20 minor specificities. Our estimate of the number of immunodominant HA peptides is a minimal estimate, because epitopes contained in overlapping peptides may represent two peptide registers rather than a single epitope shared by two overlapping peptides. For example, one of the major epitopes identified, stimulated by peptides 63 and 64, has at least two plausible registers for binding to the HLA-DR1 molecule, and preliminary data (not shown) indicate the presence of CD4 T cells from infected mice that recognized both registers. Also suggesting that our identification should be considered a conservative estimate of the diversity in the response is that our ELISPOT experiments detected an additional number of very minor specificities. These were present in sufficiently low frequencies that we did not indicate the corresponding peptides as positive for reactivity, but it was noted that the numbers of T cells specific for many additional peptides were consistently above the background level. It is possible that cell populations could become enriched with T cells with these specificities as individuals repeatedly encounter influenza virus or vaccines, a common event in human populations.
The identification of immunodominant influenza virus-derived peptides presented by a common human MHC class II molecule as we describe in this report will be extremely valuable at several levels for present efforts in vaccine design and in the study of the natural infection of humans with influenza virus. First, there is a great deal of interest in peptide-based vaccines as stable reagents to induce protection against pathogenic organisms such as influenza virus (10, 17, 33, 39, 40, 76, 83). One of the main challenges in the design of peptide-based vaccines is the extent of polymorphism in the human MHC, which in turn influences the degree of coverage that any set of peptides will have in the targeted population. Knowledge of the MHC restriction and the immunogenic peptides is essential for assembling peptides that will cover a high percentage of the targeted population. The second major value in the information gleaned from these studies is for vaccine testing. Most presently licensed influenza vaccines are tested almost exclusively for their ability to elicit antibody responses, with surprisingly few published studies on the specificity of CD4 T-cell responses to vaccines (30, 49, 60, 92). If immunodominant peptides and MHC restriction molecules are identified, lymphocytes from vaccine recipients can be tested for CD4 T cells that recognize influenza virus-derived peptides. This additional layer of analysis of vaccine recipients will add significant new insight into what factors control vaccine efficacy, how closely antibody responses correlate with T-cell responses, and which parameter is the best predictor of efficacy for any given vaccine. The identification of OR1-restricted peptides that are the focus of the immune response to influenza virus will also be extremely valuable for MHC peptide tetramer derivation. Multimers of MHC class II peptides that bind specifically to the T-cell receptors on the surfaces of antigen-specific CD4 T cells represent a powerful new technology to isolate and analyze antigen-specific T cells (reviewed in references 38, 52, 61, 71, and 78). Despite the potential power of MHC tetramer technology, at the moment there are only a few MHC class II tetramers to detect human CD4 T cells that are specific for influenza virus. A major limitation in the use of tetramers is that the construction of these reagents requires the identification not only of immunodominant influenza virus-derived peptides but also of the precise MHC molecules that present these peptides to the CD4 T cells. Collectively, the results of the studies described in this report provide a comprehensive and unbiased assessment of the specificity of CD4 T cells elicited upon the host's first confrontation with influenza virus.
One question raised by the results of this study is why the CD4 T-cell repertoire elicited by primary exposure to influenza virus is so diverse. We speculate that after natural infection with influenza virus, several events involved in endosomal peptide acquisition by class II molecules may promote a very broad reactivity of CD4 T cells to influenza virus proteins beyond the typical four to eight peptides that are the focus of the response to exogenous protein antigens. Upon viral infection, the predominant APC that primes influenza virus-specific CD4 T cells is an actively infected dendritic cell (4, 5, 34, 56, 59, 109). We hypothesize that the abundant viral protein synthesis within dendritic cells overrides some of the major forces that normally restrict the antigen specificity of CD4 T cells. The high-level synthesis of influenza virus antigens allows them continual access to class II molecules, thereby attenuating the consequences of endosomal DM editing that is thought to dramatically narrow the repertoire of peptides presented by class II molecules (reviewed in references 6, 11, 43, 81, 103, and 104). Because of the continual synthesis of the antigens within the dendritic