Abstract
Photolyases are DNA repair enzymes that use energy from blue light to repair pyrimidine dimers. We report the isolation of an Arabidopsis thaliana mutant (uvr2-1) that is defective in photorepair of cyclobutylpyrimidine dimers (CPDs). Whereas uvr2-1 is indistinguishable from wild type in the absence of UV light, low UV-B levels inhibit growth and cause leaf necrosis. uvr2-1 is more sensitive to UV-B than wild type when placed under white light after UV-B treatment. In contrast, recovery in darkness or in light lacking photoreactivating blue light results in equal injury in uvr2-1 and wild type. The uvr2-1 mutant is unable to remove CPDs in vivo, and plant extracts lack detectable photolyase activity. This recessive mutation segregates as a single gene located near the top of chromosome 1, and is a structural gene mutation in the type II CPD photolyase PHR1. This mutant provides evidence that CPD photolyase is required for plant survival in the presence of UV-B light.
Keywords: [blue light, cyclobutylpyrimidine dimer, DNA repair, pyrimidine pyrimidinone6–4dimer, uvr2-1]
Ultraviolet-B (UV-B; 280–320 nm) radiation, which is increasing in the biosphere as a result of stratospheric ozone depletion by chlorofluorocarbons (1), damages DNA and is toxic and mutagenic to cells. It is not surprising that plants, which require sunlight for photosynthesis, have developed unique strategies such as the inducible synthesis of UV-absorptive secondary metabolite sunscreens (2) to avoid the detrimental effects of UV-B. In spite of these protective filters, some UV-B penetrates into plant cells and causes DNA damage. While significant progress has been made in elucidating mechanisms of DNA repair in bacteria and mammals, our understanding of DNA repair processes in plants remains rudimentary (3).
The major type of DNA damage caused by exposure to UV-B is the formation of dimers between adjacent pyrimidines, i.e., the cyclobutylpyrimidine dimer (CPD) and the pyrimidine pyrimidinone (6–4′) dimer [(6–4) photoproduct]. Unrepaired dimers are lethal to cells because they deform the DNA helix, interfering with both replication and transcription (3). Photolyases use energy from blue light (400–500 nm) to directly reverse pyrimidine dimer bonds, accurately repairing the damage (4). Whereas most of the known photolyases are specific for the removal of CPDs, a photolyase gene that encodes a (6–4) photoproduct-specific enzyme was recently sequenced from Drosophila (5), and there is evidence for a similar enzyme activity in Arabidopsis (6), Xenopus laevis (clawed toad), and Crotalis atrox (rattlesnake) (7).
Although light-dependent DNA repair is documented in bacteria, yeast, animals, and plants, it is apparently not universal (4). While the presence of photolyase in humans remains a controversial topic (8, 9), a human homolog of the Drosophila (6–4) photolyase was recently cloned (5). The photolyase genes characterized to date fall into two classes based on sequence homology: class I “microbial” enzymes (including Escherichia coli, yeast, and Neurospora crassa) and the class II “higher eukaryotic” enzymes (10–12). Interestingly, the blue light photoreceptor of plants has strong sequence similarity to class I photolyases, presumably reflecting their common use of blue light for catalysis (13).
Mutants can help to elucidate the biochemical mechanisms that plants employ for protection from UV-B. This approach recently allowed assessment of the importance of UV-absorptive sunscreens in Arabidopsis and maize (14, 15), and identification of a UV-sensitive Arabidopsis mutant defective in dark repair of (6–4) photoproducts (16). In this study we report the isolation of a mutant of Arabidopsis that is hypersensitive to UV-B and defective in photorepair of CPDs. These results demonstrate that repair of CPDs is critical for survival of Arabidopsis, and presumably other plants as well, under UV-B radiation.
MATERIALS AND METHODS
Growth Conditions, Mutant Isolation Strategy, and Genetic Characterization.
