Abstract
Bacteriophytochromes are red/far-red photoreceptors that bacteria use to mediate sensory responses to their light environment. Here, we show that the photosynthetic bacterium Rhodopseudomonas palustris has two distinct types of bacteriophytochrome-related protein (RpBphP4) depending upon the strain considered. The first type binds the chromophore biliverdin and acts as a light-sensitive kinase, thus behaving as a bona fide bacteriophytochrome. However, in most strains, RpBphP4 does not to bind this chromophore. This loss of light sensing is replaced by a redox-sensing ability coupled to kinase activity. Phylogenetic analysis is consistent with an evolutionary scenario, where a bacteriophytochrome ancestor has adapted from light to redox sensing. Both types of RpBphP4 regulate the synthesis of light harvesting (LH2) complexes according to the light or redox conditions, respectively. They modulate the affinity of a transcription factor binding to the promoter regions of LH2 complex genes by controlling its phosphorylation status. This is the first complete description of a bacteriophytochrome signal transduction pathway involving a two-component system.
Keywords: bacteriophytochrome, light harvesting complexes, redox sensing, Rhodopseudomonas palustris
Introduction
Phytochromes are light sensors that were first discovered in plants where they mediate development and growth in response to near infrared light. More recently this class of photoreceptor has also been identified in cyanobacteria (Cphs phytochromes), in proteobacteria (BphPs phytochromes) and in fungi (Fphs phytochromes) (for review see Karniol et al, 2005). Most of these chromoproteins use a linear tetrapyrrole chromophore (bilin) and sense red and far-red light via a reversible shift from a red-absorbing form (Pr) to a far-red-absorbing form (Pfr). The discovery of prokaryotic phytochromes has provided new insights into the evolution of phytochromes and has led to a detailed biochemical and structural description of this light sensor family (for review see Rockwell et al, 2006).
Bacteriophytochromes (BphPs) have an N-terminal domain homologous to the photosensory core domain (PCD) of plant phytochromes and a C-terminal domain involved in signal transduction. The PCD, which consists of PAS, GAF and PHY sub-domains, autocatalytically binds a linear tetrapyrrole (or bilin) (Karniol et al, 2005; Rockwell et al, 2006). In most cases, BphPs use the chromophore biliverdin (BV), the simplest linear tetrapyrrole synthesized from heme by a heme oxygenase (Bhoo et al, 2001). BV is covalently bound to a Cys residue at the N-terminus of the protein (Lamparter et al, 2002). The atomic structure of the region spanning from the N-terminus to the GAF domain of DrBphP from Deinococcus radiodurans has recently been determined (Wagner et al, 2005); this revealed the key residues that form the BV binding pocket and an unusual trefoil knot that stabilizes the PAS and GAF domains.
In most cases, the BphP C-terminal domain has a two-component histidine kinase motif whose phosphorylation status is dependent on the BphP conformation. The most active phosphorylating state can be either the Pr or the Pfr form, depending on the BphP considered. In some cases, a phosphotransfer to a response regulator (RR), whose gene is often found within the BphP operon, has been evidenced (Rockwell et al, 2006). Whether or not this phosphotransfer is part of the signaling pathway is still to be demonstrated. A small-angle X-ray scattering solution structure of RpBphP2 from Rhodopseudomonas (Rps.) palustris revealed that a Y-shaped dimer forms through an interaction between the two C-terminal domains. The structure shows how photo-induced signal transduction may stimulate auto-phosphorylation followed by phosphotransfer to a RR (Evans et al, 2006).
Besides these common properties, there are interesting differences between BphPs. For the bathyBphPs, the thermal ground state is the Pfr form (Giraud et al, 2002; Karniol and Viestra, 2003), whereas for other BphPs, the Pfr form converts to the Pr form in the dark as for plant phytochromes (Butler and Lane, 1965; Lamparter et al, 2002). Unlike all BphPs studied so far, RpBphP3 from Rps. palustris converts between a Pr form and a Pnr (for near red) form, which absorbs at around 645 nm, in a light-reversible way (Giraud et al, 2005). The bacteriophytochrome Ppr of the photosynthetic bacterium Rhodospirillum centenum is a hybrid of photoactive yellow protein, bacteriophytochrome and histidine kinase domains (Jiang et al, 1999). An unusual BphP has also been identified in the photosynthetic bacterium Bradyrhizobium ORS278. This BphP binds phycocyanobilin and absorbs light maximally at around 610 nm in its dark-adapted state (Jaubert et al, 2007). This high diversity in BphPs features that most likely reflect their specific in vivo functions warrants further study.