cells, DM editing does not preclude the class II-restricted presentation of peptides that are typically removed from class II molecules during the presentation of exogenous antigens, particularly those with low-stability interactions with the MHC class II molecule (11, 51, 55, 58, 81, 88, 102). In fact, the abundant and endogenous synthesis of viral antigens by the priming APC may account partly for the relatively low predictive value of the binding algorithms tested because the strength of the binding of the peptide to the MHC class II molecule per se may not determine the epitope hierarchy expressed on the APC and, thus, may not be the driving force in shaping the immundominance hierarchy. We have found that the broad diversity of CD4 epitopes is not unique to HA but extends to viral antigens that are normally poorly presented by class II molecules, such as the cytoplasmic/nuclear protein NS1 (our unpublished results), lending support to the idea that the high-level synthesis of viral antigens by the infected APC promotes an unusually broad diversity of class II peptide complexes. Also potentially playing a role in diversifying the selected T-cell repertoire is the frequency of antigen-bearing dendritic cells that prime the CD4 T cells in the lymph node. During the course of the influenza virus infection, the number of infected dendritic cells that seed the peripheral lymph node is quite high, particularly during the early phases of the immune response (56), and is likely to exceed the typical number of dendritic cells that migrate to lymph nodes after the subcutaneous introduction of antigen. A high frequency of antigen-bearing dendritic cells would be expected to diminish the probability of CD4 T-cell competition for antigen (reviewed in reference 47). Together, the above-described factors may contribute to the generation of a large pool of primed CD4 T cells with diverse antigen specificities that is considerably less focused than the CD4 T-cell repertoire that is elicited in response to an exogenous protein antigen or an extracellular pathogen (81, 82).
It is interesting that the peptides recognized by circulating CD4 T cells span the entire HA protein and include epitopes that are likely to be unique to the infecting strain of influenza virus. CD4 T cells of these specificities are likely to be recalled in HLA-DR1-positive individuals who have received the trivalent subunit vaccines in the past decade. CD4 T cells specific for these genetically variable regions of HA may, however, decay over the course of an individual's lifetime if that individual is confronted with many different strains of influenza A virus whose HA proteins diverge substantially from the HA contained in A/New Caledonia/20/99. Decay may occur through a lack of boosting with the infecting virus or through competition in the memory pool with CD4 T cells specific for other, more highly conserved epitopes within the genetically conserved proteins encoded by the virus, such as the nuclear or polymerase proteins (19, 85, 107). Importantly, however, our study also revealed a large fraction of CD4 T cells specific for peptides that are contained within regions of HA that are genetically conserved across different strains of influenza virus. One of the most notable epitopes in this regard is the peptide epitope(s) contained within peptides 63 and 64 (shown in Fig. 7). The amino acid sequence from residues 434 to 462 is highly conserved among H1, H2, and H5 substrains and within many different isolates of those strains. In fact, in scanning more than 200 H1, H2, and H5 sequences contributed to the NCBI database hosted by the National Library of Medicine, we noted only a single amino acid difference in HA sequences from human isolates collected over the last five decades. This sequence in A/New Caledonia/20/99 virus is in fact identical to the sequence in isolates from the 1918 “Spanish flu” (77). This degree of sequence conservation suggests that HLA-DR1-positive humans that have been previously exposed to H1N1 influenza virus may have circulating CD4 T cells that recognize and can be activated by confrontation with an H5 or H2 strain of influenza virus. CD4 T cells of this specificity are likely to be recalled and their levels are likely to be boosted each time an HLA-DR1-positive individual encounters any form of immunogen that contains H1, H2, or H5 viruses and may be particularly effective in providing help for HA-specific B cells during infection with many strains of influenza virus. In terms of vaccine design, recent studies have shown that the priming of CD4 T cells and the expansion of the repertoire to include selected specificities by using peptide priming strategies can enhance the kinetics of production of neutralizing antibodies and promote viral clearance (16, 113, 114). Analyses addressing these issues for both animal and human subjects should be highly informative and are the subject of present investigations.
Acknowledgments
This work was supported by grants R21 PA-04-119 and R01 AI51542 to A. J. Sant and pilot funding to A. J. Sant by 5U19 AI062627 from the National Institutes of Health.
