Abstract
Histone H3 lysine 9 (H3K9) methylation is a crucial epigenetic mark of heterochromatin formation and transcriptional silencing. G9a is a major mammalian H3K9 methyltransferase at euchromatin and is essential for mouse embryogenesis. Here we describe the roles of G9a in germ cell development. Mutant mice in which G9a is specifically inactivated in the germ-lineage displayed sterility due to a drastic loss of mature gametes. G9a-deficient germ cells exhibited perturbation of synchronous synapsis in meiotic prophase. Importantly, mono- and di-methylation of H3K9 (H3K9me1 and 2) in G9a-deficient germ cells were significantly reduced and G9a-regulated genes were overexpressed during meiosis, suggesting that G9a-mediated epigenetic gene silencing is crucial for proper meiotic prophase progression. Finally, we show that H3K9me1 and 2 are dynamically and sex-differentially regulated during the meiotic prophase. This genetic and biochemical evidence strongly suggests that a specific set of H3K9 methyltransferase(s) and demethylase(s) coordinately regulate gametogenesis.
Keywords: G9a, histone, meiotic prophase, methylation
Introduction
In all eukaryotes, genetic information is stored as chromatin, which consists of genomic DNA, histones, and a wide array of chromosomal proteins. The most fundamental unit of chromatin is the nucleosome, which is comprised of an octamer of core histones, H2A, H2B, H3, and H4 wrapped around 147 bp of DNA (Luger et al, 1997). The N-terminal tails of core histones are subject to various chemical modifications such as phosphorylation, acetylation, methylation, ubiquitination, and ADP ribosylation. The ‘histone code' hypothesis (Strahl and Allis, 2000; Turner, 2000; Jenuwein and Allis, 2001) predicts that different modifications at specific amino-acid residues in histones or combinations of these modifications are translated into functionally distinct nuclear processes. For example, lysine methylation on histone H3 or H4 is involved in a variety of distinct biological processes including transcriptional regulation, DNA recombination and repair, heterochromatin formation, and X-chromosome inactivation (Lachner and Jenuwein, 2002; Lachner et al, 2003; Cao and Zhang, 2004).
The mechanisms by which epigenetic chromatin modification regulate development, differentiation, and many cellular functions remain unclear. In this regard, germ cell development represents an excellent system for addressing these issues. Germ-lineage cells are the only population from which parental genetic and epigenetic information can be transferred to progeny. It was shown that epigenetic cellular memories were dynamically reorganized during germ cell development (Reik et al, 2001; Seki et al, 2005; Payne and Braun, 2006). In addition, germ cells must pass through meiosis to develop into mature haploid gametes, which requires the transcriptional activation of meiosis-specific genes and the concomitant suppression of other genes that inhibit the meiotic process. Chromosomes in the meiotic prophase also must form a specific structure termed the synaptonemal complex.
Genetic analyses have revealed that several chromatin modification enzymes play essential roles in germ cell development and meiotic progression. First, the loss of Dnmt3L, which is a catalytically inactive form of DNA methyltransferase but a key regulator for de novo CpG methylation, causes a meiotic prophase defect specifically in males (Bourc'his and Bestor, 2004; Webster et al, 2005; Hata et al, 2006). Similarly, the catalytically active de novo DNA methyltransferase, Dnmt3a, is essential for male gametogenesis (Kaneda et al, 2004). In the former case at least, meiotic progression defects accompany the reactivation of retrotransposable elements, suggesting that silencing of such widely dispersed DNA elements is required for proper synapsis. Second, meiotic progression defects are also induced in two lines of knockout (KO) mice lacking histone lysine methyltransferases (HKMTases). Male germ cells in mice lacking Suv39h, which catalyzes H3K9 tri-methylation at pericentric heterochromatin, exhibit abnormal synapsis and mis-segregation of chromosomes at the male meiotic prophase (Peters et al, 2001). Meiosis-specific H3K4 tri-methyltransferase, Meisetz, which is involved in transcriptional activation of meiotic genes, is also essential for meiotic progression (Hayashi et al, 2005). Germ cells lacking Meisetz are unable to complete double-stranded break repairs at the pachytene stage in both sexes, possibly due to a defect in the appropriate activation of meiosis essential gene(s).
G9a is a major mammalian H3K9 mono- and di-methyltransferase which mainly localizes at euchromatin (Tachibana et al, 2001, 2002; Peters et al, 2003; Rice et al, 2003) and contributes to transcriptional silencing (Ogawa et al, 2002; Tachibana et al, 2002; Shi et al, 2003; Gyory et al, 2004; Nishio and Walsh, 2004; Roopra et al, 2004). Our previous biochemical studies revealed that G9a forms a multiprotein complex, which consists of at least other two polypeptides. G9a-related H3K9 methyltransferase GLP and the zinc-finger protein Wiz were integrated stoichiometrically into active G9a-complexes and formation of this tripartite complex was crucial for H3K9 methyltransferase function in vivo (Tachibana et al, 2005; Ueda et al, 2006). G9a and GLP were both crucial for embryogenesis since deletion of either G9a or GLP leads to embryonic lethality at mid-gestation. However, the ubiquitous expression profile of G9a/GLP/Wiz genes predicts important biological functions in other organs or tissues beyond embryogenesis (Tachibana et al, 2005).
To overcome the embryonic lethality of germline G9a deletion, we generated conditional mutant mice in which G9a can be inactivated by the Cre/loxP recombination system in a cell type-specific manner. Here, we focus on the role(s) of G9a in germ cell development, and report that G9a function is essential for meiotic prophase progression. We also present evidence for the genome-wide dynamics of H3K9 methylation in the meiotic prophase, and the potential involvement of specific HKMTase(s) and demethylase(s) in this reorganization of the germ cell epigenome.
Results
G9a protein expression is restricted to spermatogonia and early leptotene spermatocytes
To elucidate the functional role of G9a during germ cell development, we first analyzed G9a protein expression in testis using immunoblot analysis. As shown in Figure 1A, both G9a and GLP were abundant from postnatal day (P) 2 to P11, but signals for both gradually decreased with developmental progression. We used antibodies against PLZF as a marker for undifferentiated A-type spermatogonia (Buaas et al, 2004; Costoya et al, 2004) and the mouse VASA homologue (MVH) as a pan-germ cell marker (most strongly expressed in pachytene spermatocytes and round spermatids) (Toyooka et al, 2000). The expression profile of G9a/GLP was similar to that of PLZF. Taken collectively, these data suggest that G9a/GLP protein expression is not constant, but temporally regulated during spermatogenesis. As it was shown that formation of the G9a/GLP complex is crucial for exerting H3K9 methyltransferase activity in vivo, we examined their interactions in testicular cells and confirmed a stoichiometric interaction of G9a/GLP in testicular cells (Figure 1B).
Figure 1.
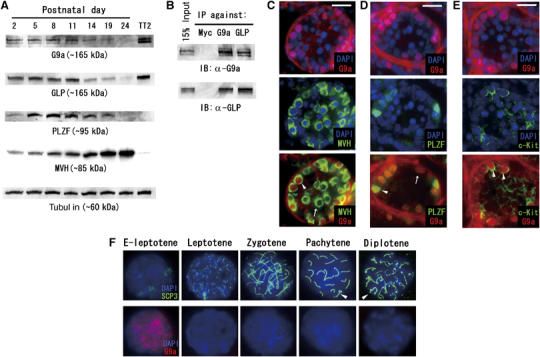
Temporal G9a/GLP expression in germ cell development. (A) Postnatal testicular cells prepared at various stages were subjected to immunoblot analysis. Total amounts of proteins loaded were normalized by determining tubulin content. ES cell lysate (TT2 strain) served as a positive control for G9a/GLP proteins. PLZF and mouse VASA homologue (MVH) were marker proteins for undifferentiated spermatogonia and pan-germ cells, respectively. (B) G9a/GLP complex formation was conserved in testicular cells. (C) Double-immunofluorescent staining profiles with anti-G9a/MVH on P10 testes sections. G9a protein is expressed in a subpopulation of male germ cells (arrowhead). The arrow represents a G9a-negative germ cell. (D) Double-immunofluorescent staining profiles with anti-G9a/PLZF on P10 testes sections. PLZF-positive spermatogonia are G9a positive (arrowhead). A subpopulation of G9a-positive cells is PLZF negative (arrow). (E) Double-immunofluorescent staining profiles with anti-G9a/c-Kit on P10 testes sections. G9a-positive germ cells are c-Kit positive (arrowhead). Scale bars in (C–E), 20 μm. (F) G9a expression profile during male meiosis. G9a protein is detected in only early leptotene (designated as E-leptotene) nuclei. Arrowheads indicate XY bodies.