Arabidopsis thaliana Landsberg erecta (Ler) was the wild-type (WT) plant used in this study. uvr1, a mutant defective in dark repair of (6–4) photoproducts (16), was used as a control in root growth assays. Plants were grown under continuous light from CW1500 cool-white fluorescent lamps (General Electric; approximately 100 μmol m−2·sec−1 photosynthetically active radiation) filtered through 0.13-mm thick mylar (AIN Plastics, Mt. Vernon, NY; removes wavelengths <310 nm) in Cornell growing mix in pots or on nutrient agar medium in plates as described (14). Supplemental UV-B light from F40UVB fluorescent lamps (Phillips, Somerset, NJ) or T12F40 UVB lamps (Ultraviolet Resources International, Cleveland), filtered through either glass or cellulose acetate (to remove wavelengths <280 nm), was monitored using a spectroradiometer (model OL 752; Optronics Laboratory, Orlando, FL) calibrated with an OL 752–150 calibration module and OL 752–10 spectral irradiance standard as described (14). For comparison with previous work, UV-B irradiance was converted to biologically effective UV-B [kilojoules (kJ) biologically effective UV-B (UV-BBE) −2·h−1] using the Caldwell action spectra normalization (17). Unweighted UV-B doses are also provided, expressed in kJ·m−2. Spectra from the F40UVB lamps with the filters described above are as described (14), and the T12F40 UVB lamps have a fluence spectrum similar to the F40UVB lamps.
To screen for UV-B-sensitive mutants, M2 plants derived from ethyl methanesulfonate mutagenesis of Ler (18) were grown in soil for 5 days under constant illumination from mylar-filtered cool-white fluorescent lamps. The plants were then subjected to supplemental UV-B treatment by turning on F40UVB lamps (1.5 kJ·m−2·h−1 unweighted UV-B and 0.4 kJ UV-BBE·m−2·h−1) for 1 or more days. Seedlings exhibiting symptoms of UV-B sensitivity, such as growth inhibition and browning or bleaching of cotyledons and leaves, were allowed to recover and self-pollinate in the absence of UV-B. The resulting M3 progeny were grown with or without UV-B as described above to test the phenotypic uniformity and specificity of the UV-sensitivity trait. More than 200 individuals were chosen for progeny testing from approximately 150,000 M2 seeds screened, and 14 met the criteria for UV-B sensitivity after the M3 screen. Because it is one of the most highly UV sensitive of the mutants identified, uvr2-1 was chosen for further characterization. The mutant line used in all experiments was derived by backcrossing once to Ler. Genetic mapping was conducted using the codominant microsatellite markers nga59 and nga63 (19) and cleaved amplified polymorphic sequence markers NCC1 and ATEAT1 (20) for an F2 population derived from a cross between uvr2-1 and the WT Columbia ecotype.
Root Sensitivity to UV-B.
Root sensitivity to UV-B was determined as described by Britt et al. (16) with the following modifications: seedlings were germinated under Sylvania F40GO gold lights (6 μmol·m−2·sec−1 photosynthetically active radiation); UV-B (1.32 kJ·m−2·min−1) was supplied by a model UVT750-M Ultraviolet Transilluminator (International Biotechnologies) with a peak output of 314 nm, filtered through 0.13-mm-thick cellulose acetate (<0.2 J·m−2·min−1 from λ < 280 nm). Following UV-B treatment, plants were either wrapped in foil or placed under CW1500 cool-white lamps with mylar filters to allow photorepair to occur in the absence of UV-B. Approximately 35 roots per experiment were measured for each treatment (except for uvr1, for which approximately 15 roots were measured per treatment).
In Vivo Dimer Repair.
Plants were grown on nutrient agar plates for 12 days under CW1500 cool-white lamps with mylar filters, and then treated with 1.32 kJ·m−2 unweighted UV-B using the transilluminator. Plates were harvested immediately (0 hr control), or allowed to recover either wrapped in aluminum foil or replaced under cool-white lamps. All plants were harvested under F40GO gold lights by pouring liquid nitrogen directly onto the plates, scraping seedlings (50–100 per sample) into chilled tubes, and freezing immediately at −80°C. DNA was extracted from frozen tissue and dimers quantified using CPD- or (6–4) photoproduct-specific monoclonal antibodies (21). Samples were measured in quadruplicate and compared by ANOVA within each experiment using the computer program statistica. The Newman–Keuls procedure was used for pairwise comparison (22). In three experiments, initial average CPD damage levels were 1.9, 1.3, and 1.7 dimers per 105 bases in Ler and 2.3, 1.2, and 1.8 dimers per 105 bases in uvr2-1.
Photolyase Activity.