We thank Snezhana Dimitrova and Tim Chapman at the University of Rochester Medical Center for advice with influenza virus growth and infection of mice, David Eckels (University of Utah) for sharing the HA peptide set with us and for advice, and Jack Gorski (Medical College of Wisconsin) and John Treanor (University of Rochester) for helpful discussions.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Al-Attiyah, R., and A. S. Mustafa. 2004. Computer-assisted prediction of HLA-DR binding and experimental analysis for human promiscuous Th1-cell peptides in the 24 kDa secreted lipoprotein (LppX) of Mycobacterium tuberculosis. Scand. J. Immunol. 59:16-24. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, D. D., and P. V. Lehmann. 2003. T-cell epitope mapping using the ELISPOT approach. Methods 29:260-269. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, B. C., D. S. Burt, C. M. Graham, A. P. Warren, J. J. Skehel, and D. B. Thomas. 1989. I-Ad restricted T cell recognition of influenza hemagglutinin. Synthetic peptides identify multiple epitopes corresponding to antibody-binding regions of the HA1 subunit. J. Immunol. 143:2663-2669. [PubMed] [Google Scholar]
- 4.Bender, A., M. Albert, A. Reddy, M. Feldman, B. Sauter, G. Kaplan, W. Hellman, and N. Bhardwaj. 1998. The distinctive features of influenza virus infection of dendritic cells. Immunobiology 198:552-567. [DOI] [PubMed] [Google Scholar]
- 5.Bhardwaj, N., A. Bender, N. Gonzalez, L. K. Bui, M. C. Garrett, and R. M. Steinman. 1994. Influenza virus-infected dendritic cells stimulate strong proliferative and cytolytic responses from human CD8+ T cells. J. Clin. Investig. 94:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, J. S., C. Ma, and S. Kovats. 1997. Antigen-presenting cells and the selection of immunodominant epitopes. Crit. Rev. Immunol. 17:411-417. [PubMed] [Google Scholar]
- 7.Brown, D. M., E. Roman, and S. L. Swain. 2004. CD4 T cell responses to influenza infection. Semin. Immunol. 16:171-177. [DOI] [PubMed] [Google Scholar]
- 8.Brown, L. R., N. R. Nygard, M. B. Graham, C. Bono, V. L. Braciale, J. Gorka, B. D. Schwartz, and T. J. Braciale. 1991. Recognition of the influenza hemagglutinin by class II MHC-restricted T lymphocytes and antibodies. I. Site definition and implications for antigen presentation and T lymphocyte recognition. J. Immunol. 147:2677-2684. [PubMed] [Google Scholar]
- 9.Brusic, V., V. B. Bajic, and N. Petrovsky. 2004. Computational methods for prediction of T-cell epitopes: a framework for modelling, testing, and applications. Methods 34:436-443. [DOI] [PubMed] [Google Scholar]
- 10.Bui, H. H., J. Sidney, K. Dinh, S. Southwood, M. J. Newman, and A. Sette. 2006. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busch, R., C. H. Rinderknecht, S. Roh, A. W. Lee, J. J. Harding, T. Burster, T. M. Hornell, and E. D. Mellins. 2005. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol. Rev. 207:242-260. [DOI] [PubMed] [Google Scholar]
- 12.Canaday, D. H., A. Gehring, E. G. Leonard, B. Eilertson, J. R. Schreiber, C. V. Harding, and W. H. Boom. 2003. T-cell hybridomas from HLA-transgenic mice as tools for analysis of human antigen processing. J. Immunol. Methods 281:129-142. [DOI] [PubMed] [Google Scholar]
- 13.Capua, I., and D. J. Alexander. 2004. Human health implications of avian influenza viruses and paramyxoviruses. Eur. J. Clin. Microbiol. Infect. Dis. 23:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Chicz, R. M., R. G. Urban, J. C. Gorga, D. A. Vignali, W. S. Lane, and J. L. Strominger. 1993. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 178:27-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couch, R. B. 2003. An overview of serum antibody responses to influenza virus antigens. Dev. Biol. 115:25-30. [PubMed] [Google Scholar]
- 16.Crowe, S. R., S. C. Miller, D. M. Brown, P. S. Adams, R. W. Dutton, A. G. Harmsen, F. E. Lund, T. D. Randall, S. L. Swain, and D. L. Woodland. 2006. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 24:457-467. [DOI] [PubMed] [Google Scholar]
