Abstract
The chlorovirus PBCV-1, like many large double-stranded DNA-containing viruses, contains several genes that encode putative proteins involved in nucleotide biosynthesis. This report describes the characterization of the PBCV-1 dCMP deaminase, which produces dUMP, a key intermediate in the synthesis of dTTP. As predicted, the recombinant protein has dCMP deaminase activity that is activated by dCTP and inhibited by dTTP. Unexpectedly, however, the viral enzyme also has dCTP deaminase activity, producing dUTP. Typically, these two reactions are catalyzed by proteins in separate enzyme classes; to our knowledge, this is the first example of a protein having both deaminase activities. Kinetic experiments established that (i) the PBCV-1 enzyme has a higher affinity for dCTP than for dCMP, (ii) dCTP serves as a positive heterotropic effector for the dCMP deaminase activity and a positive homotropic effector for the dCTP deaminase activity, and (iii) the enzymatic efficiency of the dCMP deaminase activity is about four times higher than that of the dCTP deaminase activity. Inhibitor studies suggest that the same active site is involved in both dCMP and dCTP deaminations. The discovery that the PBCV-1 dCMP deaminase has two activities, together with a previous report that the virus also encodes a functional dUTP triphosphatase (Y. Zhang, H. Moriyama, K. Homma, and J. L. Van Etten, J. Virol. 79:9945-9953, 2005), means that PBCV-1 is the first virus to encode enzymes involved in all three known pathways to form dUMP.
Members and prospective members of the family Phycodnaviridae consist of a genetically diverse, but morphologically similar, group of large double-stranded DNA-containing viruses (170 to 560 kb) that infect algae from both freshwater and marine water (4, 45). The most studied phycodnaviruses are the plaque-forming chlorella viruses that belong to the genus Chlorovirus (46). The chloroviruses infect certain freshwater, unicellular, eukaryotic, chlorella-like green algae, which normally exist as endosymbionts in various protists, such as Paramecium bursaria and Acanthocystis turfacea. The prototype chlorella virus PBCV-1 has a 331-kb genome that contains 366 putative protein-encoding genes and a polycistronic gene that encodes 11 tRNAs. PBCV-1 infects its host Chlorella NC64A by attaching rapidly, specifically, and irreversibly to the external surface of the algal cell wall (25). Attachment occurs at a unique virus vertex (30) and is followed by degradation of the host wall at the attachment point. Following host cell wall degradation, the PBCV-1 internal membrane presumably fuses with the host membrane, resulting in entry of the viral DNA and virion-associated proteins into the cell. PBCV-1 DNA replication begins 60 to 90 min after infection, and by 4 h postinfection (p.i.) the total DNA in the cell increases at least fourfold (38). Consequently, PBCV-1 DNA synthesis requires large quantities of deoxynucleotides, including dTTP, which cannot be accounted for simply by recycling deoxynucleotides from degraded host DNA. Therefore, to meet this requirement, large DNA viruses, including PBCV-1, frequently encode deoxynucleotide synthesis enzymes that either aid or replace host enzymes. PBCV-1 encodes 13 such enzymes (37).
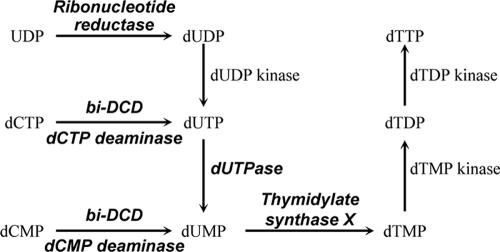
As outlined in Fig. 1, dUMP is an essential intermediate in the de novo synthesis of dTTP, and three somewhat overlapping pathways exist to synthesize dUMP: (i) deamination of dCMP to dUMP by dCMP deaminase, (ii) deamination of dCTP to dUTP by dCTP deaminase followed by hydrolysis of dUTP to dUMP by dUTPase, and (iii) reduction of UDP or UTP to dUDP or dUTP, respectively, by ribonucleotide reductase followed by hydrolysis of dUTP to dUMP by dUTPase. PBCV-1 encodes four of the enzymes listed in Fig. 1, although recombinant proteins have been characterized only for dUTPase (48) and thymidylate synthase X (11). To continue characterizing the PBCV-1 nucleotide metabolic enzymes, we report herein that the PBCV-1-encoded dCMP deaminase has the expected activity; however, unexpectedly the enzyme also has dCTP deaminase activity that converts dCTP to dUTP. To our knowledge, the PBCV-1 enzyme is the first protein to have both dCMP and dCTP deaminase activities. Furthermore, PBCV-1 encodes enzymes involved in all three known pathways that lead to dUMP synthesis.
FIG. 1.
The three known pathways for synthesis of dUMP, an intermediate in dTTP synthesis. The enzymes shown in the diagram in bold italics are encoded by chlorovirus PBCV-1 and are known to be functional, except for the ribonucleotide reductase.
MATERIALS AND METHODS
Viruses and host strains.
The growth of PBCV-1 host Chlorella NC64A on MBBM medium and Chlorella Pbi on FES medium, the production and purification of the viruses, and the isolation of virus DNAs have been described previously (32, 39, 40). Escherichia coli strains DH5MCR (E. coli Genetic Stock Center, New Haven, CT) and BL21(DE3)/pLysS (Novagen, Madison, WI) were grown in Luria-Bertani (LB) medium (34).
Cloning and expression of the dCMP deaminase gene and purification of the enzyme.
PBCV-1 open reading frame (ORF) A596R was cloned from PCR-amplified viral DNA by use of the following oligonucleotide primers: 5′ primer 5′-GGTGGTGCATATGTCAAAAGCAGAGAAG-3′ and 3′ primer 5′-TCCCTCGAGTAGGTATTCGACTTCTAT-3′. The 5′ primer contained an NdeI restriction site, and the 3′ primer contained an XhoI restriction site. The A596R gene was amplified with KOD Hot Start DNA polymerase (Novagen) in 50-μl reaction mixtures which contained 100 ng of virus DNA, 15 pM of each primer, 0.2 mM each of dATP, dGTP, dCTP, and dTTP, and 1 mM MgSO4 by 35 cycles of heating and cooling (15 s at 94°C for denaturing, 30 s at 60°C for annealing, and 1 min at 68°C for elongation). The PCR products were purified from 1.2% agarose gels by use of a QIAEX II gel extraction kit (QIAGEN, Valencia, CA), digested with NdeI and XhoI, and inserted into the NdeI/XhoI sites of the pET23a+ expression vector (Novagen). This process produced a six-His tag at the C terminus of the target protein. The construction of the recombinant expression plasmid, named pET-dCD, was confirmed by DNA sequencing. Expression of the dCMP deaminase protein was carried out in E. coli BL21(DE3)/pLysS, which contains the T7 RNA polymerase gene under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter and a plasmid, pLysS, constitutively expressing the T7 lysozyme that is the inhibitor of T7 RNA polymerase. Cells were transformed with plasmid pET-dCD and grown overnight in LB medium at 37°C. Flasks containing 400 ml LB medium which contained 100 μg/ml ampicillin and 37 μg/ml chloramphenicol were inoculated with a 1/40 volume of the overnight culture and incubated at 37°C until the absorbance at 595 nm reached 0.6 to 0.8. IPTG (Sigma, St. Louis, MO) was added to a final concentration of 0.5 mM, and incubation was continued for 2 h at 30°C. One liter of cells was harvested by centrifugation at 5,000 × g for 5 min and resuspended in 50 ml NPI-10 buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 10 mM imidazole). The cells were disrupted on ice by sonication for 4 min with a Tekmar sonic disruptor at 100% amplitude in 5-s pulses. The samples were centrifuged at 12,000 × g for 10 min to separate soluble and insoluble fractions. Then, Ni-nitrilotriacetic acid agarose beads (QIAGEN, Hilden, Germany) equilibrated with NPI-10 buffer were added to the soluble fraction. After the solution was mixed for 1 h at 4°C, the resin was loaded in a column and washed with 20 column volumes of NPI-40 buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 40 mM imidazole). The recombinant dCMP deaminase protein was eluted from the column by use of NPI-250 buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 250 mM imidazole). Glycerol was added to the sample until the final concentration was 50%, and the mixture was stored at −20°C. Protein concentrations of the purified enzyme were determined by use of a bicinchoninic acid protein assay (Pierce, Rockford, IL).