To further confirm developmental dynamics of G9a expression, we carried out immunohistochemical staining using testes sections at P10, which contained germ cells at several developmental stages ranging from undifferentiated spermatogonia to leptotene spermatocytes (Figure 1C–E). Combinational staining with anti-G9a/MVH showed that G9a protein is expressed in germ cells located near the basal lamina, suggesting its expression in spermatogonia (Figure 1C). Double staining with anti-G9a/PLZF indicated that PLZF-positive spermatogonia were all G9a positive, suggesting that G9a protein is expressed in As to Apr spermatogonia (Figure 1D). Notably, there are G9a-positive germ cells that lack PLZF signal. We performed immunostaining analysis with anti-G9a and anti-c-Kit, which is expressed from A1 spermatogonia until the leptotene spermatocyte stage (Schrans-Stassen et al, 1999; Payne and Braun, 2006). As shown in Figure 1E, some G9a-positive germ cells were c-Kit positive, suggesting that the expression of G9a protein is maintained after differentiation into A1 spermatogonia.
To examine G9a-expression during meiosis, we carried out immunostaining analysis using meiotic chromosome spreads using antibodies against G9a and synaptonemal complex protein 3 (SCP3), which is a lateral component of the synaptonemal complex. As shown in Figure 1F, expression of G9a protein is highly restricted to the early leptotene stage and the expression was hardly detectable from the leptotene through the diplotene stages of spermatocyte development. We further examined G9a expression in round or elongating spermatids from adult testes, but were unable to detect G9a-positive cells (data not shown). The expression profile of GLP was identical to that of G9a (data not shown). To summarize, G9a/GLP protein is expressed from the undifferentiated spermatogona until the early leptotene spermatocyte stage during spermatogenesis. We also examined G9a/GLP expression during the female meiotic prophase and the kinetics were similar to those observed for male germ cells (Supplementary Figure S1).
Germ-lineage specific deletion of G9a produces infertility accompanied with a drastic loss of germ cells in adult gonads
To generate germ-lineage-specific conditional G9a-KO mice, we generated a conditionally defective G9a allele containing target sites for the Cre/loxP recombination system (Supplementary Figure S2). Mice carrying the G9aflox mutation were crossed with tissue-nonspecific alkaline phosphatase (TNAP)-Cre knock-in mice, which express the Cre recombinase in primordial germ cells from E9.5 to late gestation (Lomeli et al, 2000).
The germ-lineage-specific G9a-KO mice (genotyped as TNAP-Cre, G9aflox/Δ) were obtained by crossing G9aflox/flox females with TNAP-Cre, G9a+/flox males. TNAP-Cre, G9aflox/Δ mice were born in a slightly lower Mendelian ratio, that is, we obtained 30 TNAP-Cre, G9aflox/Δ mice out of 200 progeny, while the expected number was 50. As leaky Cre expression in somatic tissues during embryogenesis is observed for this TNAP-Cre line (Kaneda et al, 2004), we speculate that some of the TNAP-Cre, G9aflox/Δ embryos died due to the inactivation of G9a in non-germ cells. However, the delivered germ-lineage G9a-KO mice were indistinguishable from wild-type (WT) mice in appearance, reached normal adult body size, and lived more than 2 years.
To examine fertility in the KO mice, we crossed them with WT animals. There were no apparent defects for the mice genotyped as G9aflox/flox (both alleles active, referred to as WT hereafter), G9aflox/Δ (single allele active), and TNAP-Cre, G9a+/flox mice (single allele active, referred to as heterozygous hereafter) in both sexes, and they were all fertile. In contrast, when the germ-lineage G9a-KO male mice (n=8) were crossed with WT females, no pregnancy features were observed in the partner females. In the case of females, only three out of 22 germ-lineage G9a-KO females were pregnant and delivered pups, whereas the other 19 females did not. The fertility of the three G9a-KO females is listed in Table I. The average number of the pups per a delivery was smaller (3.0) than in WT females (7.1). Moreover, we did not observe any deliveries from these G9a-KO females after 7 months old age, suggesting that germ-lineage-specific G9a-KO females had a paucity of oocytes, even when they were fertile. Importantly, some of the progeny from the G9a-KO females carried the G9aflox allele, indicating that TNAP-Cre-mediated depletion of the floxed G9a locus was not complete during female germ cell development (see legend of Table I).
Table 1.
Fertility of germ cell G9a-KO female mice
| Pup numbers (mother age at deliverya) | |||||||
|---|---|---|---|---|---|---|---|
| #1 | 7 (2) | 3 (3) | 4 (4) | 3 (5) | |||
| #2 | 1 (3) | 3 (4) | 1 (5) | 1 (6) | |||
| #3 | 4 (2) | ||||||
| Wild-type ♀ | 8 (3) | 10 (4) | 7 (5) | 9 (6) | 4 (8) | 6 (9) | 6 (10) |
| Natural mating was started using germ KO female mice at 1 month old age and wild-type fertile males at 4 months old age, and then the mating was kept until the female reached to 1 year old age. Out of 22 females, only three delivered pups described above. | |||||||
| Average numbers per a delivery are 3.0 for KO (27/9) and 7.1 for wild-type (50/7) mice. | |||||||
| Out of 27 offspring from KO females, five carried G9aflox allele. | |||||||
To examine G9a protein expression in the germ-lineage of these animals, we performed immunocytochemical analyses on embryonic or postnatal gonads (Figure 2A and B). Although G9a protein was ablated in the vast majority of germ cells in E12.5 females and males (Figure 2A and not shown) and P7 males (Figure 2B), we observed a subpopulation of germ cells that remained G9a positive. The ratio of G9a-positive versus -negative germ cells is summarized in Table II. The efficiencies of G9a depletion were 80–90% in both sexes at E12.5. In contrast, the depletion efficiencies of P7-spermatogonia reached nearly 100%. The high efficiency of G9a depletion at P7 compared with E12.5 may derive from prolonged exposure of the conditional allele to Cre enzyme.
Figure 2.