Three-week-old plants grown on nutrient agar plates were either maintained under CW1500 cool-white lamps filtered through mylar, immediately wrapped in foil, or treated with supplemental UV-B from F40UVB lamps (1.5 kJ·m−2h·−1 unweighted UV-B; 0.4 kJ·UV-BBE m−2 ·h−1) for 24 hr and then harvested using liquid nitrogen. Extracts were prepared as described (23), and a chromatographic assay for removal of CPDs from heavily irradiated bacterial DNA substrate was performed as described (24). Assay incubation times (up to 19 hr) were chosen such that activity was proportional to the amount of extract (typically 100–600 μg) for at least three time points. Dark-incubated control extracts showed no CPD removal.
RESULTS
Isolation and Genetic Characterization of uvr2-1.
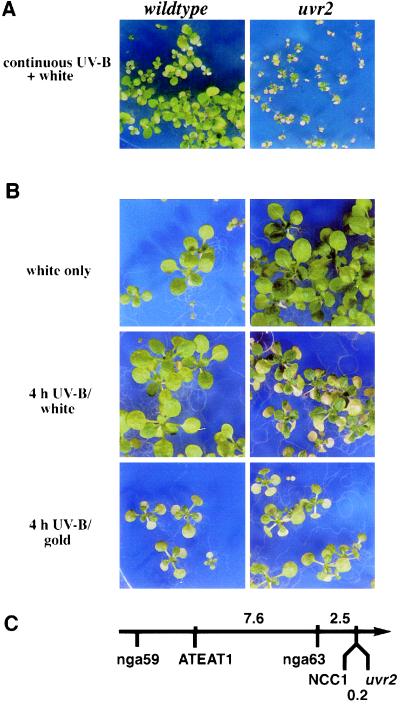
The uvr2-1 mutant was identified in a screen for plants whose growth is strongly inhibited when treated with supplemental UV-B. uvr2-1 is very sensitive to UV-B: chronic exposure to daily UV-B doses lower than that experienced by plants under clear-sky sunlight is sufficient to dramatically inhibit growth of uvr2-1 seedlings (Fig. 1A). Symptoms of UV-B sensitivity are also observed following short-term UV-B treatment: as little as 4 hr of irradiation produces withering and necrosis of expanded leaves within 6 days after UV-B (Fig. 1B Middle). UV-B sensitivity is observed throughout development, from early vegetative growth through flowering. The mutant is indistinguishable from the WT when grown in the absence of UV-B (Fig. 1B Top), demonstrating that the injury is caused by UV-B.
Figure 1.
Phenotypes and map location of uvr2-1. (A) Seed were germinated on nutrient agar plates; beginning on day 3 seedlings were exposed to continuous UV-B from F40UVB lamps (0.13 kJ·UV-BBE m−2·h−1; 0.64 kJ·m−2·h−1 unweighted UV-B) and photographed 10 days later. This daily UV-BBE dose is approximately one-half that of a clear mid-summer day in Ithaca, New York. (B) Thirteen-day-old, light-grown plants with two to four fully expanded true leaves were treated with UV-B from two T12F40 (1.7 kJ·UV-BBE·m−2·h−1; 5.0 kJ·m−2·h−1 unweighted UV-B) for 0.5, 1, 4, 8, or 16 hr (4-hr treatment shown), and then either returned to grow under light from CW1500 cool-white lamps filtered through mylar (Middle) or placed under F40GO gold fluorescent lights (Bottom) for 7 days. Plants of the same age grown continuously under CW1500 cool-white lamps with mylar filters are shown for comparison (Top). (C) Map locations of uvr2-1 and nearby markers on the top of chromosome 1. Distances are in centimorgans. The arrow points toward the centromere; the relative map position of nga59 is shown for orientation. Map distances to ATEAT1 and nga63 were determined using 100 uvr2-1/uvr2-1 F2 plants.
The uvr2-1 recessive mutation defines a single genetic locus. When uvr2-1 and WT were crossed, F1 progeny were UV-resistant; self-pollinated F2 progeny segregated in a 3:1 ratio of resistant:sensitive plants (n = 715, χ2 = 0.029). The mutation mapped to the top of chromosome 1, with only one recombination event occurring between uvr2-1 and NCC1 in 266 F2 plants. This places the two loci within 0.2 centimorgans of each other (Fig. 1C). One hundred of the uvr2-1/uvr2-1 F2 plants were also analyzed for segregation of nga63 and ATEAT1 (Fig. 1C). Although a previously reported UV-B sensitivity mutation (uvh6) is also located at the top of chromosome 1, it is not tightly linked to uvr2, mapping 12 centimorgans from NCC1 (M. Jenkins and D. Mount, personal communication) and 1.6 centimorgans from nga59 (25).
uvr2-1 UV-B Hypersensitivity Depends on Light Conditions.