dCMP deaminase assay.
Both dCMP deaminase activity and dCTP deaminase activity were assayed by using the difference in the molar extinction coefficients between deoxycytidine and deoxyuridine compounds of 10.3 × 103 at 290 nm (pH 2.0), modified from a method described earlier (23). The standard reaction mixture contained 50 mM CAPSO (3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid sodium salt), pH 9.5, 2 mM 2-mercaptoethanol, 1 mM MgCl2, 1 mM dCMP, 0.1 mM dCTP, and 5 μg/ml enzyme. The reaction cuvette contained all ingredients minus the enzyme equilibrated to 42°C in a Beckman DU-530 spectrophotometer; the reaction was started by addition of the enzyme, with rapid mixing. Enzyme activity was monitored at A290 with 5-s acquisition intervals in a temperature-equilibrated reaction chamber by use of a Peltier temperature control module (Beckman).
The determination of optimal temperature was performed at 25°C, 37°C, 42°C, 50°C, and 55°C. To evaluate the optimal pH range for these activities, a mixture of buffering compounds was used {PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], HEPES, Tris, and CAPSO}, each at 10 mM. This mixture was adjusted to the desired pH and used in a range of 6.0 to 11.0. Divalent-cation-dependent experiments were performed in the same reaction system by replacing MgCl2 with 1 mM MnCl2, CaCl2, NiCl2, CoCl2, CuCl2, or ZnSO4. Kinetic studies of dCMP deaminase were performed under standard assay conditions with various substrate concentrations, ranging from 0.1 to 2 mM dCMP, in the presence and absence of dCTP. The reaction mixtures used in the kinetic studies were incubated at 42°C and evaluated from their initial velocities.
dCTP deaminase assay.
The standard reaction mixture contained 50 mM Tris-HCl, pH 7.0, 2 mM 2-mercaptoethanol, 1 mM MgCl2, 1 mM dCTP, and 5 μg/ml enzyme. The reactions were started by addition of the enzyme. As with the dCMP deaminase assay, the dCTP deaminase activity of the enzyme was determined by the spectrophotometric method. The optimal temperature, pH, and cation requirements were determined as described for characterizing dCMP deaminase activity. Kinetic studies of dCTP deaminase were performed under standard assay conditions with various substrate concentrations, ranging from 0.1 to 1.6 mM dCTP.
TLC assays.
For thin-layer chromatography (TLC) assays, the reaction mixtures consisted of 50-μl volumes and the optimal conditions for dCMP deaminase and dCTP deaminase activity were used. At intervals, 5-μl samples were withdrawn from the reaction mixture and applied to PEI TLC plates (Aldrich, Milwaukee, WI). Twenty nmol each of dCTP, dCMP, dUTP, and dUMP was applied to the plates as standards. The plates were developed stepwise in the following solutions: (i) 1 M acetic acid for ∼1 min, (ii) 0.9 M acetic acid and 0.3 M LiCl to 3 cm above the starting line, and (iii) 1 M acetic acid and 1.3 M LiCl to 14 cm above the starting line. After development, the plates were washed in 100% methanol for 5 min and dried with hot air, and the nucleotide spots were identified by UV light. The retardation factor values for dCTP, dCMP, dUTP, and dUMP were 0.3, 0.74, 0.26, and 0.63, respectively.
Inhibition assay of the enzyme activities.
The inhibition reaction rates were monitored continuously with time by determining the decrease in absorption at 290 nm after addition of enzyme at 30°C by use of a Gilford 250 spectrophotometer, as described previously (20). For inhibition of dCMP deaminase activity, the reaction mixtures contained 1 mM dCMP, 50 mM CAPSO, pH 9.5, 1 mM MgCl2, 2 mM 2-mercaptoethanol, 0.1 mM dCTP, and 2.2 μg of the enzyme in a final volume of 1.0 ml. For inhibition of dCTP deaminase activity, the reaction mixtures contained 1 mM dCTP, 50 mM Tris-HCl, pH 7.0, 1 mM MgCl2, 2 mM 2-mercaptoethanol, and 2.2 μg of the enzyme in a final volume of 1.0 ml. Each inhibitor, pyrimidin-2-one deoxyribotide (PDRP) (2) and H4dUMP (14), was present at concentrations from 2 μM to 10 μM.
Sedimentation velocity studies.
The PBCV-1 enzyme in NPI-250 buffer was dialyzed against two changes of 100 ml each of a solution containing 50 mM potassium phosphate, pH 7.5, 100 mM NaCl, and 2 mM dithiothreitol at 4°C and then diluted to an A280 of 0.32. The viscosity and density of the buffers and partial specific volume (Vbar) of the protein were obtained from SEDNTERP software (http://www.jphilo.mailway.com). The Vbar of the enzyme was calculated from the amino acid content to be 0.7313 ml/g at 25°C or 0.7291 ml/g at 20°C. The protein (0.4 ml) and 0.42 ml dialysis buffer were centrifuged at 50,000 rpm in 12-mm Epon charcoal-filled double-sector centerpieces in a Beckman XL-I analytical ultracentrifuge and an An-60 Ti rotor at 25°C. Absorption measurements were made at 280 nm. A single sample was run with zero time between scans; the Rmin was set at 6.0, and the samples were scanned from the earliest time until the boundaries were at the cell bottom so as to obtain a large number of scans. Scans were set at a spacing of 0.003-cm radial step size in a continuous scan mode. Care was taken to have the samples at thermal equilibrium before starting. The centrifuge was accelerated directly to the speed of the experiment. The data were analyzed using the c(s) method found in SEDFIT, a program developed by P. Schuck (http://www.analyticalultracentrifugation.com). The experimentally calculated sedimentation coefficients from the SEDFIT program were converted to s20,W values by use of the transform within the SEDFIT software.
Northern and dot blot analyses.