- 17.Dawson, D. V., M. Ozgur, K. Sari, M. Ghanayem, and D. D. Kostyu. 2001. Ramifications of HLA class I polymorphism and population genetics for vaccine development. Genet. Epidemiol. 20:87-106. [DOI] [PubMed] [Google Scholar]
- 18.Depil, S., G. Angyalosi, O. Morales, M. Delacre, N. Delhem, V. Francois, B. Georges, J. Hammer, B. Maillere, C. Auriault, and V. Pancre. 2006. Peptide-binding assays and HLA II transgenic Abeta degrees mice are consistent and complementary tools for identifying HLA II-restricted peptides. Vaccine 24:2225-2229. [DOI] [PubMed] [Google Scholar]
- 19.Doherty, P. C., D. J. Topham, and R. A. Tripp. 1996. Establishment and persistence of virus-specific CD4+ and CD8+ T cell memory. Immunol. Rev. 150:23-44. [DOI] [PubMed] [Google Scholar]
- 20.Doytchinova, I. A., and D. R. Flower. 2005. In silico identification of supertypes for class II MHCs. J. Immunol. 174:7085-7095. [DOI] [PubMed] [Google Scholar]
- 21.Draenert, R., M. Altfeld, C. Brander, N. Basgoz, C. Corcoran, A. G. Wurcel, D. R. Stone, S. A. Kalams, A. Trocha, M. M. Addo, P. J. Goulder, and B. D. Walker. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19-29. [DOI] [PubMed] [Google Scholar]
- 22.Fleischer, B., H. Becht, and R. Rott. 1985. Recognition of viral antigens by human influenza A virus-specific T lymphocyte clones. J. Immunol. 135:2800-2804. [PubMed] [Google Scholar]
- 23.Geginat, G., S. Schenk, M. Skoberne, W. Goebel, and H. Hof. 2001. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 166:1877-1884. [DOI] [PubMed] [Google Scholar]
- 24.Gelder, C., M. Davenport, M. Barnardo, T. Bourne, J. Lamb, B. Askonas, A. Hill, and K. Welsh. 1998. Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int. Immunol. 10:211-222. [DOI] [PubMed] [Google Scholar]
- 25.Gelder, C. M., J. R. Lamb, and B. A. Askonas. 1996. Human CD4+ T-cell recognition of influenza A virus hemagglutinin after subunit vaccination. J. Virol. 70:4787-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelder, C. M., K. I. Welsh, A. Faith, J. R. Lamb, and B. A. Askonas. 1995. Human CD4+ T-cell repertoire of responses to influenza A virus hemagglutinin after recent natural infection. J. Virol. 69:7497-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdil, C. 2003. The annual production cycle for influenza vaccine. Vaccine 21:1776-1779. [DOI] [PubMed] [Google Scholar]
- 28.Gerdil, C. 2003. Using the strains and getting the vaccine licensed: a vaccine manufacturer's view. Dev. Biol. (Basel) 115:17-21. [PubMed] [Google Scholar]
- 29.Gerhard, W. 2001. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 260:171-190. [DOI] [PubMed] [Google Scholar]
- 30.Gillim-Ross, L., and K. Subbarao. 2006. Emerging respiratory viruses: challenges and vaccine strategies. Clin. Microbiol. Rev. 19:614-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillis, J. S. 2006. An avian influenza vaccine for humans targeting the polymerase B2 protein inside the capsid instead of hemagglutinin or neuramidase on the virus surface. Med. Hypotheses 66:975-977. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Nunez, M., J. Pinilla-Ibarz, T. Dao, R. J. May, M. Pao, J. S. Jaggi, and D. A. Scheinberg. 2006. Peptide binding motif predictive algorithms correspond with experimental binding of leukemia vaccine candidate peptides to HLA-A*0201 molecules. Leuk. Res. 30:1293-1298. [DOI] [PubMed] [Google Scholar]
- 33.Greenstein, J. L., V. C. Schad, W. H. Goodwin, A. B. Brauer, B. K. Bollinger, R. D. Chin, and M. C. Kuo. 1992. A universal T cell epitope-containing peptide from hepatitis B surface antigen can enhance antibody specific for HIV gp120. J. Immunol. 148:3970-3977. [PubMed] [Google Scholar]
- 34.Hamilton-Easton, A., and M. Eichelberger. 1995. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. J. Virol. 69:6359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansson, L., H. Rabbani, J. Fagerberg, A. Osterborg, and H. Mellstedt. 2003. T-cell epitopes within the complementarity-determining and framework regions of the tumor-derived immunoglobulin heavy chain in multiple myeloma. Blood 101:4930-4936. [DOI] [PubMed] [Google Scholar]