Because it is known that repair of pyrimidine dimers by photolyase requires blue light, we asked whether light influences the UV-B sensitivity of uvr2-1. WT and uvr2-1 were grown without UV-B until the seedlings had two to four fully-expanded true leaves and then were treated with 1.7 kJ·UV-BBE·m−2·h−1 (5.0 kJ·m−2·h−1 unweighted UV-B) from two T12F40 UVB lamps for 0.5, 1, 4, 8, or 16 hr. When plants recovered in the light, cotyledons and leaves of uvr2-1 that had previously been exposed to UV-B for ≥ 4 hr turned brown and died, whereas WT was unaffected by this treatment (Fig. 1B Middle). The shorter treatments had no effect on either genotype. Eight- and 16-hr UV-B treatments caused injury to WT leaves, yet uvr2-1 was more severely damaged than WT. In contrast, both WT and uvr2-1 were injured by 4 hr UV-B when allowed to recover under gold lights, which lack photoreactivating wavelengths (Fig. 1B Bottom). Under these nonphotoreactivating conditions, uvr2-1 was indistinguishable from WT for all UV-B doses tested.
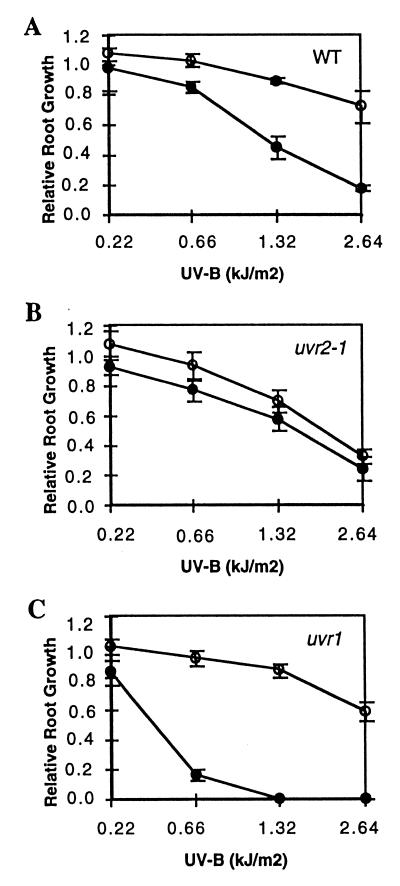
Although roots receive only a low fluence of UV-B and blue light in soil, uvr2-1 roots also exhibit light-dependent UV-B sensitivity (Fig. 2). Consistent with a defect in a light-dependent repair process, uvr2-1 did not exhibit the light-stimulated recovery seen in the WT. Following treatment with 2.64 kJ·m−2 unweighted UV-B, new growth of uvr2-1 roots was approximately 50% that of WT. As expected, the light-independent (6–4) photoproduct repair mutant uvr1 (16) was more sensitive to UV-B than WT in the dark, but not in the light (Fig. 2).
Figure 2.
Root growth of uvr2-1 is affected by UV-B during recovery in the light, but not in the dark. Three-day-old seedlings were treated with 0.22–2.62 kJ·m−2 unweighted UV-B (1.32 kJ·m−2·min−1) from a transilluminator followed by 24 hr incubation under CW1500 cool-white lamps with mylar filtration or in the dark, to allow new root growth. (A) WT. (B) uvr2-1. (C) uvr1. Open symbols, light; solid symbols, dark. Standard error is indicated with vertical lines. Data are the means of three experiments with root growth normalized to that of untreated plants.
uvr2-1 Is Defective in CPD Repair.