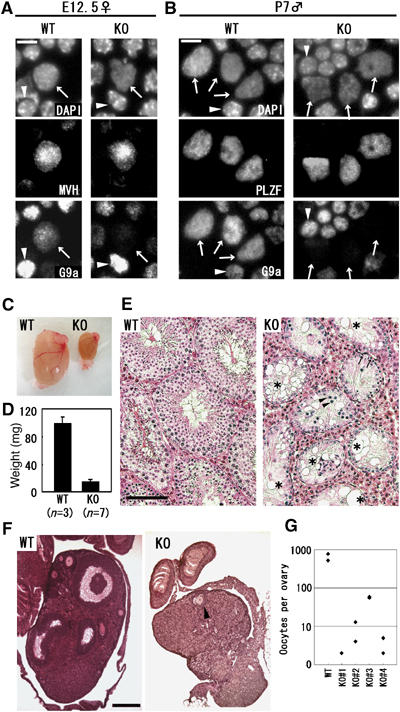
Loss of germ cells in G9a-KO gonads. (A–B) Depletion of G9a protein in G9a-KO gonads. Enzymatically dissociated cells from E12.5 female gonads (A) and P7 testes (B) were squashed onto slide glass and stained with the indicated antibodies. Calculated efficiencies of G9a depletion are shown in Table II. Arrows and arrowheads indicate germ cells and control somatic cells, respectively. Bars, 10 μm. (C) Gross morphology of wild-type (WT) and germ cell G9a-KO (KO) testes at 3-month old age. (D) Testes weight of WT and KO mice at 3 months of age. The average weights were 98 and 15 mg, respectively. Error bars represent standard deviation. (E) Histological examination of WT and KO testes from 3-month-old mice. Paraffin-embedded sections were stained with hematoxylin and eosine. Seminiferous tubules devoid of germ cells were indicated by an asterisk. Leptotene-like spermatocytes and spermatogonia were indicated by arrowheads and arrows, respectively. Scale bars, 100 μm. (F) Histological examination of WT and KO ovaries at 2 months of age. Paraffin-embedded sections were stained with hematoxylin and eosine. The KO ovary was significantly smaller than that of WT mice. Only one oocyte was recognized in a section of KO ovary (arrowhead). Scale bar, 200 μm. (G) Gross numbers of oocytes per ovary at 2 months of age.
Table 2.
Efficiency of G9a depletion by TNAP-Cre at several developmental stages
| Genotype | Stage sex | Cells examined | Ratio of depletion (%) | |
|---|---|---|---|---|
| MVH+/G9a+ | MVH+/G9a− | |||
| Het #1 | E12.5 ♀ | 50 | 0 | — |
| Het #2 | E12.5 ♀ | 50 | 0 | — |
| KO #1 | E12.5 ♀ | 9 | 41 | 82 |
| KO #2 | E12.5 ♀ | 7 | 43 | 86 |
| MVH+/G9a+ | MVH+/G9a− | |||
| Het #3 | E12.5 ♂ | 50 | 0 | — |
| Het #4 | E12.5 ♂ | 50 | 0 | — |
| KO #3 | E12.5 ♂ | 5 | 45 | 90 |
| KO #4 | E12.5 ♂ | 8 | 42 | 84 |
| PLZF+/G9a+ | PLZF+/G9a− | |||
| Het #5 | P7 ♂ | 300 | 0 | — |
| Het #6 | P7 ♂ | 300 | 0 | — |
| KO #5 | P7 ♂ | 2 | 298 | 99.3 |
| KO #6 | P7 ♂ | 7 | 293 | 97.7 |
| Enzymatically dissociated cells from embryonic or postnatal gonads were squashed onto slide glass, and then were stained with antibodies against germ cell marker protein and G9a as described in Figure 2A and B. | ||||
To assess the cause of infertility, we dissected gonads from the G9a-KO adult mice. As shown in Figure 2C and D, adult testes of KO males were markedly smaller than those of WT. Histological examinations showed that the majority (∼70%) of seminiferous tubules in the KO testes lacked any type of germ cells, and the remainder (∼30%) contained only a few spermatogonia and leptotene-like cells (Figure 2E). A typical result from histological analysis of a KO female ovary is shown in Figure 2F. The numbers of maturating and primordial oocytes were greatly reduced (only one oocyte per section) in comparison with those of WT (more than 10 per section). To evaluate these defects quantitatively, we counted the gross numbers of oocytes by scanning serial sections of adult ovaries (Figure 2G). The WT female contained 520 and 777 ooctyes per ovary, whereas those from four independent KO females were one- or two orders fewer in magnitude (2 and 2, 13 and 4, 55 and 59, 2 and 5). Thus, we conclude that infertility in the germ-lineage-specific G9a-KO mice stems from a complete loss or a drastic reduction of sperm or oocytes, respectively.
Developmental defects at the pachytene stage in G9a-KO male germ cells
To determine the development stage of germ cells perturbed by the G9a deficiency, we examined the first round of spermatogenesis using juvenile testes (Bellve et al, 1977) (Figure 3A). In P7 testes, difference between heterozygous and KO tubules was not apparent, suggesting that differentiation into spermatogonia was normal. Importantly, we observed nearly complete depletion of G9a in the PLZF-positive spermatogonia at this stage (Figure 2B and Table II). In contrast, seminiferous tubules in the KO testes were apparently abnormal at P14 and P21, exhibiting a consistent increase in apoptotic cell death by TUNEL analysis (data not shown). A subpopulation of heterozygous spermatocytes at P14 had differentiated into the pachytene stage of meiotic prophase, while apparent pachytene spermatocytes were hardly detectable in KO tubules. In P21 heterozygous testes, subpopulations of germ cells had completed meiosis and differentiated into haploid spermatids; however, KO tubules at P21 completely lacked spermatids. These data strongly suggest that the G9a mutation affects meiotic progression. To determine more precisely when meiosis was blocked in KO testes, we examined several time points for the appearance of apoptotic cells. As shown in Figure 3B, apoptotic nuclei were frequently detected in KO tubules in which spermatocytes developed into the early pachytene stage based on their light microscopic characteristics. These data indicate that meiosis was aborted during the early pachytene stage in G9a-KO male germ cells.
Figure 3.
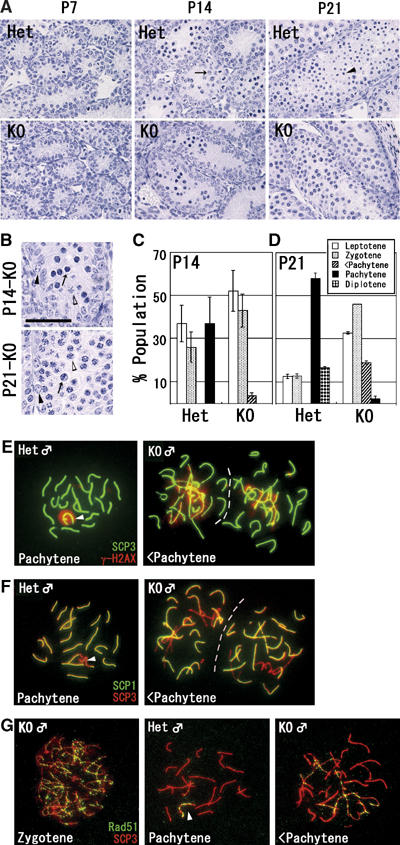
Developmental defects at the pachytene stage in G9a-KO male germ cells. (A) Paraffin-embedded sections (3 μm) of testes at various developmental time points (postnatal day (P) 7, and 14, and 21) were prepared and stained with hematoxylin. Pachytene spermatocytes at P14 and haploid spermatids at P21 in heterozygous testes were indicated by arrow and arrowhead, respectively. Scale bar, 50 μm. (B) Magnification of KO tubules. Apoptotic nuclei were detected both at P14 (top) and P21 (bottom) in KO tubules (arrows). Black and white arrowheads indicate Intermediate spermatogonia and early pachytene spermatocytes. Scale bar, 50 μm. (C–D) Histogram of male meiotic prophase stages at P14 (C) and P21 (D) between heterozygous and KO spread preparations. Developmental stages were classified by staining profiles with anti-SCP3/γH2AX. Bars represent the average of two samples per genotype (>150 cells were analyzed per sample). <Pachytene represents incomplete pachytene spermatocyte. (E–F) Perturbed synapsis formation in KO spermatocyte. P14 spermatocytes were stained with a combination of anti-SCP3/γH2AX (E) and anti-SCP1/SCP3 antibodies (F). Typical pachytene spermatocytes from heterozygous testes and incomplete pachytene spermatocytes (<Pachytene) from KO testes were represented. Arrowheads indicate XY bodies. (G) Immunofluorescence staining profile with anti-Rad51 antibodies in KO spermatocytes. At the pachytene stage, the signals of Rad51 protein were retained only at the XY body in heterozygous nuclei (arrowhead). In contrast, the signals were still retained on asynapsed chromosomes in KO spermatocyte (right).