- 36.He, X., E. F. Rosloniec, L. K. Myers, W. L. McColgan III, M. Gumanovskaya, A. H. Kang, and J. M. Stuart. 2004. T cell receptors recognizing type II collagen in HLA-DR-transgenic mice characterized by highly restricted V beta usage. Arthritis Rheum. 50:1996-2004. [DOI] [PubMed] [Google Scholar]
- 37.Hilleman, M. R. 2002. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine 20:3068-3087. [DOI] [PubMed] [Google Scholar]
- 38.Hobeika, A. C., M. A. Morse, T. Osada, M. Ghanayem, D. Niedzwiecki, R. Barrier, H. K. Lyerly, and T. M. Clay. 2005. Enumerating antigen-specific T-cell responses in peripheral blood: a comparison of peptide MHC Tetramer, ELISpot, and intracellular cytokine analysis. J. Immunother. 28:63-72. [DOI] [PubMed] [Google Scholar]
- 39.Hunziker, I. P., R. Zurbriggen, R. Glueck, O. B. Engler, J. Reichen, W. J. Dai, W. J. Pichler, and A. Cerny. 2001. Perspectives: towards a peptide-based vaccine against hepatitis C virus. Mol. Immunol. 38:475-484. [DOI] [PubMed] [Google Scholar]
- 40.Iwai, L. K., M. Yoshida, J. Sidney, M. A. Shikanai-Yasuda, A. C. Goldberg, M. A. Juliano, J. Hammer, L. Juliano, A. Sette, J. Kalil, L. R. Travassos, and E. Cunha-Neto. 2003. In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasiliensis frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals. Mol. Med. 9:209-219. [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen, E. M., N. M. Droin, E. E. Lemmens, M. J. Pinkoski, S. J. Bensinger, B. D. Ehst, T. S. Griffith, D. R. Green, and S. P. Schoenberger. 2005. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature 434:88-93. [DOI] [PubMed] [Google Scholar]
- 42.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421:852-856. [DOI] [PubMed] [Google Scholar]
- 43.Jensen, P. E., D. A. Weber, W. P. Thayer, X. Chen, and C. T. Dao. 1999. HLA-DM and the MHC class II antigen presentation pathway. Immunol. Res. 20:195-205. [DOI] [PubMed] [Google Scholar]
- 44.Joseph, T., and K. Subbarao. 2005. Human infections with avian influenza viruses. Maryland Med. 6:30-32. [PubMed] [Google Scholar]
- 45.Karlsson, A. C., J. N. Martin, S. R. Younger, B. M. Bredt, L. Epling, R. Ronquillo, A. Varma, S. G. Deeks, J. M. McCune, D. F. Nixon, and E. Sinclair. 2003. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J. Immunol. Methods 283:141-153. [DOI] [PubMed] [Google Scholar]
- 46.Katz, J. M., J. Plowden, M. Renshaw-Hoelscher, X. Lu, T. M. Tumpey, and S. Sambhara. 2004. Immunity to influenza: the challenges of protecting an aging population. Immunol. Res. 29:113-124. [DOI] [PubMed] [Google Scholar]
- 47.Kedl, R. M., J. W. Kappler, and P. Marrack. 2003. Epitope dominance, competition and T cell affinity maturation. Curr. Opin. Immunol. 15:120-127. [DOI] [PubMed] [Google Scholar]
- 48.Khanolkar, A., M. J. Fuller, and A. J. Zajac. 2004. CD4 T cell-dependent CD8 T cell maturation. J. Immunol. 172:2834-2844. [DOI] [PubMed] [Google Scholar]
- 49.Kitler, M. E., P. Gavinio, and D. Lavanchy. 2002. Influenza and the work of the World Health Organization. Vaccine 20(Suppl. 2):S5-S14. [DOI] [PubMed] [Google Scholar]
- 50.Kobasa, D., and Y. Kawaoka. 2005. Emerging influenza viruses: past and present. Curr. Mol. Med. 5:791-803. [DOI] [PubMed] [Google Scholar]
- 51.Kropshofer, H., A. B. Vogt, G. Moldenhauer, J. Hammer, J. S. Blum, and G. J. Hammerling. 1996. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 15:6144-6154. [PMC free article] [PubMed] [Google Scholar]
- 52.Kwok, W. W., N. A. Ptacek, A. W. Liu, and J. H. Buckner. 2002. Use of class II tetramers for identification of CD4+ T cells. J. Immunol. Methods 268:71-81. [DOI] [PubMed] [Google Scholar]
- 53.Lamb, J. R., D. D. Eckels, M. Phelan, P. Lake, and J. N. Woody. 1982. Antigen-specific human T lymphocyte clones: viral antigen specificity of influenza virus-immune clones. J. Immunol. 128:1428-1432. [PubMed] [Google Scholar]