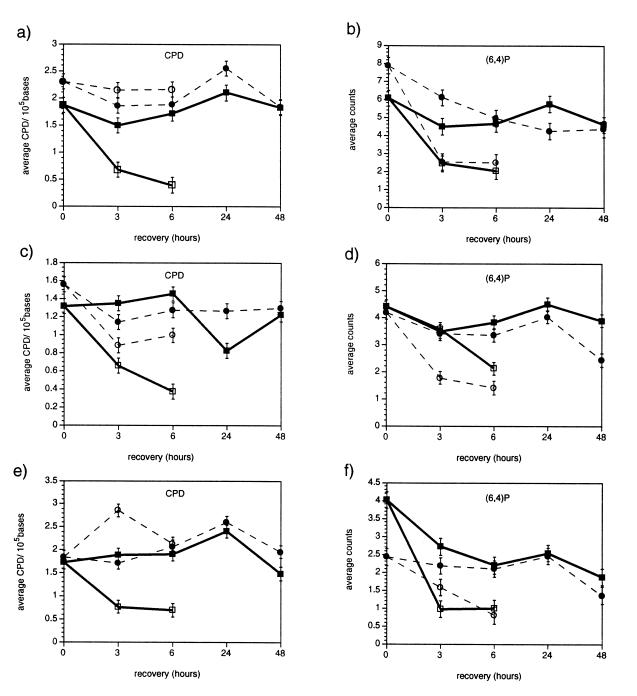
The light dependence of differences in UV-B sensitivity between WT and uvr2-1 was consistent with the hypothesis that this mutant is defective in photorepair of UV-induced DNA damage. To test this, CPDs and (6–4) photoproducts were measured in plants treated with an acute dose of UV-B, followed by recovery in the light or dark. Fig. 3 illustrates that, in contrast to WT, uvr2-1 showed no statistically significant removal of CPDs in the light (P > 0.05 in each experiment after 6 hr of recovery). The observation that both WT and uvr2-1 removed (6–4) photoproducts in the light indicates that uvr2-1 specifically affects light repair of CPDs. Repair, rather than shielding, is implicated because UV-B generated a similar number of CPDs in WT and uvr2-1 tissues. Furthermore, HPLC analysis of flavonoids and hydroxycinnamic acids also indicated that UV-B sunscreens are normal in uvr2-1 (L.G.L. and R.L.L., unpublished results). Neither WT nor uvr2-1 showed statistically significant repair of CPDs or (6–4) photoproducts in the dark.
Figure 3.
CPD and (6–4) photoproduct levels in Ler and uvr2-1 during recovery. WT (squares/solid lines) and uvr2-1 (circles/dashed lines) plants were irradiated with 1.32 kJ·m−2 UV-B from a transilluminator and allowed to recover under CW1500 cool-white lamps with mylar filtration (open symbols) or in the dark (solid symbols). The data show three experiments with monoclonal antibody detection for CPDs (a, c, and e) and (6–4) photoproducts (b, d, and f). Values are the means of four measurements for each sample; vertical bars are standard error.
uvr2-1 Lacks Photolyase Activity.
Persistence of CPDs in irradiated uvr2-1 tissue suggested a defect in photolyase. This activity was assayed in extracts from plants adapted to darkness, maintained in the light, or treated with UV-B (Table 1). As predicted, extracts from uvr2-1 plants had no detectable photolyase activity when assayed under saturating levels of both dimer substrate and photoreactivating photons. Light-grown wild-type plants had approximately 4-fold higher photolyase activity than those adapted to the dark for 24 hr. UV-B treatment apparently did not increase photolyase activity, although it is possible that we underestimated activities of UV-B treated extracts, i.e., from competition between dimers in endogenous plant DNA and the irradiated bacterial DNA substrate. In an independent experiment, mixing equal volumes of WT and uvr2-1 extracts yielded 93% (dark adapted), 100% (light grown), and 102% (UV-B treated) of the expected WT values with uvr2-1 samples contributing no activity or inhibition. These data indicate that uvr2-1 does not contain a photolyase inhibitor and that WT extracts do not supply a cofactor missing in uvr2-1, consistent with a defect in the photolyase apoprotein.
Table 1.
Photolyase activities in plant extracts
| Extract | Dimers (106) removed per hr per μg of protein*
|
||
|---|---|---|---|
| Dark† | Light† | Light + UV-B† | |
| WT | 215 (±24) | 783 (±54) | 750 (±66) |
| uvr2-1 | <14 | <10 | <12 |
Three-week-old seedlings were either wrapped in foil for 24 hr (dark), maintained under CW1500 cool-white lamps with mylar filters (light), or supplemented with UV-B from F40UVB lamps (2 kJ·m−2·h−1 unweighted UV-B; 0.6 kJ·UV-BBE·m−2·h−1; light + UV-B) for 24 hr.
Means for four trials each. Standard deviations for WT given in parentheses.
Using a second set of extracts, mixing WT and uvr2-1 (1:1), yielded 93% (dark), 100% (light), and 102% (+UV-B) of the expected WT values with uvr2-1 samples contributing no activity or inhibition.