Chlorella cells (1 × 109 cells) were collected at various times after PBCV-1 infection, frozen in liquid nitrogen, and stored at −80°C. RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), denatured with formaldehyde, separated on a 1.2% agarose denaturing gel, and transferred to nylon membranes (Micron Separations, Inc., Westborough, MA), as described previously (10). The membrane was subsequently photographed under UV illumination to visualize transferred RNA. The RNA was hybridized with a double-stranded 32P-A596R gene probe at 65°C in 50 mM sodium phosphate, 1% bovine serum albumin, and 2% sodium dodecyl sulfate (SDS), pH 7.2. The probe was labeled with 32P by use of a random-primer DNA labeling system (Invitrogen). After hybridization, radioactivity bound to the membranes was detected and quantified using a Storm 840 PhosphorImager and ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, CA). To monitor possible loading differences between samples, the relative amount of the 3.6-kb rRNA in each lane was determined by converting the photographs of stained membranes to digital images with a Hewlett-Packard ScanJet 4C scanner and analyzing the images with ImageQuant software.
Viral DNAs used for dot blots were denatured, applied to nylon membranes fixed by UV cross-linker, and hybridized with the same probes used for the Northern analysis.
Phylogenetic analyses.
A BLASTP search with the amino acid sequence of PBCV-1 A596R (NCBI NP_048952) was conducted using default settings in the Biology Workbench (http://workbench.sdsc.edu/). Twenty-one taxa, including archea, prokaryotes, and eukaryotes, as well as several viruses, were selected. The 22 taxa, which included PBCV-1, were aligned with ClustalW by use of the default setting in the Biology Workbench. The alignment was changed to NEXUS format and imported into PAUP 4.0b10 (36) for phylogenetic analysis. Trees were constructed using the following three methods: maximum-parsimony heuristic, neighbor joining, and maximum-parsimony bootstrap (1,000 replicates). The DNA binding protein from Streptococcus thermophilus (NCBI YP_142074), which has some sequence common to dCMP deaminase sequences, was used as the outgroup to root the trees.
Other procedures.
DNA and putative protein sequences were analyzed with the University of Wisconsin Computer Group version 10.1 package of programs (Genetics Computer Group, 2000).
RESULTS
PBCV-1 ORF A596R encodes a dCMP deaminase.
Chlorella virus PBCV-1 encodes a 142-codon ORF (A596R) that has 30 to 40% amino acid identity with dCMP deaminases from eukaryotes, prokaryotes, and T4 phage. The predicted dCMP deaminase has an inferred molecular mass of ∼16 kDa. Like dCMP deaminases from other organisms, A596R contains a conserved domain that includes a putative zinc-binding domain, an allosteric regulator binding site, and residues that form the catalytic site (1, 22, 26) (Fig. 2).
FIG. 2.
Amino acid alignment of dCMP deaminases. The amino acid sequences from human (P32321), Lactococcus lactis (E86767), Pyrococcus abyssi (CAB49495), Saccharomyces cerevisiae (S46762), T4 phage (AAD42546), and PBCV-1 (O41078) were aligned by use of the Wisconsin GCG Pileup program. (Accession numbers are from the NCBI database.) The most conserved domain of dCMP deaminases, which forms the catalytic site, is boxed. The amino acid residues indicated with a solid square correspond to one zinc ion-binding site of T4 phage dCMP deaminase; the amino acid residues indicated with a solid inverted triangle correspond to the regulator (dCTP and dTTP) binding site of T4 phage dCMP deaminase. A black box around an individual residue indicates identity; a gray box indicates similarity.
The G+C content of the A596R gene is 39%, which is close to the 40% G+C content of the PBCV-1 genome (41) and significantly less than the 67% G+C content of Chlorella NC64A DNA. Typically, the 50 nucleotides preceding PBCV-1 translational start codons are at least 70% A+T (37). The A596R gene fulfills this requirement, with 78% A+T. Three possible TATAAT-like sequences are located within the 15 nucleotides prior to the AUG translation start codon (data not shown). Consequently, we expected the A596R gene to encode a functional dCMP deaminase.
Expression and purification of the protein.
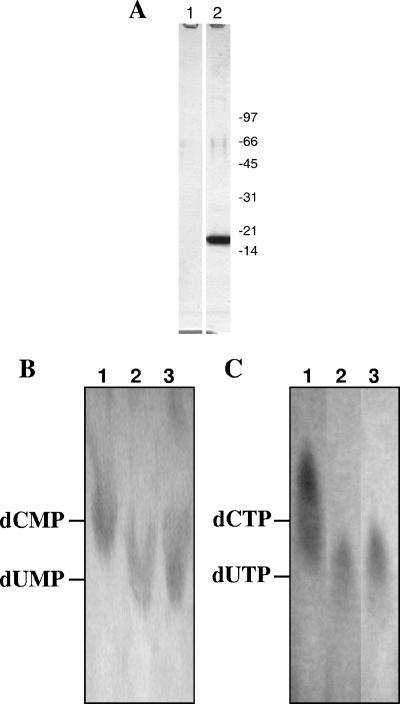
Plasmid pET-dCD, containing the PBCV-1 A596R gene, was constructed, and the protein was expressed as a 17-kDa fusion protein containing a six-His tag at the C terminus in E. coli strain BL21(DE3)/pLysS. The His tag allowed the recombinant protein to be purified by Ni-nitrilotriacetic acid affinity chromatography. About 20 mg of soluble recombinant protein was obtained per liter of E. coli culture, which quickly precipitates out of solution when the concentration of the protein is >2 mg/ml. However, if the concentration is less than 2 mg/ml, the protein could be stored for at least 1 year in 50% glycerol without precipitation. As described below, the isolated protein deaminates dCMP to dUMP and, unexpectedly, the protein also deaminates dCTP to dUTP. Therefore, we named the protein bifunctional dCMP-dCTP deaminase (bi-DCD). Elution of the His-tagged recombinant protein produced a single Coomassie brilliant blue staining band on SDS-polyacrylamide gels (Fig. 3A), which contained both the dCMP and the dCTP deaminase activities. Empty vector control cells (pET-23a+) produced no measurable dCMP or dCTP deaminase activity.
FIG. 3.
Expression and characterization of bi-DCD. (A) SDS-polyacrylamide gel electrophoresis of purified recombinant PBCV-1 bi-DCD (lane 2). Lane 1 is from an empty vector, pET-23a+, that was cultured, expressed, and fractionated in parallel with pET-dCD. Molecular mass markers (in kilodaltons) are from Bio-Rad (low-range standards). (B) TLC analysis. Lanes 1 and 3 are dCMP and dUMP standards, respectively; lane 2 is the product of dCMP deamination by bi-DCD. (C) Lanes 1 and 3 are dCTP and dUTP standards, respectively; lane 2 is the product of dCTP deamination by bi-DCD.
Typically, dCMP deaminases function as hexamers (1), and a sedimentation velocity analysis yielded an S20,W of 6.06 for bi-DCD (results not shown). Based on a subunit mass of 17 kDa, the PBCV-1 enzyme sedimented as a hexamer.
Characterization of the bi-DCD dCMP deaminase activity.