- 54.Latham, K. A., K. B. Whittington, R. Zhou, Z. Qian, and E. F. Rosloniec. 2005. Ex vivo characterization of the autoimmune T cell response in the HLA-DR1 mouse model of collagen-induced arthritis reveals long-term activation of type II collagen-specific cells and their presence in arthritic joints. J. Immunol. 174:3978-3985. [DOI] [PubMed] [Google Scholar]
- 55.Lazarski, C. A., F. A. Chaves, and A. J. Sant. 2006. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J. Exp. Med. 203:1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Legge, K. L., and T. J. Braciale. 2003. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity 18:265-277. [DOI] [PubMed] [Google Scholar]
- 57.Lewis, D. B. 2006. Avian flu to human influenza. Annu. Rev. Med. 57:139-154. [DOI] [PubMed] [Google Scholar]
- 58.Lich, J. D., J. A. Jayne, D. Zhou, J. F. Elliott, and J. S. Blum. 2003. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J. Immunol. 171:853-859. [DOI] [PubMed] [Google Scholar]
- 59.Lopez, C. B., A. Fernandez-Sesma, S. M. Czelusniak, J. L. Schulman, and T. M. Moran. 2000. A mouse model for immunization with ex vivo virus-infected dendritic cells. Cell. Immunol. 206:107-115. [DOI] [PubMed] [Google Scholar]
- 60.Luke, C. J., and K. Subbarao. 2006. Vaccines for pandemic influenza. Emerg. Infect. Dis. 12:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mallone, R., and G. T. Nepom. 2004. MHC class II tetramers and the pursuit of antigen-specific T cells: define, deviate, delete. Clin. Immunol. 110:232-242. [DOI] [PubMed] [Google Scholar]
- 62.Marsh, S. G. E., P. Parham, and L. D. Barber. 2000. The factsbook series. The HLA factsbook. Academic Press, New York, NY.
- 63.McHeyzer-Williams, L. J., L. P. Malherbe, and M. G. McHeyzer-Williams. 2006. Helper T cell-regulated B cell immunity. Curr. Top. Microbiol. Immunol. 311:59-83. [DOI] [PubMed] [Google Scholar]
- 64.McHeyzer-Williams, L. J., and M. G. McHeyzer-Williams. 2005. Antigen-specific memory B cell development. Annu. Rev. Immunol. 23:487-513. [DOI] [PubMed] [Google Scholar]
- 65.Mori, M., P. G. Beatty, M. Graves, K. M. Boucher, and E. L. Milford. 1997. HLA gene and haplotype frequencies in the North American population: the National Marrow Donor Program Donor Registry. Transplantation 64:1017-1027. [DOI] [PubMed] [Google Scholar]
- 66.Mustafa, A. S., and F. A. Shaban. 2006. ProPred analysis and experimental evaluation of promiscuous T-cell epitopes of three major secreted antigens of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 86:115-124. [DOI] [PubMed] [Google Scholar]
- 67.Myers, L. K., Y. Sakurai, E. F. Rosloniec, J. M. Stuart, and A. H. Kang. 2004. An analog peptide that suppresses collagen-induced arthritis. Am. J. Med. Sci. 327:212-216. [DOI] [PubMed] [Google Scholar]
- 68.Myers, L. K., Y. Sakurai, B. Tang, X. He, E. F. Rosloniec, J. M. Stuart, and A. H. Kang. 2002. Peptide-induced suppression of collagen-induced arthritis in HLA-DR1 transgenic mice. Arthritis Rheum. 46:3369-3377. [DOI] [PubMed] [Google Scholar]
- 69.Neumann, G., and Y. Kawaoka. 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg. Infect. Dis. 12:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Northrop, J. K., R. M. Thomas, A. D. Wells, and H. Shen. 2006. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 177:1062-1069. [DOI] [PubMed] [Google Scholar]
- 71.Novak, E. J., A. W. Liu, G. T. Nepom, and W. W. Kwok. 1999. MHC class II tetramers identify peptide-specific human CD4+ T cells proliferating in response to influenza A antigen. J. Clin. Investig. 104:R63-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pajot, A., M. L. Michel, M. Mancini-Bourgine, M. N. Ungeheuer, D. M. Ojcius, Q. Deng, F. A. Lemonnier, and Y. C. Lone. 2006. Identification of novel HLA-DR1-restricted epitopes from the hepatitis B virus envelope protein in mice expressing HLA-DR1 and vaccinated human subjects. Microbes Infect. 8:2783-2790. [DOI] [PubMed] [Google Scholar]