DISCUSSION
uvr2-1 is a UV-B-sensitive mutant of Arabidopsis that completely lacks detectable photolyase activity and light-dependent repair of CPDs. uvr2-1 is defective in the light-mediated repair of CPDs, but not (6–4) photoproducts, and is more sensitive to UV-B than WT in the light, but not in the dark. These characteristics distinguish uvr2-1 from uvr1, which defines a gene involved in dark repair of the (6–4) photoproduct (16). uvr2-1 appears to be a novel mutation because it is genetically unlinked to both uvr1 and the previously reported UV-sensitivity mutations uvh1 (25) and uvh6 (M. Jenkins and D. Mount, personal communication.).
Previous reports demonstrated photolyase activity in higher plants (for examples see refs. 3, 23, and 26), but this is the first direct evidence that CPD removal is necessary for survival. Although plant genes with homology to class I photolyase have been described from Arabidopsis and mustard, all available data indicate that these encode blue-light photoreceptors (13, 27–29). Recently, an Arabidopsis gene with strong similarity to higher eukaryotic class II photolyases was cloned (PHR1) (30), and recent results indicated that uvr2-1 is allelic to this photolyase gene. For example, PHR1 is genetically linked to uvr2-1, the uvr2-1 mutant has a frameshift mutation in PHR1 (30) with a concomitant reduction of PHR1 mRNA (L.G.L., J.L., and R.L.L., unpublished results).
The absence of photolyase activity in plant extracts (Table 1) and the lack of in vivo CPD repair (Fig. 3) in uvr2-1 suggest that there is only one CPD-specific photolyase in Arabidopsis. While the existence of independent photolyases for nuclear and chloroplastic DNA was suggested by a Chlamydomonas mutant deficient in photolyase activity (31), recent experiments indicate that young Arabidopsis seedlings do not repair CPDs in mitochondrial or chloroplastic DNA (32), and may require only a nuclear CPD photolyase. uvr2-1 is fully capable of light repair of the second major form of pyrimidine dimer, the (6–4) photoproduct (Fig. 3), suggesting that like class I photolyases, the class II Arabidopsis enzyme only repairs CPDs.
Although light-independent repair of (6–4) photoproducts was reported in Arabidopsis and wheat (16, 33), our results suggest that this did not occur under our experimental conditions. While a great deal of variability was seen for light-independent (6–4) photoproduct repair, there was no statistically significant repair in WT, nor significant differences observed between uvr2-1 and WT. While this discrepancy with published results may reflect technical differences [for example, our transilluminator emitted a significant amount of 314 nm light, which could affect the proportion of (6–4) dimers in the Dewar isomer form], the disparity in plant developmental stage may also be relevant. Published studies demonstrating dark repair of (6–4) photoproducts used germinating seedlings, whereas we used older plants. It is possible that the relative amount of DNA repair completed by dark- vs. light-dependent mechanisms is developmentally regulated in Arabidopsis.
Differences in the quality or quantity of UV-B radiation may also be responsible for the differences reported with respect to dark repair of CPDs. For example, in agreement with our results, previously published work with Arabidopsis (16) showed no repair of CPDs in the absence of light when plants were treated with 1.3 kJ·m−2 unweighted UV-B from a transilluminator (peak output at 305 nm). In contrast, dark repair of CPDs was detected in Arabidopsis plants treated with 1 kJ·m−2 UV from a germicidal lamp (peak output 254 nm; ref. 23), and wheat seedlings treated with approximately 4 kJ·m−2 UV-BBE from a fluorescent lamp (33). It was previously demonstrated that UV-B dose can affect the amount of CPDs repaired in the dark: at low UV-B doses dark repair was undetectable in alfalfa, but at higher doses dark repair of CPDs was readily observed (26). It thus is likely that the spectral quality of UV as well as the total dose, may affect induction of dark repair mechanisms. These complexities illustrate that much remains to be learned about the repair of DNA in plants. The uvr2-1 mutant, in addition to showing for the first time that light repair is essential for the survival of a higher plant, will enhance these studies, because this mutant must rely solely on dark repair to remove CPDs.