In this context, the purple photosynthetic bacterium Rps. palustris is a very interesting organism to study. It has a wide distribution and exhibits extraordinary metabolic versatility. Moreover, its photosystem is one of the most elaborate in terms of light harvesting (LH) complexes. In addition to the LH1 complexes that absorb light at around 870 nm and surround the reaction center, peripheral LH2 and LH4 complexes have been described (Tadros and Waterkamp, 1989; Evans et al, 1990). The LH2 complexes, which are encoded by three distinct pucBA operons (a, b and e), absorb 800 and 850 nm light, while the LH4 complexes, encoded by the pucBA.d operon, only absorb at around 800 nm (Evans et al, 1990; Hartigan et al, 2002). The relative proportion of these LH complexes varies according to environmental factors such as light and oxygen tension (Evans et al, 1990; Hartigan et al, 2002). Finally, the genomes of five distinct strains of Rps. palustris have been sequenced (Larimer et al, 2004; http://spider.jgi-psf.org/index.html). The genome of the model strain CGA009 contains six putative BphP genes scattered throughout the genome, four of which are close to photosynthesis genes (Larimer et al, 2004). The role of these genes has recently been described: (i) RpBphP1 triggers the synthesis of the entire photosynthetic apparatus (Giraud et al, 2002, 2004), and (ii) RpBphP2 and RpBphP3 act in tandem to control the expression of the pucBA.d operon (Giraud et al, 2005). The fourth putative BphP (RpBphP4) gene is in the vicinity of the pucBA.e operon, suggesting it could be involved in the control of LH2 complex synthesis. Surprisingly, the RpBphP4 from strain CGA009 does not have the conserved N-terminal Cys residue involved in bilin attachment, and a purified recombinant RpBphP4 homologue isolated from another Rps. palustris strain (2.1.6) does not bind any chromophore (Evans et al, 2005). Comparison of the CGA009 genome with the genomes of four other Rps. palustris strains reveals that this gene is only present in two other strains, BisB5 and HaA2. Unlike RpBphP4s in the CGA009 and 2.1.6 strains, RpBphP4s from BisB5 and HaA2 do have the N-terminal Cys residue responsible for bilin attachment. Can this last form of RpBphP4 capture light? And more generally, how do these different RpBphP4s function and what is their role?
In this study, a phylogenetic analysis of RpBphP4 highlights an organization into two sister clades. By combining biophysical, biochemical and genetic approaches, we demonstrate that RpBphP4 acts either as a bona fide BphP or as a redox sensor depending on the clade. In addition, we show that both types of RpBphP4 use the same signal transduction pathway to control the synthesis of the LH2 complexes.
Results
Phylogenetic analysis of RpBphP4 from Rps. palustris strains
The RpBphP4 (rpa1490) gene of strain CGA009 encodes a protein with the classical bacteriophytochrome architecture (Figure 1B). This gene is flanked by a RR encoding gene (rpa1489), and by the pucBA.e operon encoding the LH2 complex apoproteins (Figure 1A). To investigate RpBphP4 gene diversity, polymerase chain reaction (PCR) was used to screen for the presence of this gene in the genome of seven additional Rps. palustris strains, using sets of primers specific to the pucB.e and rpa1489 genes, respectively. The RpBphP4 gene was found in all the strains tested (CEA001, LMG4316, LMG4317, DSM126, DSM131, DSM8283 and DSM7375). The amino-acid (AA) sequences of the newly found RpBphP4s have a very high level of identity (>93%) with RpBphP4 from CGA009, and also lack the canonical Cys residue used as the bilin attachment site (Figure 1C).
Figure 1.