To further confirm the block in meiotic prophase, the frequency of the meiotic stage from leptotene until diplotene spermatocytes in juvenile testes were calculated (Figure 3C and D). Spermatocytes were classified by their immnunofluorescent staining profiles with antibodies against SCP3 and histone H2AX phosphorylated on serine 139 (γH2AX), which marks sites of double-strand breaks (Mahadevaiah et al, 2001). As shown in Figure 3C, we could not detect full pachytene spermatocytes in P14 KO testes, whereas more than 30% of spermatocytes reached the full pachytene stage in heterozygous testes. Despite the lack of full pachytene spermatocytes, incomplete pachytene spermatocytes were frequently observed in KO testes. Diplotene spermatocytes emerged at P21 in heterozygous testes but were not observed in KO testes (Figure 3D).
Next, we examined details of the meiotic defects in G9a-KO testis (P14) using testicular cell spreads. At the leptotene and zygotene stages, there was no apparent difference in staining profiles between the G9a heterozygous and KO spermatocytes using anti-SCP3/γH2AX (data not shown). However, perturbed synapsis formation was easily detected in G9a-KO spermatocytes at more advanced pachytene stage (Figure 3E). The autosomes in heterozygous pachytene spermatocytes were fully synapsed and characterized as entire thick SCP3 distribution and negative for γH2AX (left panel). In contrast, G9a-KO spermatocytes contained a mixture of fully synapsed, partially synapsed, and asynapsed chromosomes (the asynapsed portion is characterized as thin SCP3 signal with massive γH2AX accumulation) (right panel). We further confirmed these incomplete synaptonemal complex (SC) formation by double staining with antibodies against SCP3/SCP1, which is a central component of the SC localized at synapsed regions of chromosomes. As shown in Figure 3F, G9a-KO spermatocytes again contained both synapsed (SCP3/1 double positive) and asynapsed chromosomes (SCP3 single positive). We rarely identified spermatocytes with such incomplete SC formation in heterozygous mice (summarized in Figure 3C and D).
To examine whether the double-stranded break (DSB) repair pathway progresses normally in KO spermatocytes, we performed double staining analysis with anti-SCP3/Rad51 (Figure 3G). Signals corresponding to Rad51 protein were detected as numerous foci along all meiotic chromosomes in KO spermatocytes at the zygotene stage (left panel), which was indistinguishable from that observed in heterozygotes (not shown). These data suggest that the loading of Rad51 protein onto early meiotic chromosomes occurs normally in the KO spermatocytes. However, Rad51 protein persisted on asynapsed chromosomes in incomplete pachytene spermatocytes from KO testes (right panel). These data as well as γH2AX staining profiles suggest that the processing of DSB repair is incomplete in asynapsed meiotic chromosomes of KO spermatocytes. Collectively, our data indicate that the loss of G9a arrests meiosis in male germ cells at the early pachytene stage and is characterized by disordered progression of SC formation.
G9a deficiency leads to a global loss of H3K9me2 and 1 but not H3K9me3 at male meiotic prophase
To elucidate the contribution of G9a to H3K9 methylation during the meiotic prophase, we examined the levels and kinetics of H3K9 mono-, di-, and tri-methylation in male meiotic nuclei. As shown in Figure 4A and C, G9a-heterozygous spermatocytes from the early leptotene until the zygotene stage displayed disperse nuclear staining profile for H3K9me2 and me1. Unexpectedly, these signals disappeared in pachytene spermatocytes even in the heterozygous testes (right panels in Figure 4A and C). As the meiotic prophase proceeds without an S phase, the rapid loss of H3K9me marks at this stage might be induced by an active mechanism rather than a passive one such as histone dilution accompanied by DNA synthesis. Importantly, H3K9me2/1 signals were hardly detectable in the G9a-KO nucleus at any stages of the meiotic prophase (Figure 4B and D).
Figure 4.
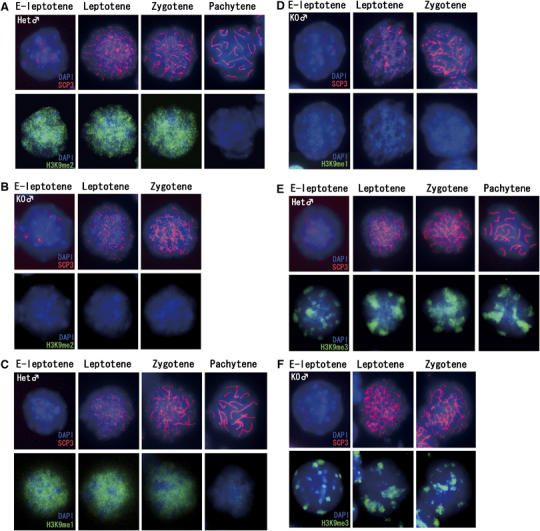
The kinetics of H3K9 methylation during male meiotic prophase. (A–B) Kinetics of H3K9me2 in male meiotic prophase. Meiotic nuclear spreads were prepared from P14 testes of heterozygous (A) and KO (B) male mice, and the status of H3K9me2 was monitored in combination with anti-SCP3 antibody. (C–D) Kinetics of H3K9me1 in male meiotic prophase in nuclei of heterozygous (C) and KO spermatocytes (D). (E–F) Kinetics of H3K9me3 in male meiotic prophase in nuclei of heterozygous (E) and KO spermatocytes (F).
Considering the restricted expression of G9a/GLP proteins in germ cells from spermatogonia to early leptotene spermatocytes (Figure 1), it seems likely that the H3K9me2/1 marks are added before initiation of synapsis and are maintained up to the completion of synapsis. Subsequently, these marks are rapidly and actively removed at the pachytene stage. In contrast, H3K9me3 kinetics was distinct from those of me2/1. Large blocks of H3K9me3 signals were not erased until the pachytene stage (Figure 4E), as shown previously (Peters et al, 2001). The H3K9me3 signals in G9a-KO nuclei were indistinguishable from heterozygous nuclei (Figure 4F), indicating that most H3K9me3 is G9a independent in spermatocytes, similar to what was observed previously in ES cells (Peters et al, 2003; Rice et al, 2003).
Loss of germ cells during meiotic prophase in G9a-KO female gonads
To investigate whether the loss of G9a also affects meiosis in females, we assessed the reduction of germ cells at several developmental stages in G9a-KO embryonic ovaries. Figure 5A shows ovarian sections stained with anti-MVH antibodies at different developmental periods. Primordial oocytes at the day of birth (DOB) were very few in KO ovaries (squares in right panels), suggesting that the lack of maturating oocytes in adults ovaries derives, at least in part, from a profound reduction in germ cells during embryogenesis. The ratio of germ cell numbers in KO versus heterozygous ovaries is summarized in Figure 5B. The reduction in germ cells was modest until E16.5 (∼20%). However, an approximate 90% loss was observed from E17.5 onwards.
Figure 5ae.