Acknowledgments
We thank Anne Britt for providing the uvr1 mutant and comments on the manuscript, and Mike Jenkins and David Mount for sharing unpublished uvh6 mapping data. This research was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Postdoctoral Fellowship 94–37100-0315 to L.G.L., National Science Foundation Presidential Young Investigator Award DMB-9058134 and U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 90–37280-5677 to R.L.L., and by Environmental Protection Agency Grant R82-0008 to V.W.
Footnotes
References
- 1.Caldwell M M, Teramura A H, Tevini M. Trends Ecol Evol. 1989;4:363–367. doi: 10.1016/0169-5347(89)90100-6. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Ou-Lee T-M, Raba R, Amundson R G, Last R L. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britt A B. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:75–100. doi: 10.1146/annurev.arplant.47.1.75. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 5.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 6.Chen J J, Mitchell D L, Britt A B. Plant Cell. 1994;6:1311–1317. doi: 10.1105/tpc.6.9.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S-T, Malhotra K, Taylor J-S, Sancar A. Photochem Photobiol. 1996;63:292–295. doi: 10.1111/j.1751-1097.1996.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 8.Li Y F, Kim S-T, Sancar A. Proc Natl Acad Sci USA. 1993;90:4389–4393. doi: 10.1073/pnas.90.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutherland B M, Bennett P V. Proc Natl Acad Sci USA. 1995;92:9732–9736. doi: 10.1073/pnas.92.21.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuhira S, Yasui A. J Biol Chem. 1992;267:25644–25647. [PubMed] [Google Scholar]
- 11.Yasui A, Eker A P M, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. EMBO J. 1994;13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato T, Todo T, Ayaki H, Ishizaki K, Morita T, Mitra S, Ikenaga M. Nucleic Acids Res. 1994;22:4119–4124. doi: 10.1093/nar/22.20.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad M, Cashmore A R. Nature (London) 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 14.Landry L G, Chapple C C S, Last R L. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stapleton A E, Walbot V. Plant Physiol. 1994;105:881–889. doi: 10.1104/pp.105.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britt A B, Chen J-J, Wykoff D, Mitchell D. Science. 1993;261:1571–1574. doi: 10.1126/science.8372351. [DOI] [PubMed] [Google Scholar]
- 17.Caldwell M M. In: Photophysiology. Giese A C, editor. Vol. 6. New York: Academic; 1971. pp. 131–177. [Google Scholar]
- 18.Barczak A J, Zhao J, Pruitt K D, Last R L. Genetics. 1995;140:303–313. doi: 10.1093/genetics/140.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 20.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 21.Stapleton A E, Mori T, Walbot V. Plant Mol Biol Rep. 1993;11:230–236. [Google Scholar]
- 22.Keuls M. Euphytica. 1952;1:112–122. [Google Scholar]
- 23.Pang Q, Hays J B. Plant Physiol. 1991;95:536–543. doi: 10.1104/pp.95.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaustein A R, Hoffman P D, Hokit D G, Kiesecher J M, Walls S C, Hays J B. Proc Natl Acad Sci USA. 1994;91:1791–1795. doi: 10.1073/pnas.91.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins M E, Harlow G R, Liu Z, Shotwell M A, Ma J, Mount D W. Genetics. 1995;140:725–732. doi: 10.1093/genetics/140.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quaite F E, Takayanagi S, Ruffini J, Sutherland J C, Sutherland B M. Plant Cell. 1994;6:1635–1641. doi: 10.1105/tpc.6.11.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batschauer A. Plant J. 1993;4:705–709. doi: 10.1046/j.1365-313x.1993.04040705.x. [DOI] [PubMed] [Google Scholar]
- 28.Malhotra K, Kim S-T, Batschauer A, Dawut L, Sancar A. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman, P. D., Batschauer, A. & Hays, J. B. (1996) Mol. Gen. Genet., in press. [DOI] [PubMed]
- 30.Ahmad, M., Jarillo, J. A., Klimczak, L. J., Landry, L. G., Peng, T., Last, R. L. & Cashmore, A. R. (1997) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 31.Cox J L, Small G D. Mutat Res. 1985;146:249–255. doi: 10.1016/0167-8817(85)90065-3. [DOI] [PubMed] [Google Scholar]
- 32.Chen J-J, Jiang C-Z, Britt A B. Plant Physiol. 1996;111:19–25. doi: 10.1104/pp.111.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor R M, Nikaido O, Jordan B R, Rosamond J, Bray C M, Tobin A K. Plant Cell Environ. 1996;19:171–181. [Google Scholar]