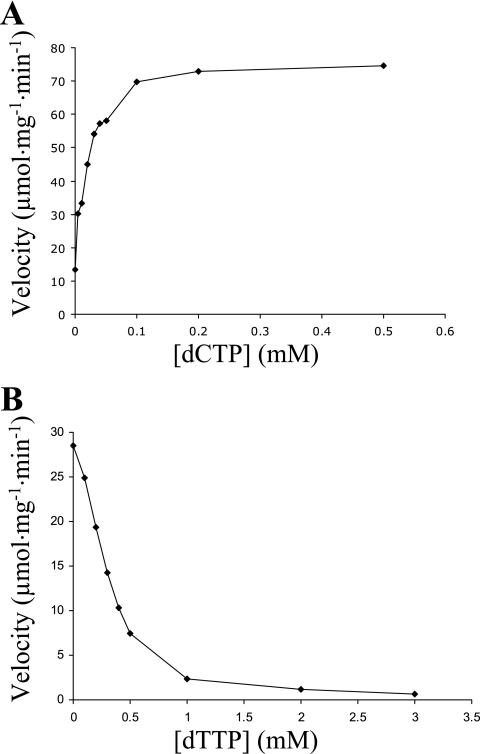
As shown in Fig. 3B, lane 2, TLC experiments established that bi-DCD deaminates dCMP to dUMP. Characterization of the enzyme using dCMP as the substrate was conducted at different temperatures, pHs, and cations. The enzyme is active from 25°C to 55°C, with an optimum of 42°C, which is higher than the 25°C optimum temperature for growing the host and the virus. The enzyme is active from pH 6.0 to 11.0, with an optimal pH at 9.5. The enzyme prefers Mg2+ but retains some activity in the presence of Ca+2, Mn2+, and Ni2+. Although the protein contains a putative zinc-binding site, its enzymatic activity in Zn2+ is less than 6% of the activity obtained with Mg2+. Assays for Zn2+ carried out using the metallochromic indicator 4-(2-pyridylazo) resorcinol, as described previously for T4 dCMP deaminase (27), revealed little or no Zn2+ in the protein. Like activities of dCMP deaminases from other organisms (24), the bi-DCD dCMP deaminase activity is activated by dCTP and feedback inhibited by dTTP, which is the end product of dTTP synthesis. The activity increased ∼7-fold in the presence of 0.005 to 0.1 mM dCTP (Fig. 4A) and was reduced ∼12-fold in the presence of 1 mM dTTP (Fig. 4B). Without dCTP, the enzyme exhibited very weak activity at high concentrations of dCMP (results not shown).
FIG. 4.
Effects of dCTP (A) and dTTP (B) on bi-DCD dCMP deaminase activity. The reactions were performed under standard dCMP deaminase assay conditions, with variations in dCTP or dTTP concentrations as indicated.
Characterization of the bi-DCD dCTP deaminase activity.
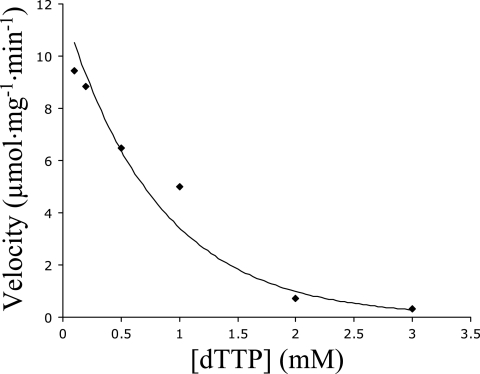
Unexpectedly, TLC revealed that bi-DCD also has dCTP deaminase activity that converts dCTP to dUTP (Fig. 3C, lane 2). Therefore, dCTP may serve as either a substrate or a positive heterotropic effector (or activator) of the enzyme. Like the dCMP deaminase activity, the dCTP deaminase activity was highest at 42°C and enzyme activity occurred from 25°C to 55°C. The enzyme prefers Mg2+ and retains some activity in the presence of Ca+2, Mn2+, or Ni2+; essentially no activity occurred with Zn2+. However, the optimum pH for the dCTP deaminase activity was 7.0, which differs from the pH optimum of 9.5 for dCMP deaminase activity. The dCTP deaminase activity was also feedback inhibited by dTTP, decreasing about 10-fold in the presence of 2 mM dTTP (Fig. 5).
FIG. 5.
Effect of dTTP on bi-DCD dCTP deaminase activity. The reactions were performed under standard dCTP deaminase assay conditions, with variations in dTTP concentrations as indicated.
Kinetic analyses of bi-DCD.
To characterize the kinetic properties of bi-DCD, initial rates for both dCMP and dCTP deaminase activities were measured at various substrate concentrations. dCMP deaminase activity exhibited a small sigmoidal relationship between the rate of dCMP deamination and the concentration of substrate (dCMP), with a Hill number of 1.8 (Table 1). At 0.1 mM dCTP, the Km of the dCMP deaminase activity was 0.76 mM (Table 1), which is slightly higher than the Km of 0.64 mM for dCTP deaminase activity. Like dCMP deaminase activity, dCTP deaminase activity exhibited a sigmoidal relationship between the rate of dCTP deamination and the concentration of substrate (dCTP), with a Hill number of 2.5 (Table 1). These results imply that dCTP alters the catalytic properties of the enzyme by binding to the enzyme at a distinct allosteric site when it serves as a positive heterotropic effector for the dCMP deaminase activity; dCTP also acts as a positive homotropic effector for the dCTP deaminase activity that increases the activity of the enzyme by binding either to the substrate binding site or to the allosteric effector site. As a substrate, dCTP has a higher binding affinity to the enzyme than dCMP. However, the enzymatic efficiency of the dCMP deaminase is about four times higher than that of the dCTP deaminase (see kcat/Km in Table 1).
TABLE 1.
Kinetic constants of bi-DCD for two substrates
| Substrate | Km (mM) | kcat (s−1) | Vmax (μmol min−1 mg−1) | kcat/Km (103 M−1 s−1) | Hill coefficient |
|---|---|---|---|---|---|
| dCMPa | 0.76 | 24.9 | 87.9 | 32.8 | 1.8 |
| dCTP | 0.64 | 5.7 | 20.2 | 8.9 | 2.5 |
With 0.1 mM dCTP.
Inhibitor studies.
The effects of two well-known inhibitors of dCMP deaminases, PDRP and H4dUMP, on both bi-DCD activities were tested. Both compounds inhibited dCMP deaminase activity in the absence and presence of dCTP and also inhibited dCTP deaminase activity (Table 2). These results suggest that the same active site is involved in both dCMP and dCTP deaminations.
TABLE 2.
Effects of inhibitors on bi-DCD dCMP and dCTP deaminase activities
| Substrate | Amt of inhibitor (μm) | % Inhibition
|
|
|---|---|---|---|
| PDRP | H4dUMP | ||
| dCMPa | 0 | ||
| 2 | 74 | 59 | |
| 5 | 84 | 77 | |
| 10 | 97 | 85 | |
| dCMPb | 0 | ||
| 2 | 36 | 7 | |
| 5 | 64 | 38 | |
| 10 | 77 | 60 | |
| dCTP | 0 | ||
| 2 | 56 | 49 | |
| 5 | 82 | 85 | |
| 10 | 94 | 97 | |
With 0.1 mM dCTP.