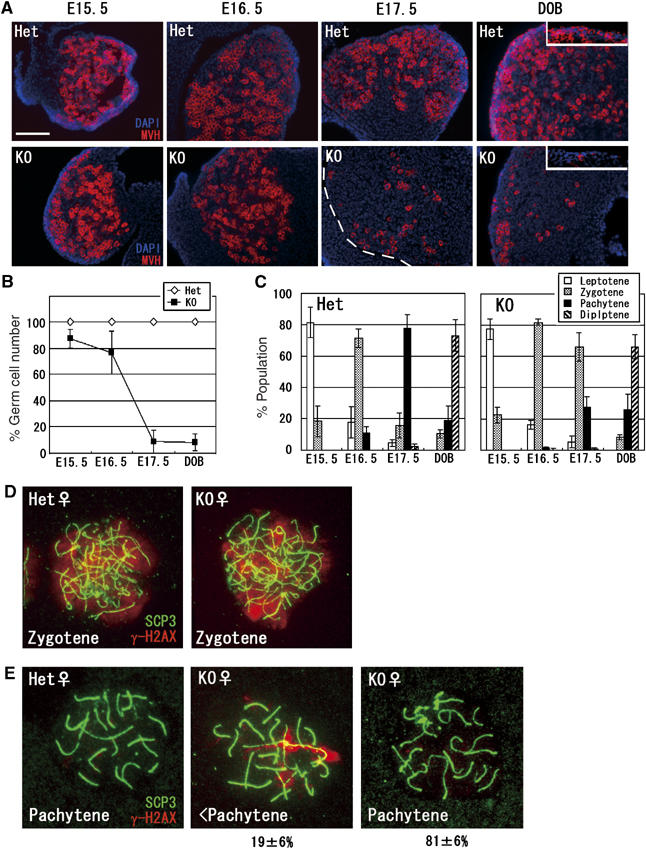
Developmental defects in G9a-KO female germ cells. (A) Germ cell development during meiotic prophase in KO ovaries. Frozen sections of heterozygous and KO ovaries at various developmental time points were prepared and stained with anti-MVH. Primordial oocytes were represented in the box of DOB panels. Scale bar, 100 μm. (B) Kinetics of germ cell reduction in KO ovaries. MVH-positive cells per 0.01 mm2 of ovary section were counted and the numbers of germ cells in heterozygous ovaries are normalized as 100%. Data represent the average of two samples per genotype (>100 cells were counted per ovary). (C) Developmental dynamics of female germ cells. Ovaries at various developmental stages were collected and oocytes spreads were stained with anti-SCP3/γH2AX antibodies for determining meiotic stages. Three independent ovaries were used for calculation at every embryonic stage (>50 cells were analyzed per ovary). (D–E) Perturbed synapsis formation in G9a-KO oocytes. Oocytes at the zygotene (D) and the pachytene stages (E) were prepared from E17.5 ovaries and stained with anti-SCP3/γH2AX antibodies. In the pachytene stage, γH2AX signals disappeared from heterozygous nuclei (E, left panel). In contrast, a subpopulation of KO oocytes exhibited synapsis perturbation similar to males, in which γH2AX signals were still retained along certain axial elements (E, middle panel). The other population of pachytene oocytes from KO ovaries exhibited γH2AX-negative staining profiles (E, right panel). Percentages represent population distributions (>30 cells were analyzed per ovary). (F–G) Kinetics of H3K9me2
To further determine the developmental stage when meiosis was perturbed in G9a-KO embryonic ovaries, we prepared oocyte cell spreads. Meiotic stages were determined by immunocytological staining profiles with antibodies against SCP3/γH2AX and the frequency distribution of meiotic prophase is summarized in Figure 5C. The meiotic prophase in G9a-KO ovaries progressed normally until the zygotene stage around E16.5. Indeed, the status of γH2AX in G9a-KO zygotene oocytes was indistinguishable from that observed in heterozygous animals (Figure 5D). However, at E17.5, ∼80% of the SCP3-positive germ cells reached the pachytene stage in heterozygote females, while a majority of the G9a-KO germ cells remained at the zygotene stage and only ∼30% were pachytene. Considering that the total number of germ cells was drastically reduced at E17.5 (Figure 5A and B), it seems most likely that G9a inactivation also aborts developmental progression around the pachytene stage in female meiosis. In keeping with this notion, a significant population of KO pachytene oocytes exhibited perturbed synaptic progression (∼20%), in which γH2AX signals were retained along certain axial elements, similar to that observed in G9a-KO males (Figure 5E). In contrast, such incomplete pachytene cells were hardly detectable in heterozygous pachytene oocytes (left panel). Taken together, it seems likely that the mechanism of G9a mutation-induced meiotic arrest is at least partly conserved between males and females.
We next checked the H3K9me status in the female meiotic prophase in heterozygous samples. H3K9me2/1 signals until the zygotene stage were similar to those of males. However, these signals were persistent in the pachytene stage onwards, as was H3K9me3 (Figure 5F and G and data not shown). These data indicate that active removal of H3K9me2/1 at the pachytene stage is a feature of male but not female meiosis. Because a subpopulation of premeiotic germ cells in E12.5 KO ovaries still contained G9a protein and some pups derived form G9a-KO oocytes retained the G9aflox allele (Tables I and II), it seems likely that some female germ cells might enter meiosis without losing the G9aflox allele. Indeed, we detected both types of meiotic nuclei, with and without H3K9me2 signals, from individual KO ovary preparations, strongly suggesting incomplete G9a inactivation during female meiosis (Figure 5F–H). Most zygotene oocytes (∼95%) at E16.5 were H3K9me2 negative, but the remainder (∼5%) were H3K9me2 positive (described in the bottom of Figure 5F). The H3K9me2-positive population in G9a-KO ovaries was significantly increased (>30%) from the pachytene stage onwards, presumably due to the increase in death among H3H9me2-negative oocytes around this stage. However, some oocytes lacking H3K9me2 were detectable even at the diplotene stage at DOB (Figure 5H, left), indicating that a subpopulation of G9a-depleted oocytes could develop at least until the diplotene stage. Oocytes in KO ovaries were all H3K9me3 positive and had similar H3K9me3 distribution patterns as seen in the heterozygous controls (Figure 5F and not shown).
Figure 5fj.
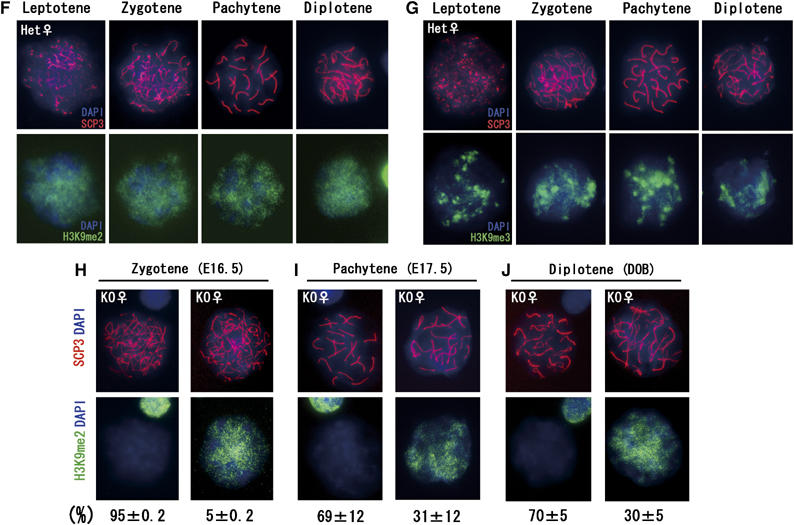
(F) and me3 (G) in female heterozygous nuclei during the meiotic prophase. (H–J) Mosaicism of H3K9me2 staining profiles of female KO germ cells during meiotic prophase. Staining profiles of H3K9me2-positive and -negative oocytes at the zygotene stage at E16.5 (H), the pachytene stage at E17.5 (I), and the diplotene stage at DOB (J) were represented, and their population distributions are described below. The data were obtained from at least two independent KO female mice (>50 cells were analyzed per ovary).