- 73.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10:S82-S87. [DOI] [PubMed] [Google Scholar]
- 74.Rammensee, H., J. Bachmann, N. P. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 75.Reche, P. A., J. P. Glutting, H. Zhang, and E. L. Reinherz. 2004. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56:405-419. [DOI] [PubMed] [Google Scholar]
- 76.Reche, P. A., and E. L. Reinherz. 2005. PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands. Nucleic Acids Res. 33:W138-W42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reijonen, H., and W. W. Kwok. 2003. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods (Duluth) 29:282-288. [DOI] [PubMed] [Google Scholar]
- 79.Rojas, J. M., S. E. McArdle, R. B. Horton, M. Bell, S. Mian, G. Li, S. A. Ali, and R. C. Rees. 2005. Peptide immunisation of HLA-DR-transgenic mice permits the identification of a novel HLA-DRbeta1*0101- and HLA-DRbeta1*0401-restricted epitope from p53. Cancer Immunol. Immunother. 54:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosloniec, E. F., K. B. Whittington, D. M. Zaller, and A. H. Kang. 2002. HLA-DR1 (DRB1*0101) and DR4 (DRB1*0401) use the same anchor residues for binding an immunodominant peptide derived from human type II collagen. J. Immunol. 168:253-259. [DOI] [PubMed] [Google Scholar]
- 81.Sant, A. J., F. Chaves, S. Jenks, K. Richards, P. Menges, J. M. Weaver, and C. L. Lazarski. 2005. The relationship between immundominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol. Rev. 207:261-278. [DOI] [PubMed] [Google Scholar]
- 82.Sant, A. J., F. C. Chaves, F. Krafcik, C. A. Lazarski, P. R. Menges, K. A. Richards, and J. Weaver. Immunodominance in CD4 T cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev. Vaccines, in press. [DOI] [PubMed]
- 83.Schipper, R. F., C. A. van Els, J. D'Amaro, and M. Oudshoorn. 1996. Minimal phenotype panels. A method for achieving maximum population coverage with a minimum of HLA antigens. Hum. Immunol. 51:95-98. [DOI] [PubMed] [Google Scholar]
- 84.Schirle, M., T. Weinschenk, and S. Stevanovic. 2001. Combining computer algorithms with experimental approaches permits the rapid and accurate identification of T cell epitopes from defined antigens. J. Immunol. Methods 257:1-16. [DOI] [PubMed] [Google Scholar]
- 85.Selin, L. K., and R. M. Welsh. 2004. Plasticity of T cell memory responses to viruses. Immunity 20:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singer, E. 2005. Pandemic fears hatch new methods in flu vaccine industry. Nat. Med. 11:4. [DOI] [PubMed] [Google Scholar]
- 87.Singh, H., and G. P. Raghava. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236-1237. [DOI] [PubMed] [Google Scholar]
- 88.Sloan, V. S., P. Cameron, G. Porter, M. Gammon, M. Amaya, E. Mellins, and D. M. Zaller. 1995. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375:802-806. [DOI] [PubMed] [Google Scholar]
- 89.Southwood, S., J. Sidney, A. Kondo, M. F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363-3373. [PubMed] [Google Scholar]
- 90.Stephenson, I., K. G. Nicholson, J. M. Wood, M. C. Zambon, and J. M. Katz. 2004. Confronting the avian influenza threat: vaccine development for a potential pandemic. Lancet Infect. Dis. 4:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stohr, K., and M. Esveld. 2004. Public health. Will vaccines be available for the next influenza pandemic? Science 306:2195-2196. [DOI] [PubMed] [Google Scholar]
- 92.Subbarao, K., B. R. Murphy, and A. S. Fauci. 2006. Development of effective vaccines against pandemic influenza. Immunity 24:5-9. [DOI] [PubMed] [Google Scholar]
- 93.Sun, J. C., and M. J. Bevan. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun, J. C., M. A. Williams, and M. J. Bevan. 2004. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 5:927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Swain, S. L., J. N. Agrewala, D. M. Brown, D. M. Jelley-Gibbs, S. Golech, G. Huston, S. C. Jones, C. Kamperschroer, W. H. Lee, K. K. McKinstry, E. Roman, T. Strutt, and N. P. Weng. 2006. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol. Rev. 211:8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tamura, S., and T. Kurata. 2004. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn. J. Infect. Dis. 57:236-247. [PubMed] [Google Scholar]