Without dCTP.
Expression of the A596R gene in PBCV-1-infected chlorella cells.
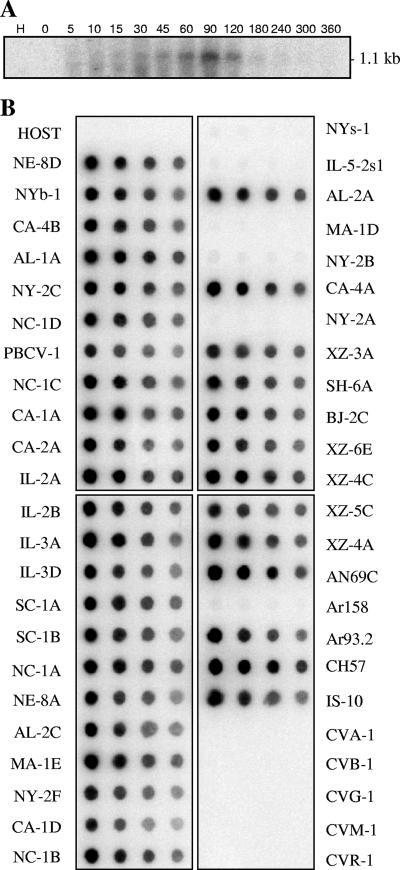
RNA was extracted from virus-infected cells at various times after infection and hybridized to an A596R gene probe to determine when the gene is transcribed during PBCV-1 replication. The probe bound to an ∼1.1-kb RNA transcript extracted from cells beginning at ∼45 min p.i. The intensity of the hybridization increased until 90 min p.i., after which it decreased rapidly (Fig. 6A). PBCV-1 DNA synthesis begins 60 to 90 min after virus infection (37), indicating that the bi-DCD gene is expressed as an early-late gene. The 1.1-kb RNA transcript was slightly larger than expected for a PBCV-1-encoded 142-amino-acid protein.
FIG. 6.
Transcription of PBCV-1-encoded bi-DCD gene and distribution of the bi-DCD genes among chlorella viruses. (A) Total RNAs were isolated from uninfected (H) and PBCV-infected Chlorella NC64A cells at the indicated times (in minutes) and hybridized with a 32P-labeled A596R gene probe. (B) Hybridization of the PBCV-1 bi-DCD gene to DNAs isolated from the host Chlorella NC64A, 42 viruses that infect Chlorella NC64A, and five viruses (CVA-1, CVB-1, CVG-1, CVM-1, and CVR-1) that infect Chlorella Pbi. The DNAs were hybridized with a 32P-labeled A596R gene probe. The blots contain 1, 0.5, 0.25, and 0.12 μg DNA from left to right, respectively.
Occurrence of the bi-DCD gene in other chlorella viruses.
To determine if the bi-DCD gene is common among the chlorella viruses, genomic DNAs from 47 chlorella viruses isolated from diverse geographical regions, as well as the host chlorella, were hybridized to the PBCV-1 A596R gene probe used in the Northern analyses (Fig. 6B). DNAs from 36 of the viruses that infect Chlorella NC64A (NC64A viruses) hybridized strongly with the probe, while no hybridization was observed with the other six NC64A viruses, NYs-1, IL-5-2s1, MA-1D, NY-2B, NY-2A, and Ar158. Typically, the nucleotide sequences of gene homologs in these six viruses differ sufficiently from that of PBCV-1, such that hybridization signals are barely detectable (e.g., see the RNase III gene [47]). No hybridization was detected with DNA from the Chlorella NC64A host or DNAs from five viruses that infect Chlorella Pbi (Pbi viruses). This lack of hybridization is not surprising since the nucleotide identity of homologous genes between the NC64A and Pbi viruses is typically 60 to 65% (37). The genomes of viruses NY-2A (an NC64A virus) and MT325 (a Pbi virus) have been sequenced recently (6, 7), and computer analysis indicates that both viruses encode a dCMP deaminase ORF, with 81% and 63% amino acid identity, respectively, to PBCV-1 bi-DCD. Furthermore, recombinant proteins from the two viruses have both dCMP and dCTP deaminase activities (results not shown).
Phylogenetic analysis.
Maximum-parsimony (heuristic), neighbor-joining (distance method), and maximum-parsimony using bootstrap (1,000 replicates) analyses produced nearly identical tree topologies. The tree shown in Fig. 7 was constructed using the neighbor-joining algorithm in PAUP (36). The values on the branches are the percentages of bootstrap support (1,000 replicates). In order to root the tree, the sequence of a Streptococcus thermophilus late competence protein required for DNA binding (contains a dCMP deaminase domain) (NCBI YP_142074) was used as the outgroup in the phylogenetic analyses. The taxon closest to PBCV-1 was from another virus, bacteriophage phiJL001, whose host is a marine proteobacterium that associates with a sponge (19). The next two most closely related taxa in the same clade are the proteobacteria Vibrio parahaemolyticus and Magnetospirillum magnetotacticum. The long length of the horizontal lines in the tree indicates extensive evolutionary divergence. The dCMP deaminases from the six eukaryotes in the analysis are members of a different clade (Fig. 7).
FIG. 7.
Rooted phylogram of dCMP deaminase. A protein-protein BLAST search was conducted using the dCMP deaminase gene of PBCV-1 (NP_048952). The sequence for Streptococcus thermophilus (YP_142074), a late competence protein required for DNA binding, was used as the outgroup in the phylogenetic analyses (contains a deoxycytidylate deaminase domain). ClustalW was used to align the sequences. Neighbor joining (PAUP) was used to construct the tree. Similar tree topologies were produced by maximum-parsimony and bootstrap analyses. The values on the branches are the percentages of bootstrap support (1,000 replicates). Color coding is as follows: Bacteroidetes, blue; Firmicutes, dark teal; Fusobacterium sp., violet; proteobacteria, teal; archaea, rust; metazoa, red; plants, green; double-stranded DNA viruses, black. Accession numbers (from the NCBI database unless otherwise indicated) are as follows: Arabidopsis thaliana, NP_190423; Archaeoglobus fulgidus, NP_070592; Bacteroides thetaiotaomicron, NP_813171; bacteriophage 44RR2.8, NP_932553; bacteriophage KVP40, NP_899367; bacteriophage phi, YP_223954; bacteriophage T4, Protein Data Bank no. 1VQ2A; Caenorhabditis elegans, NP_498980; Clostridium perfringens, NP_561982; Drosophila melanogaster, NP_649197; Fusobacterium nucleatum, ZP_00143514; Homo sapiens, NP_001912; Magnetospirillum magnetotacticum, ZP_00052863; Mesoplasma florum, YP_053628; Mycoplasma pulmonis, NP_326109; Oryza sativa, BAD87146; PBCV-1, NP_048952; Pongo pygmaeus, CAH90638; Pyrococcus abyssi, NP_126264; Streptococcus thermophilus, YP_142074; Thermococcus kodakarensis, BAD86446; Vibrio parahaemolyticus, NP_800702.