Reactivation of several G9a-regulated genes in G9a-KO germ cells
To address whether the loss of G9a and H3K9 me2/1 has an impact on transcription, we assessed gene expression profiles in the G9a-deficient male germ cells. Microarray analysis using total RNA from whole P10 testes identified several genes up- or downregulated in the G9a-KO context (listed in Supplementary Table S1). Among them, we re-checked the RNA expression levels of 10 genes (six genes upregulated and four genes downregulated in G9a-KO testes) by RT–PCR. We standardized each reverse-transcribed sample by measuring the amounts of SCP3 transcripts, as SCP3 is a meiotic prophase-specific gene. The expression of SCP3 in leptotene spermatocytes was indistinguishable between G9a-KO and heterozygous samples (Figure 4A–C and not shown). The G9a downregulated genes, termed Akr1c13, Tnmd, Defb42, Ptgds, and Chst11, were overexpressed in G9a-KO testes compared with controls (Figure 6A). The G9a upregulated genes were mostly germ cell or meiosis-specific genes. However, meiotic progression was mildly affected in P10 G9a-KO testes (data not shown). Therefore, the smaller number of spermatocytes in the G9a-KO testes might explain a reduction in the relative abundance of these spermatocyte-specific gene transcripts per testis. Consistent with this notion, we could not reproduce the transcriptional repression of these four genes once RT–PCR signals were normalized for SCP3 (see Supplementary Table S1).
Figure 6.
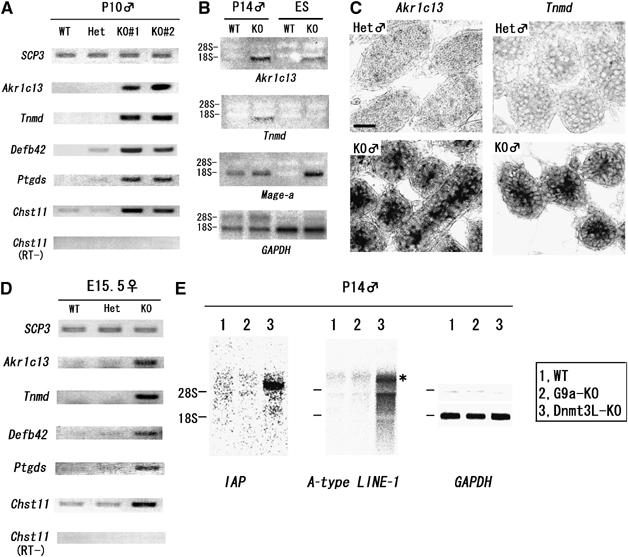
G9a suppresses specific genes during meiotic prophase. (A) De-repression of genes in G9a-KO testes. Transcripts of Akr1c13, Tnmd, Defb42, Ptgds, and Chst11 were amplified by using cDNAs from independently prepared testes at P10 with the indicated genotype. (B) Comparison of transcriptional perturbation between G9a-KO germ cells and ES cells by Northern blot analysis. (C) RNA in situ hybridization analysis of Akr1c13 and Tnmd in P10 testes sections from heterozygous (top) and KO animals (bottom). Bar, 20 μm. (D) Several G9a-regulated genes were conserved during meiotic prophase in both sexes. cDNA was prepared from total RNA from E15.5 ovaries and amplified using specific primers as shown in (A). (E) Retrotransposons were suppressed in G9a-KO testes. Total RNAs from P14 whole testes from the indicated genotypes were blotted with specific probes as indicated. Transcripts of IAP (∼5.4 kb transcripts of IΔ1 type IAP) and LINE-1 (dominant ∼7 kb transcripts, shown as asterisk) were upregulated in Dnmt3L-KO but not in G9a-KO testes.
To address whether some G9a-regulated genes were common between testicular and ES cells, we compared expression by Northern blot analysis using P14 testes and ES cells corresponding to each genotype. As shown in Figure 6B, Akr1c13 was a common G9a-regulated gene but Tnmd served as a gene regulated only in testicular cells. Our previous analyses indicated that G9a deficiency reactivates the Mage-a gene in ES cells (Tachibana et al, 2002, 2005). In contrast to ES cells, Mage-a transcripts were robustly detected even in WT testes (Clotman et al, 2000). These observations suggest that G9a might regulate distinct sets of genes in a cell type-dependent manner. To further confirm whether such de-repression occurs in the G9a-KO germ cell population, we performed RNA in situ hybridization for Akr1c13 and Tnmd RNA. As shown in Figure 6C, Akr1c13 and Tnmd transcripts accumulated in cells located inside of tubules from P10 G9a-KO testes, where spermatocytes differentiate up to the leptotene stage. In contrast, these transcripts were hardly detectable in heterozygous testes. These data suggest that de-repression of the ‘G9a negatively regulated' genes, at least Akr1c13 and Tnmd, occurs in the G9a-KO germ cells at the meiotic prophase.
To evaluate whether G9a-mediated gene regulation during meiosis was similarly controlled in males and females, we examined the expression of certain G9a-regulated genes in E15.5 ovaries. As shown in Figure 6D, all five of the examined genes were upregulated in KO ovaries and testes, suggesting a conservation of G9a-specific gene regulation in both sexes during meiosis.
As reactivation of the genome-wide dispersed retrotransposable elements is linked with a synapsis disorder in Dnmt3L-deficient testes (Bourc'his and Bestor, 2004), we examined the expression of A-type LINE1 and IAP in G9a-KO testes. As shown in Figure 6E, reactivation of these retrotransposable elements was evident in Dnmt3L-KO testes as described (Bourc'his and Bestor, 2004), but not in G9a-KO testes.
Correlation of JHDM2A expression and loss of G9a-mediated H3K9 methylation in pachytene spermatocytes
As shown in Figure 4, we found a novel dynamic of H3K9 methylation during male meiotic prophase in which G9a-mediated H3K9me2/1 is lost in pachytene spermatocytes. Recently, JHDM2A was characterized as an H3K9me2/1 demethylating enzyme (Yamane et al, 2006). Moreover, the rat JHDM2A counterpart was originally identified as a testis-specific gene (Hoog et al, 1991). Thus, we speculated that JHDM2A is a candidate for the H3K9me2/1 demethylase in pachytene spermatocytes. We generated specific antibodies against mouse JHDM2A (Figure 7A) and analyzed its expression in spermatocytes or oocytes (Figure 7B–D). The nuclei of pachytene spermatocytes were clearly JHDM2A positive (Figure 7C, arrowhead), but those of leptotenes/zygotenes were negative. It should be noted that the nuclear distribution of JHDM2A in male pachytene cells and that of H3K9me2 in zygotene cells was quite similar (compare Figure 7B and C). JHDM2A signals could be detected in nuclei until the diplotene stage but were weak or absent in round spermatids (data not shown). On the other hand, female nuclei of pachytene as well as leptotene/zygotene oocytes lacked JHDM2A signals (Figure 7D and not shown). In conclusion, our analyses of JHDM2A expression in early meiotic cells strongly suggests that sex- and stage-specific reduction of H3K9me2/1 in the meiotic prophase is mediated, at least in part, by JHDM2A.
Figure 7.