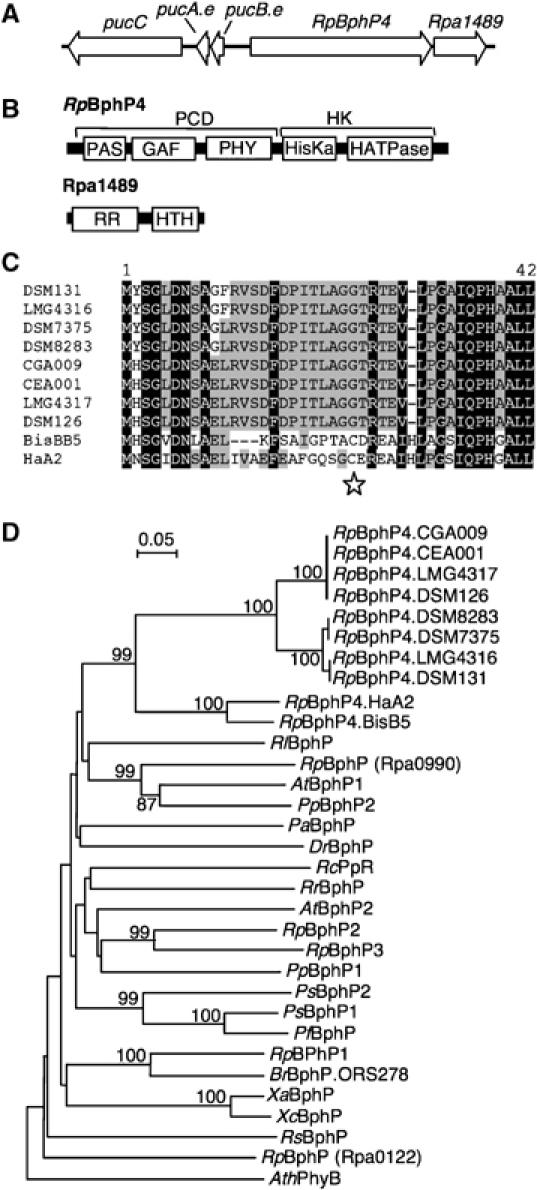
Molecular characterization of RpBphP4. (A) Arrangement of genes around RpBphP4 (rpa1490 from CGA009). (B) Predicted domain structure of RpBphP4 and Rpa1489. PCD: photosensory core domain; HK: histidine kinase domain; HisKa: phosphoacceptor domain; HATPase: ATP binding domain; RR: response regulator domain; HTH: helix–turn–helix domain. (C) AA sequence alignment of the N-termini of RpBphP4 proteins from different Rps. palustris strains. Residues conserved at more than 50 and 90% are highlighted in gray and black, respectively. Star indicates the Cys residue used as BV binding site. (D) Phylogenetic analysis of the BphP family based on an alignment of the GAF domains. The sequences were aligned by CLUSTALX and the tree was generated by the neighbor-joining method and displayed using NJPLOT. Bootstrap values, expressed as percentages of 1000 replications, are shown at the branching points. Species abbreviations: At, Agrobacterium tumefaciens; Ath, Arabidopsis thaliana; Br, Bradyrhizobium sp.; Dr, Deinococcus radiodurans; Pa, Pseudomonas aeruginosa; Pf, Pseudomonas fluorescens; Pp, Pseudomonas putida; Ps, Pseudomonas syringae; Rc, Rhodospirillum centenum; Rl, Rhizobium leguminosarum; Rp, Rhodopseudomonas palustris; Rr, Rhodospirillum rubrum; Rs, Rhodobacter sphaeroides; Xa, Xanthomonas axonopodis; Xc, Xanthomonas campestris.
A phylogenetic analysis of the GAF domains showed that all RpBphP4s are related to the BphP family and belong to the same monophyletic group, with a very high bootstrap value of 99% (Figure 1D). Similar clustering patterns were obtained using the PHY and PAS domains (data not shown). Interestingly, two sister clades can be distinguished: a large clade containing most of the RpBphP4s, which all lack the conserved Cys residue involved in bilin attachment, and a smaller clade formed by two representatives (HaA2 and BisB5), which have the Cys residue (Figure 1C). The presence or the absence of this canonical Cys residue suggests there are fundamental differences in the physico-chemical properties of RpBphP4s from each clade. In order to confirm such a hypothesis, three RpBphP4s representative of these two clades (CEA001, HaA2 and BisB5) were studied in more detail.
Expression and purification of recombinant RpBphP4 homologues
RpBphP4s from CEA001, HaA2 and BisB5 were coexpressed with a heme oxygenase (required for chromophore synthesis) in Escherichia coli and then purified by affinity chromatography. As shown in Figure 2A, the purified recombinant CEA001-RpBphP4 is colorless as is RpBphP4 from the 2.1.6 strain (Evans et al, 2005). The absence of bound bilin was confirmed by spectral analysis and zinc-induced fluorescence (Figure 2A). In addition, several attempts of in vitro reconstitution by incubation of the purified CEA001-RpBphP4 apoprotein with various bilins (BV, phycocyanobilin and phycoerythrobilin) were unsuccessful to produce a holo-protein. In contrast, RpBphP4s purified from HaA2 and BisB5 are colored and their absorption spectra are characteristic of a bona fide BphP (Figure 2A for HaA2, data not shown for BisB5). Zn-induced fluorescence of proteins separated by SDS–PAGE confirmed the presence of a covalently bound chromophore (Figure 2A).
Figure 2.