The ClustalW amino acid sequence alignment of the 22 taxa (see the legend for Fig. 7 for a listing of taxa used in the alignment) showed high divergence among taxa, with only 11 of 142 amino acids being identical. The taxon closest to PBCV-1 was bacteriophage phiJL001 (NCBI YP_223954), with 63 identical and 27 similar amino acids out of 142. Together, the four taxa in the PBCV-1 clade differed considerably, with only 33 identical and 24 conserved amino acids. Except for another phycodnavirus infecting the alga Emiliani huxleyi (44), the gene is absent in other viruses infecting eukaryotes.
DISCUSSION
In deoxynucleotide metabolism, dUMP is the immediate precursor in the endogenous de novo synthesis of dTTP (Fig. 1). There are three known pathways to synthesize dUMP: (i) deamination of dCMP to dUMP by dCMP deaminase, (ii) deamination of dCTP to dUTP by dCTP deaminase followed by hydrolysis of dUTP to dUMP by dUTPase, and (iii) reduction of UDP or UTP to dUDP or dUTP, respectively, by ribonucleotide reductase followed by hydrolysis of dUTP to dUMP by dUTPase. The dCMP deaminase route is the most common pathway for synthesizing dUMP in eukaryotes and prokaryotes (21, 31). However, dCMP deaminase is not ubiquitous. In the gram-negative bacteria E. coli and Salmonella enterica serovar Typhimurium, dUMP is synthesized from dCTP that is produced sequentially by dCTP deaminase and dUTPase (12, 28, 29).
Rapidly multiplying DNA viruses place a huge demand on their host to supply deoxynucleotides for genome replication; the problem is even more acute in nonproliferating cells. To overcome this problem, many large DNA viruses encode some nucleotide synthetic enzymes, including both subunits of ribonucleotide reductase. Chlorella virus PBCV-1 follows this pattern and encodes at least 13 putative enzymes involved in nucleotide metabolism (46). Recombinant proteins have been made from 5 of these 13 genes (and shown to be functional): aspartate transcarbamylase (18), glutaredoxin (Y. Zhang et al., unpublished results), dUTPase (48), thymidylate synthase X (11), and, as described here, dCMP deaminase. Unexpectedly, however, the PBCV-1 dCMP deaminase is bifunctional and has both dCMP and dCTP deaminase activities. Thus, assuming that the PBCV-1-encoded ribonucleotide reductase is active, the virus encodes enzymes in all three known pathways to synthesize dUMP, indicating that dTTP synthesis is well supported during PBCV-1 DNA replication (Fig. 1). Presumably the dTTP pool size in host cells (the chlorella host DNA is ∼33% A+T) is not large enough to support rapid growth of PBCV-1, whose DNA is 60% A+T. In addition, the DNA concentration in PBCV-1-infected cells increases at least fourfold by 4 h p.i.; this amount cannot be accounted for simply by recycling deoxynucleotides from degraded host DNA (37). The bi-DCD gene is transcribed shortly after infection (30 min p.i.), and the transcripts are present during the early phase of DNA synthesis (60 to 90 min p.i.) (Fig. 6A). During this time, the virus is preparing a cellular environment for optimal viral DNA synthesis, and as an allosteric regulatory enzyme, bi-DCD probably plays a central role in adjusting the nucleotide pools. The fact that all four PBCV-1-encoded enzymes that contribute to dTTP synthesis are present in five more sequenced chlorella viruses (6-8) supports the importance of dTTP synthesis to chlorovirus replication.
The discovery that the PBCV-1 dCMP deaminase also has dCTP deaminase activity was a major surprise because, although their substrates are similar, dCMP deaminase and dCTP deaminase belong to separate enzyme classes that have different mechanisms of deamination, i.e., the former requires Zn2+ and the latter does not. dCMP deaminase is most closely related to cytidine deaminase, which catalyzes the same reaction but uses unphosphorylated cytidine as a substrate. Both dCMP and cytidine deaminases have similar molecular topologies and have two similar domains; the second domain contains the residues that form the catalytic site, suggesting similarities in the catalytic mechanisms of the enzymes (1, 33). In contrast, dCTP deaminase and dUTPase share conserved motifs, one of which is the uridine-binding site that is also common in pseudouridine synthases, suggesting that these functionally connected enzymes have evolved from a common ancestor (17).
An amino acid sequence analysis of bi-DCD indicates that it contains motifs characteristic of dCMP deaminases from other organisms (Fig. 2). In contrast, bi-DCD has no amino acid similarity to E. coli dCTP deaminase (15). A comparison of the PBCV-1 enzyme with dCMP deaminases from human and T4 phage, which have been studied extensively and represent two distinct classes of the enzyme, indicates that PBCV-1 bi-DCD has more similarity to human dCMP deaminase than to the T4 phage enzyme. Human dCMP deaminase, which has no dCTP deaminase activity (F. Maley, unpublished results), contains a single zinc ion per monomer that is required for catalytic activity. In contrast, the T4 enzyme contains two zinc ions per monomer and both are required for activity, but only one appears to be involved in the catalytic reaction mechanism (1). PBCV-1 bi-DCD has one putative zinc-binding site that could be formed by amino acid residues Cys95, Cys98, and His67 (Fig. 2); however, the activity of bi-DCD obtained with Zn2+ is less than 6% of the activity obtained with Mg2+, suggesting the enzyme does not require Zn2+ for activity. Furthermore, analysis of bi-DCD for Zn2+ indicated that Zn2+ is not bound to the protein.
Like activities of dCMP deaminases from other organisms, the bi-DCD dCMP deaminase activity is activated by dCTP and inhibited by dTTP. Thr75 and Arg78 correspond to the dCTP and dTTP binding sites in T4 phage dCMP deaminase (Phe112 and Arg115) (16). PBCV-1 bi-DCD, like human dCMP deaminase, retains some activity in the absence of dCTP, while T4 dCMP deaminase is completely dependent on dCTP for activity. As a bifunctional enzyme, PBCV-1 bi-DCD has similar reaction requirements for the two substrates, except for pH; dCMP deaminase activity is highest at pH 9.5, and dCTP deaminase activity is highest at pH 7.0. Two well-known dCMP deaminase inhibitors, PDRP and H4dUMP, inhibited both bi-DCD activities, suggesting that the same active site is involved in dCMP and dCTP deamination. Taken together, the results suggest that bi-DCD uses the same mechanism to deaminate both dCMP and dCTP. bi-DCD may have a more flexible active site than other dCMP deaminases; this flexibility could allow the site to interact with either dCMP or dCTP. This hypothesis is supported by the finding that bi-DCD also converts dCDP to dUDP (results not shown). The conformation of the enzyme may change slightly at different pHs and allow the active sites to accommodate the three substrates.