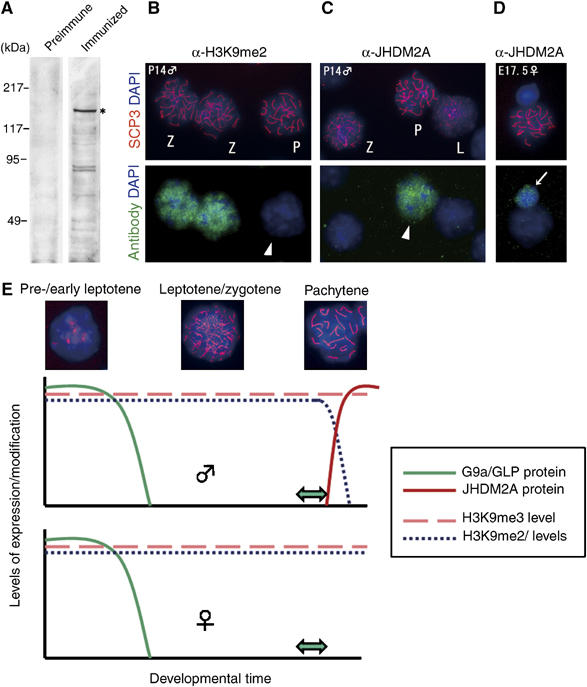
Strong correlation between JHDM2A expression and stage- and sex-dependent removal of H3K9 methylation. (A) Generation of specific antibodies against mouse JHDM2A. Antiserum against mouse JHDM2A (details were described in Supplementary data) was used for immunoblot analysis of whole-cell extracts of TT2-ES cells (left, preimmune serum; right, immunized serum). Expression of the JHDM2A in ES cells was confirmed by RT–PCR analysis beforehand (data not shown). Specific signals around 150 kDa (asterisk) are in accordance with the molecular weight of endogenous JHDM2A protein as described previously (Yamane et al, 2006). (B–D) Male pachytene-specific expression of JHDM2A protein. (C) Immunostaining profiles of IgG fractions from antiserum against JHDM2A on G9a-heterozygous spermatocyte spreads (P14). The same preparations were also stained with anti-H3K9me2 antibodies as a control (B). L, leptotene; Z, zygotene; P, pachytene. (D) Oocytes spreads were prepared from G9a-heterozygous ovaries (E17.5) and then stained with the anti-JHDM2A antibodies. Arrow indicates a somatic cell in which JHDM2A was expressed weakly. (E) The kinetics of H3K9 methylation in meiotic prophase. In spermatogenesis, G9a/GLP proteins were detected from spermatogonia until early leptotene spermatocytes and then rapidly extinguished. This suggests that H3K9me2/1 marks were added within this stage. Predeposited H3K9me2/1 marks were persistent until the late zygotene stage even without catalytic enzymes. However, after the completion of synapsis, the marks were rapidly and actively removed. The ‘active' removal of H3K9me2/1 marks seems to be catalyzed by JHDM2A protein at least in part, which is expressed after the completion of synapsis. As the ‘active' removal mechanism was absent in females at least until early diplotene stage, H3K9me2/1 modification was maintained throughout meiotic prophase. Double arrows indicate the meiotic catastrophic points by the loss of G9a and H3K9me2/1.
Discussion
Using a conditional KO system, we have described an essential function for the H3K9 methyltransferase G9a during germ cell development. Germ-lineage-specific deletion of the G9a gene led to a drastic loss of H3K9me2/1 during meiotic prophase in both sexes. Furthermore, synapsis formation was perturbed in KO meiotic germ cells. As proposed in previous studies (Ogawa et al, 2002; Tachibana et al, 2002; Shi et al, 2003; Gyory et al, 2004; Nishio and Walsh, 2004; Roopra et al, 2004), one important function of G9a and G9a-mediated H3K9me2/1 is transcriptional silencing of specific genes, and this could be essential for proper meiotic prophase progression in both sexes. Unexpectedly and interestingly, we found that the kinetics of H3K9 methylation were not static but dynamic during male meiotic prophase. Here we discuss further these novel findings.
To date, two types of HKMTases have been demonstrated to posses a critical function during meiosis in mammals. First, loss of the H3K9 tri-methylating enzymes Suv39h1/2 led to a male meiotic disorder (Peters et al, 2001). Second, loss of the H3K4 tri-methylating enzyme Meisetz also led to meiotic defects in both sexes (Hayashi et al, 2005). Thus, G9a is the third HKMTase that plays an important role in meiosis. Among the three, Suv39h-KO germ cells undergo apoptosis during the pachytene stage accompanied by a synaptic disorder. Meisetz-KO spermatocytes and oocytes exhibit impaired DSB repair during the pachytene stage and undergo cell death before completing meiosis. The developmental defect observed in G9a-deficient germ cells was also at the pachytene stage and subsequent post-pachytene germ cells were absent or drastically reduced (Figures 3 and 5, respectively). Taken together, meiotic prophase progression, especially during the pachytene stage, must be strictly controlled by a variety of epigenetic modifiers. The G9a-KO phenotype in germ cells was manifested as increased incomplete pachytene cells in both sexes, which contained a mixture of asynapsed to fully synapsed chromosomes within the same nucleus (Figures 3E and 5E). This aborted synapsis may result from a failure to search for a homologous chromosome. As G9a-deposited H3K9me2/1 was maintained in germ cells during synapsis, the presence of H3K9me2/1 on entire chromosomes may play a role(s) in the efficient search for homologous one. As methylated H3K9 acts as a scaffold for many chromatin bound proteins (Kim et al, 2006), it is possible that such binding proteins act as landmarks for homologous chromosome searching.
In contrast to male germ cells, few oocytes escaped cell death at the pachytene stage and survived until the diplotene stage even though they lacked any detectable H3K9me2 (Figure 5I–J). The impact of G9a deficiency and consequent loss of H3K9me2/1 upon meiotic progression is partly different between the sexes. Similarly, mutant mice for several meiotic genes display sex-biased phenotypes (Hunt and Hassold, 2002).
It was shown that the reactivation of genome-wide dispersed retrotransposons is linked with male meiotic arrest in several mutant mice, such as Dnmt3L-KO (Bourc'his and Bestor, 2004; Webster et al, 2005; Hata et al, 2006) and Miwi2-KO mice (Carmell et al, 2007). As the developmental defects of these KO testes are similar to those observed for the G9a-KO mice, we examined reactivation of A-type LINE1 and IAP elements and found that neither were reactivated in G9a-deficient testicular cells (Figure 6E). It was speculated that reactivation of genome-wide dispersed retrotransposons in male germ cells represents a possible mechanism for the spermatogenetic defect seen in Dnmt3L-KO and Miwi2-KO mice. However, the G9a-KO phenotype demonstrates that reactivation of retrotransposons can be dissociated from synapsis disorder phenotypes.
We identified a cohort of genes that are reactivated in meiocytes in G9a-KO testes and also upregulated in G9a-KO ovaries during meiosis (Figure 6A–D), suggesting that there are some common G9a-regulating genes during meiosis in both sexes. It is remains unclear whether de-repression of these genes directly leads to perturbations of germ cell development. One possible mechanism is that enhanced hydroxysteroid dehydrogenase activity caused by the upregulation of akr1c13/12 may affect hormone metabolism and subsequent germ cell development (Yee et al, 2006). We also speculate that either deregulation of transcription or irregular formation of chromatin structure (described above), or both, leads to synaptic disorder in G9a-KO gonads.
Gene expression microarray analysis also showed that a relatively small set of genes was upregulated in G9a-KO testicular cells (eight genes were >2 × upregulated, Supplementary Table S1) although a global reduction of H3K9me2/1 was evident. These data suggest that G9a and G9a-mediated H3K92/1 methylation is mostly dispensable for gene silencing even though H3K9 methylation is a general transcriptional silencing mark. Recent G9a inhibitor work also suggests that H3K9me2/1 is probably part of G9a-mediated repressive mechanisms (Kubicek et al, 2007). In accordance with this notion, we examined whether H3K4me and H3K9ac (general transcriptional active histone marks) were affected in G9a-KO spermatocytes, but could not find significant differences in H3K4me3 and H3K9ac signals between KO and heterozygous cells (data not shown).