- 97.Tamura, S., T. Tanimoto, and T. Kurata. 2005. Mechanisms of broad cross-protection provided by influenza virus infection and their application to vaccines. Jpn. J. Infect. Dis. 58:195-207. [PubMed] [Google Scholar]
- 98.Teo, S. S., J. S. Nguyen-Van-Tam, and R. Booy. 2005. Influenza burden of illness, diagnosis, treatment, and prevention: what is the evidence in children and where are the gaps? Arch. Dis. Child. 90:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thomas, P. G., R. Keating, D. J. Hulse-Post, and P. C. Doherty. 2006. Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 12:48-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tobery, T. W., and M. J. Caulfield. 2004. Identification of T-cell epitopes using ELISpot and peptide pool arrays. Methods Mol. Med. 94:121-132. [DOI] [PubMed] [Google Scholar]
- 101.Tobery, T. W., S. Wang, X. M. Wang, M. P. Neeper, K. U. Jansen, W. L. McClements, and M. J. Caulfield. 2001. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J. Immunol. Methods 254:59-66. [DOI] [PubMed] [Google Scholar]
- 102.van Ham, S. M., U. Gruneberg, G. Malcherek, I. Broker, A. Melms, and J. Trowsdale. 1996. Human histocompatibility leukocyte antigen (HLA)-DM edits peptides presented by HLA-DR according to their ligand binding motifs. J. Exp. Med. 184:2019-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vogt, A. B., S. O. Arndt, G. J. Hammerling, and H. Kropshofer. 1999. Quality control of MHC class II associated peptides by HLA-DM/H2-M. Semin. Immunol. 11:391-403. [DOI] [PubMed] [Google Scholar]
- 104.Vogt, A. B., H. Kropshofer, and G. J. Hammerling. 1997. How HLA-DM affects the peptide repertoire bound to HLA-DR molecules. Hum. Immunol. 54:170-179. [DOI] [PubMed] [Google Scholar]
- 105.Wang, X., and T. Mosmann. 2001. In vivo priming of CD4 T cells that produce interleukin (IL)-2 but not IL-4 or interferon (IFN)-gamma, and can subsequently differentiate into IL-4- or IFN-gamma-secreting cells. J. Exp. Med. 194:1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Webby, R. J., and R. G. Webster. 2003. Are we ready for pandemic influenza? Science 302:1519-1522. [DOI] [PubMed] [Google Scholar]
- 107.Welsh, R. M., L. K. Selin, and E. Szomolanyi-Tsuda. 2004. Immunological memory to viral infections. Annu. Rev. Immunol. 22:711-743. [DOI] [PubMed] [Google Scholar]
- 108.Wood, J. M. 2002. Selection of influenza vaccine strains and developing pandemic vaccines. Vaccine 20(Suppl. 5):B40-B44. [DOI] [PubMed] [Google Scholar]
- 109.Woodland, D. L., and T. D. Randall. 2004. Anatomical features of anti-viral immunity in the respiratory tract. Semin. Immunol. 16:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woods, A., H. Y. Chen, M. E. Trumbauer, A. Sirotina, R. Cummings, and D. M. Zaller. 1994. Human major histocompatibility complex class II-restricted T cell responses in transgenic mice. J. Exp. Med. 180:173-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.World Health Organization. 2005. New WHO influenza vaccine recommendations for 2005-2006 season, northern hemisphere. Euro Surveill. 10:E050217.1. [PubMed] [Google Scholar]
- 112.Yewdell, J., and A. Garcia-Sastre. 2002. Influenza virus still surprises. Curr. Opin. Microbiol. 5:414-418. [DOI] [PubMed] [Google Scholar]
- 113.Zhao, J., Q. Huang, W. Wang, Y. Zhang, P. Lv, and X. M. Gao. 2007. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 81:6079-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong, W., D. Marshall, C. Coleclough, and D. L. Woodland. 2000. CD4+ T cell priming accelerates the clearance of Sendai virus in mice, but has a negative effect on CD8+ T cell memory. J. Immunol. 164:3274-3282. [DOI] [PubMed] [Google Scholar]