As an activator, dCTP increased dCMP binding to bi-DCD and increased dCMP deaminase activity, suggesting that dCTP alters the conformation of the enzyme to make the active site of the enzyme more suitable for dCMP and more efficient for the deamination reaction. This result suggests that activator dCTP may bind to a putative regulatory site that causes a conformation change in the enzyme. However, when dCTP serves as a substrate at high concentrations, dCTP is deaminated at the active site of the enzyme. Therefore, the enzyme presumably has two dCTP binding sites, one involved in activation and the other serving as a substrate in the catalytic site.
Accumulating evidence suggests that the chlorella viruses have a long evolutionary history (4), possibly dating back to the time that prokaryotic and eukaryotic organisms separated, ca. 2.0 to 2.7 billion years ago (3, 5, 9, 13). Phylogenetic analysis of DNA polymerases places the phycodnavirus enzymes near the root of all eukaryotic δ DNA polymerases (42, 43). A couple of observations in the current manuscript are consistent with the proposed ancient history of the chlorella viruses. bi-DCD is the second known bifunctional enzyme encoded by PBCV-1. Previously, it was reported that the PBCV-1-encoded ornithine decarboxylase decarboxylates arginine more efficiently than ornithine (35). Maybe progenitor enzymes, like the two PBCV-1 bifunctional enzymes, are more precocious than the highly evolved enzymes in present-day organisms, where two separate enzymes carry out the function of one PBCV-1 enzyme. (Note: the bifunctional activity does not result from a fusion between two separate catalytic domains.) The versatility of bi-DCD is also reflected in its ability to function in a broad range of environmental conditions, e.g., temperatures (25 to 55°C) and pHs (6 to 11). Finally, the phylogenetic analyses indicate that PBCV-1 bi-DCD is more closely related to a bacteriophage and two proteobacteria than it is to enzymes from eukaryotes (Fig. 7). One thing in common among these four organisms is their aquatic origins. PBCV-1's host, Chlorella, is common in freshwater, as is Magnetospirillum magnetotacticum. Vibrio parahaemolyticus is found in brackish water, and the host for bacteriophage phiJL001 is a marine bacterium.
In summary, (i) chlorella virus PBCV-1 encodes a bifunctional enzyme (bi-DCD) that deaminates dCMP and dCTP to dUMP and dUTP, respectively; (ii) dCTP can serve as either a substrate or an activator of bi-DCD; (iii) dTTP inhibits both bi-DCD activities; (iv) although the enzyme has a putative Zn2+ binding site, no detectable Zn2+ is associated with the protein (the best enzyme activity occurs with Mg2+); (v) the native structure of bi-DCD is a hexamer; (vi) transcription of the bi-DCD gene begins 45 min after PBCV-1 infection, and the mRNAs disappear quickly after 120 min; (vii) the bi-DCD gene is widely distributed in the chlorella viruses; and (viii) to date, the gene has been found only in viruses infecting algae.
Acknowledgments
We thank Leslie Eisle of the Wadsworth Center biochemistry core facility for performing the sedimentation analysis, Sebastien Graziani of the Ecole Polytechnique, Palaiseau, France, for analyzing the kinetic data, and Tony Fehr for helping with some of the cloning.
This investigation was supported in part by Public Health Service grant GM32441 (to J.L.V.E.), NIH grant P20RR15635 from the COBRE program of the National Center for Research Resources (to J.L.V.E.), Layman award 26-6235-0104-001 from the University of Nebraska—Lincoln (to Y.Z.), and NIH INBRE grant P20RR016469 (to G.D.).
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.Almog, R., F. Maley, G. F. Maley, R. Maccoll, and P. Van Roey. 2004. Three-dimensional structure of the R115E mutant of T4-bacteriophage 2′-deoxycytidylate deaminase. Biochemistry 43:13715-13723. [DOI] [PubMed] [Google Scholar]
- 2.Barchi, J. J., Jr., D. A. Cooney, Z. Hao, Z. H. Weinberg, C. Taft, V. E. Marquez, and H. Ford, Jr. 1995. Improved synthesis of zebularine [1-(beta-D-ribofuranosyl)-dihydropyrimidin-2-one] nucleotides as inhibitors of human deoxycytidylate deaminase. J. Enzyme Inhib. 9:147-162. [DOI] [PubMed] [Google Scholar]
- 3.Brocks, J. J., G. A. Logan, R. Buick, and R. E. Summons. 1999. Archean molecular fossils and the early rise of eukaryotes. Science 285:1033-1036. [DOI] [PubMed] [Google Scholar]
- 4.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 5.Feng, D. F., G. Cho, and R. F. Doolittle. 1997. Determining divergence times with a protein clock: update and reevaluation. Proc. Natl. Acad. Sci. USA 94:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, J. Hartigan, and J. L. Van Etten. 2007. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology 358:459-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, W. C. Nierman, and J. L. Van Etten. 2007. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 358:472-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, L. A., M. V. Graves, X. Li, J. Hartigan, A. J. P. Pfitzner, E. Hoffart, and J. L. Van Etten. 2007. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocytis turfacea. Virology 362:350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glansdorff, N. 2000. About the last common ancestor, the universal life-tree and lateral gene transfer: a reappraisal. Mol. Microbiol. 38:177-185. [DOI] [PubMed] [Google Scholar]
- 10.Graves, M. V., C. T. Bernadt, R. Cerny, and J. L. Van Etten. 2001. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285:332-345. [DOI] [PubMed] [Google Scholar]
- 11.Graziani, S., Y. Xia, J. R. Gurnon, J. L. Van Etten, D. Leduc, S. Skouloubris, H. Myllykallio, and U. Liebl. 2004. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J. Biol. Chem. 279:54340-54347. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, G. R., R. L. Somerville, and S. Dewolf. 1962. Resolution of phage-initiated and normal host thymidylate synthetases of Escherichia coli. Proc. Natl. Acad. Sci. USA 48:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, T. M., and B. Runnegar. 1992. Megascopic eukaryotic algae from the 2.1-billion-year-old negaunee iron-formation, Michigan. Science 257:232-235. [DOI] [PubMed] [Google Scholar]
- 14.Heinemann, V., and W. Plunkett. 1989. Modulation of deoxynucleotide metabolism by the deoxycytidylate deaminase inhibitor 3,4,5,6-tetrahydrodeoxyuridine. Biochem. Pharmacol. 38:4115-4121. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, E., M. Fano, J. H. Bynck, J. Neuhard, S. Larsen, B. W. Sigurskjold, U. Christensen, and M. Willemoes. 2005. Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J. Biol. Chem. 280:3051-3059. [DOI] [PubMed] [Google Scholar]