We describe novel findings for dynamic alterations in H3K9 methylation during the meiotic prophase (Figure 7E). G9a/GLP proteins exist only in the early stages of the male meiotic prophase and then disappear rapidly. Previous studies showed that H3K9me2/1 signals were absent in A-type spematogonia and emerged at the Intermediate spermatogonia stage (Payne and Braun, 2006). Taken together, we suggest that the G9a/GLP complex deposits methyl marks in a specific developmental window ranging from intermediate spematogonia until the pre-/early leptotene stage in males. In the case of females, deposition of the H3K9me2 marks also seems to be completed by the pre-/early leptotene stage (see Supplementary Figure S1). The H3K9me2 marks persist during synapsis and then rapidly removed after completion of synapsis in males. On the other hand, in female, the methyl marks were maintained until the diplotene stage at least. Immunocytochemical staining showed that the JHDM2A protein, which catalyses demethylation of H3K9me2/1 (but not H3K9me3) (Yamane et al, 2006), was expressed specifically in male germ cells at the pachytene stage (Figure 7A–C). As two other JHDM2A-related proteins, JHDM2B and C, exist in mice and humans, these JHDM2 family proteins may also contribute to the male pachytene-specific demethylation. After the pachytene stage, developmental processes of male germ cells are drastically distinct from those of female cells. Post pachytene male germ cells rapidly complete meiosis, and differentiate into sperm, whereas female meiosis arrests at the diplotene stage and persists for an extended period until ovulation. The male-specific turnover of H3K9me2/1 at the pachytene stage may explain the sex difference in post-pachytene germ cell development.
In conclusion, our findings strongly suggest that H3K9 methylation during meiotic prophase is dynamically regulated by a combination of G9a(/GLP) HKMTase and JHDM2(A) histone lysine demethylase. Current and further studies of the H3K9 methylating and demethylating enzymes and their functional effects on meiosis could provide many valuable insights into how other histone methyl marks are dynamically regulated and how such regulation impacts development, differentiation, and cellular functions.
Materials and methods
Generation of G9a-conditional KO mice and genotyping
The strategy for generating G9a-conditional KO mice, and functional analysis of the conditional KO ES cells are described in Supplementary Figure S2.
Histological analysis
Reproductive organs were fixed in Bouin's fixative, dehydrated through graded ethanol, embedded in paraffin wax and sectioned. Sections were stained with hematoxylin–eosin or hematoxylin only as described (Nozaki et al, 1999). For immunohistological analyses, the organs were fixed in 4% paraformaldehyde for 2 h, washed in PBS, incubated in serially concentrated sucrose solution, embedded with TISSUE-TEK® (Miles) at liquid nitrogen, and then were sectioned (16 μm thick). Immunofluorescence analyses were performed using antibodies or serum diluted as follows: anti-mVASA rabbit serum, anti-SCP3 rabbit serum, anti-PLZF antibodies (Santa Cruz, H-300), anti-c-Kit antibodies (Santa Cruz, C-19), and anti-G9a (Perseus Proteomix, A8620A). When mouse monoclonal antibodies were used as a primary staining reagent, tissues were pretreated with mouse IgG blocking solution (Vector).
Meiotic chromosome spreads
Spread chromosomes were prepared as described (Peters et al, 2001). Meiotic stages were determined according to the criteria described previously (Mahadevaiah et al, 2001). Antibodies used were listed in Supplementary data.
Immunoblot analysis
Testicular single cells were isolated as described (Bellve et al, 1977) and incubated with 10% FBS containing DMEM at 37°C on gelatin-coated dish for plating adherent somatic cells. After a 6 h incubation, suspended cells were collected and subjected to immunoblot analysis. Protein concentration was normalized with anti-Tubulin antibody (Ab-1, Oncogene). Immnunoprecipitation analysis with anti-G9a and GLP was described previously (Tachibana et al, 2005).
Northern blot analysis
Total RNA for Northern blot analysis was isolated from P14 whole testis with Sepasol RNA I (Nakarai), incubated with DNase I (Promega) at 37°C for 15 min, and separated on a 1% agarose gel. The sequences of probes used were described in Supplementary data.
RNA in situ hybridization
Testis (P10) were prepared for in situ RNA hybridization as described previously (Ohsawa et al, 2005). The sequences of probes used were described in Supplementary data.
RT–PCR analysis
In total, 1 μg of total RNA from whole testes (P10) was treated with DNase I as described above and then used for cDNA synthesis with oligo d(T) primer and Superscript II reverse transcriptase (Invitrogen). Primers used for PCR amplification were listed in Supplementary data.
Supplementary Material
Supplementary Figure S1
Supplementary Table S1
Supplementary Information
Supplementary Figure S2
Legend to Supplementary Figures and Information
Acknowledgments
We are particularly grateful to E Oltz for critically reading the manuscript. We thank T Noce for rabbit serum against MVH, S Chuma and N Nakatsuji for rabbit serum against SCP3, K Hata for providing Dnmt3L-KO testes, A Nagy for TNAP-Cre mice, K Yamane and Y Zhang for anti-JHDM2A antibody, and S Kuramochi-Miyagawa for probes to detect reactivation of retrotransposons. We also thank R Kageyama, R Ohsawa, and I Imayoshi for technical advice, K Hayashi and Y Seki for critical adivice, Kazusa DNA research institute for mKIAA0742 cDNA (a variant of mouse JHDM2A), and our laboratory members for assistance. This work was supported by a Grant-in Aid from the Ministry of Education, Science, Technology and Culture of Japan.
References
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M (1977) Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74: 68–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE (2004) Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Clotman F, De Backer O, De Plaen E, Boon T, Picard J (2000) Cell- and stage-specific expression of mage genes during mouse spermatogenesis. Mamm Genome 11: 696–699 [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP (2004) Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL (2004) PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5: 299–308 [DOI] [PubMed] [Google Scholar]
- Hata K, Kusumi M, Yokomine T, Li E, Sasaki H (2006) Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Mol Reprod Dev 73: 116–122 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378 [DOI] [PubMed] [Google Scholar]
- Hoog C, Schalling M, Grunder-Brundell E, Daneholt B (1991) Analysis of a murine male germ cell-specific transcript that encodes a putative zinc finger protein. Mol Reprod Dev 30: 173–181 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ (2002) Sex matters in meiosis. Science 296: 2181–2183 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T (2002) The many faces of histone lysine methylation. Curr Opin Cell Biol 14: 286–298 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124 [DOI] [PubMed] [Google Scholar]
- Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A (2000) Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26: 116–117 [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Nishio H, Walsh MJ (2004) CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA 101: 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J (1999) Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest 79: 293–299 [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R (2005) Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci 25: 5857–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C, Braun RE (2006) Histone lysine trimethylation exhibits a distinct perinuclear distribution in Plzf-expressing spermatogonia. Dev Biol 293: 461–472 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF (2004) Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell 14: 727–738 [DOI] [PubMed] [Google Scholar]
- Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM (1999) Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 140: 5894–5900 [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y (2005) Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309–25317 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T (2000) Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 93: 139–149 [DOI] [PubMed] [Google Scholar]
- Turner BM (2000) Histone acetylation and an epigenetic code. BioEssays 22: 836–845 [DOI] [PubMed] [Google Scholar]
- Ueda J, Tachibana M, Ikura T, Shinkai Y (2006) Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281: 20120–20128 [DOI] [PubMed] [Google Scholar]
- Webster KE, O'Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, Meachem SJ, Antonarakis SE, de Kretser DM, Hedger MP, Peterson P, Carroll BJ, Scott HS (2005) Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci USA 102: 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Yee DJ, Balsanek V, Bauman DR, Penning TM, Sames D (2006) Fluorogenic metabolic probes for direct activity readout of redox enzymes: selective measurement of human AKR1C2 in living cells. Proc Natl Acad Sci USA 103: 13304–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Table S1
Supplementary Information
Supplementary Figure S2
Legend to Supplementary Figures and Information