- 16.Keefe, R. G., G. F. Maley, R. L. Saxl, and F. Maley. 2000. A T4-phage deoxycytidylate deaminase mutant that no longer requires deoxycytidine 5′-triphosphate for activation. J. Biol. Chem. 275:12598-12602. [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 24:2411-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landstein, D., M. Mincberg, S. Arad, and J. Tal. 1996. An early gene of the chlorella virus PBCV-1 encodes a functional aspartate transcarbamylase. Virology 221:151-158. [DOI] [PubMed] [Google Scholar]
- 19.Lohr, J. E., F. Chen, and R. T. Hill. 2005. Genomic analysis of bacteriophage ΦJL001: insights into its interaction with a sponge-associated alpha-proteobacterium. Appl. Environ. Microbiol. 71:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maley, F. 1967. Doxycytidylate deaminase, p. 170-182. In L. Grossman and K. Moldave (ed.), Methods in enzymology, vol. 12. Academic Press, New York, NY. [Google Scholar]
- 21.Maley, F., and G. F. Maley. 1990. A tale of two enzymes, deoxycytidylate deaminase and thymidylate synthase. Prog. Nucleic Acid Res. Mol. Biol. 39:49-80. [DOI] [PubMed] [Google Scholar]
- 22.Maley, G. F., A. P. Lobo, and F. Maley. 1993. Properties of an affinity-column-purified human deoxycytidylate deaminase. Biochim. Biophys. Acta 1162:161-170. [DOI] [PubMed] [Google Scholar]
- 23.Maley, G. F., and F. Maley. 1959. Nucleotide interconversions in embryonic and neoplastic tissues. I. The conversion of deoxycytidylic acid to deoxyuridylic acid and thymidylic acid. J. Biol. Chem. 234:2975-2980. [PubMed] [Google Scholar]
- 24.Maley, G. F., and F. Maley. 1962. Nucleotide interconversions. IX. The regulatory influence of deoxycytidine 5′-triphosphate and deoxythymidine 5′-triphosphate on deoxycytidylate deaminase. J. Biol. Chem. 237:PC3311-PC3312. [PubMed] [Google Scholar]
- 25.Meints, R. H., K. Lee, D. E. Burbank, and J. L. Van Etten. 1984. Infection of a chlorella-like alga with the virus, PBCV-1: ultrastructural studies. Virology 138:341-346. [DOI] [PubMed] [Google Scholar]
- 26.Moore, J. T., J. M. Ciesla, L. M. Changchien, G. F. Maley, and F. Maley. 1994. Identification of a site necessary for allosteric regulation in T4-phage deoxycytidylate deaminase. Biochemistry 33:2104-2112. [DOI] [PubMed] [Google Scholar]
- 27.Moore, J. T., R. E. Silversmith, G. F. Maley, and F. Maley. 1993. T4-phage deoxycytidylate deaminase is a metalloprotein containing two zinc atoms per subunit. J. Biol. Chem. 268:2288-2291. [PubMed] [Google Scholar]
- 28.Neuhard, J., and E. Thomassen. 1971. Deoxycytidine triphosphate deaminase: identification and function in Salmonella typhimurium. J. Bacteriol. 105:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Donovan, G. A., G. Edlin, J. A. Fuchs, J. Neuhard, and E. Thomassen. 1971. Deoxycytidine triphosphate deaminase: characterization of an Escherichia coli mutant deficient in the enzyme. J. Bacteriol. 105:666-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onimatsu, H., K. Suganuma, S. Uenoyama, and T. Yamada. 2006. C-terminal repetitive motifs in Vp130 present at the unique vertex of the Chlorovirus capsid are essential for binding to the host chlorella cell wall. Virology 353:433-442. [DOI] [PubMed] [Google Scholar]
- 31.Prangishvili, D., H. P. Klenk, G. Jakobs, A. Schmiechen, C. Hanselmann, I. Holz, and W. Zillig. 1998. Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV. J. Biol. Chem. 273:6024-6029. [DOI] [PubMed] [Google Scholar]
- 32.Reisser, W., D. E. Burbank, S. M. Meints, R. H. Meints, B. Becker, and J. L. Van Etten. 1988. A comparison of viruses infecting two different chlorella-like green algae. Virology 167:143-149. [DOI] [PubMed] [Google Scholar]
- 33.Reizer, J., S. Buskirk, A. Bairoch, A. Reizer, and M. H. Saier, Jr. 1994. A novel zinc-binding motif found in two ubiquitous deaminase families. Protein Sci. 3:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Shah, R., C. S. Coleman, K. Mir, J. Baldwin, J. L. Van Etten, N. V. Grishin, A. E. Pegg, B. A. Stanley, and M. A. Phillips. 2004. Paramecium bursaria Chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases. J. Biol. Chem. 279:35760-35767. [DOI] [PubMed] [Google Scholar]
- 36.Swofford, D. 2000. Phylogenetic analysis with parsimony (and other methods). Sinauer Associates, Sunderland, MA.
- 37.Van Etten, J. L. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37:153-195. [DOI] [PubMed] [Google Scholar]
- 38.Van Etten, J. L., D. E. Burbank, J. Joshi, and R. H. Meints. 1984. DNA synthesis in a chlorella-like alga following infection with the virus PBCV-1. Virology 134:443-449. [DOI] [PubMed] [Google Scholar]
- 39.Van Etten, J. L., D. E. Burbank, Y. Xia, and R. H. Meints. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126:117-125. [DOI] [PubMed] [Google Scholar]
- 40.Van Etten, J. L., R. H. Meints, D. E. Burbank, D. Kuczmarski, D. A. Cuppels, and L. C. Lane. 1981. Isolation and characterization of a virus from the intracellular green algae symbiotic with Hydra viridis. Virology 113:704-711. [DOI] [PubMed] [Google Scholar]
- 41.Van Etten, J. L., A. M. Schuster, L. Girton, D. E. Burbank, D. Swinton, and S. Hattman. 1985. DNA methylation of viruses infecting a eukaryotic chlorella-like green alga. Nucleic Acids Res. 13:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villarreal, L. P. 1999. DNA viruses: their influence on host evolution. In E. Domingo, R. Webster, J. J. Holland, and T. Pickett (ed.), Origin and evolution of viruses. Academic Press, New York, NY.
- 43.Villarreal, L. P., and V. R. DeFilippis. 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 74:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, W. H., D. C. Schroeder, M. J. Allen, M. T. Holden, J. Parkhill, B. G. Barrell, C. Churcher, N. Hamlin, K. Mungall, H. Norbertczak, M. A. Quail, C. Price, E. Rabbinowitsch, D. Walker, M. Craigon, D. Roy, and P. Ghazal. 2005. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309:1090-1092. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, W. H., J. L. Van Etten, D. S. Schroeder, K. Nagasaki, C. Brussaard, N. Delaroque, G. Bratbak, and C. Suttle. 2005. Phycodnaviridae, p. 163-175. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the international committee on taxonomy of viruses, vol. 8. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 46.Yamada, T., H. Onimatsu, and J. L. Van Etten. 2006. Chlorella viruses, p. 293-366. In K. Maramorosch and A. J. Shatkin (ed.), Advances in virus research, vol. 66. Elsevier Academic Press, San Diego, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., I. Calin-Jageman, J. R. Gurnon, T. J. Choi, B. Adams, A. W. Nicholson, and J. L. Van Etten. 2003. Characterization of a chlorella virus PBCV-1 encoded ribonuclease III. Virology 317:73-83. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., H. Moriyama, K. Homma, and J. L. Van Etten. 2005. Chlorella virus-encoded deoxyuridine triphosphatases exhibit different temperature optima. J. Virol. 79:9945-9953. [DOI] [PMC free article] [PubMed] [Google Scholar]