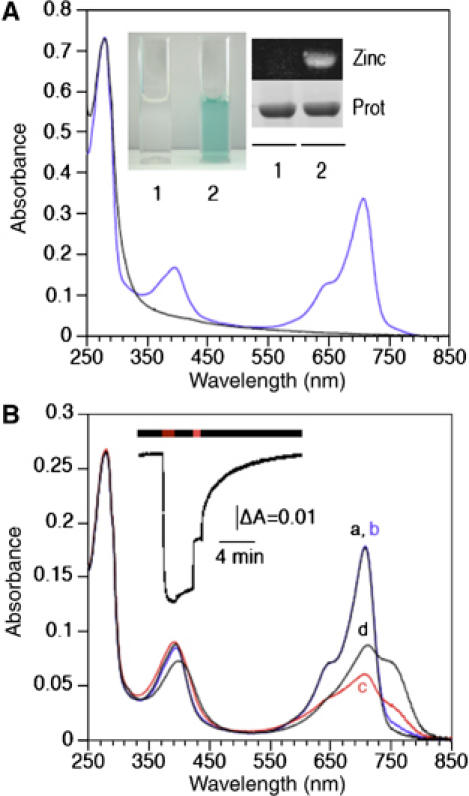
Absorption spectra of RpBphP4 proteins. (A) Absorption spectra of recombinant RpBphP4 proteins from strains CEA001 (achromo-RpBphP4, black line) and HaA2 (chromo-RpBphP4, blue line). The inset shows (1) achromo- and (2) chromo-RpBphP4 proteins. The covalent binding of BV to RpBphP4 proteins was visualized by zinc-induced fluorescence or Coomassie staining. (B) Absorption spectra of purified HaA2 RpBphP4. The spectra were recorded immediately after: 770 nm illumination (a, black line) followed by 15 min of darkness (b, blue line), 705 nm illumination (c, red line) followed by 15 min of darkness (d, black line). The inset shows the kinetics of changes in absorbance at 750 nm induced by 770 nm light, followed by 2 min of darkness, 1 min of 705 nm light and then a long dark period.
In an attempt to generate a functional chromophore binding site in CEA001-RpBphP4, a Cys residue was introduced at the conserved position using site-directed mutagenesis (the G24C mutant). We also exchanged the first 33 AA residues with the N-terminal sequence (residues 1–29) of the holo-BphP RpBphP1. In both cases, the purified mutated proteins were colorless (data not shown). This implies that the lack of BV binding in CEA001-RpBphP4 is not just due to the absence of the N-terminal Cys but is also the result of additional AA differences. There are possible candidates for this in the chromophore binding pocket of DrBphP (Wagner et al, 2005). First, Tyr216, which is involved in hydrogen bonding of the chromophore, is not conserved in the CEA001-RpBphP4 sequence. In addition, four AA (Ile29, Ala212, Phe203 and Met267) out of the 15 involved in Van der Waals' contacts with BV are not conserved. Are these subtle changes sufficient to prevent chromophore binding? Further site-directed mutagenesis and structural studies would be necessary to prove this.
Altogether, these data indicate that there are two distinct types of RpBphP4 in Rps. palustris, depending on the strain considered. In most strains, RpBphP4 does not bind BV, whereas the second RpBphP4 type does bind this chromophore. To distinguish between these two RpBphP4 types, the prefixes ‘chromo' or ‘achromo' (indicating the presence or the absence of a chromophore, respectively) will be used from now onward.
Achromo-RpBphP4 is redox sensitive
When freshly prepared achromo-RpBphP4 from CEA001 is analyzed by non-reducing SDS–PAGE, a major band of around 87 kDa, which corresponds to its expected molecular mass (Figure 3A, lane a) is detected. When the same sample was analyzed after ultrafiltration or dialysis, the 87 kDa band was almost undetectable, although several very high-molecular-weight bands were detected (Figure 3A, lane b). This is likely to reflect the formation of aggregates of an unknown number of subunits. A similar effect was observed when the protein sample was exposed to air overnight (data not shown). We therefore assumed that the oligomerization state of the protein was related to the oxygenation, that is, the redox state of the sample. In support of this hypothesis, incubating the protein for 30 min with a reducing agent (DTT) before SDS–PAGE analysis restored the 87 kDa band, while subsequent treatment with an oxidant (potassium ferricyanide) reinduced the formation of aggregates (Figure 3A, lanes c and d). Therefore, the change in the oligomerization state of achromo-RpBphP4 is reversible and redox sensitive. Such an effect was not observed for chromo-RpBphP4s from HaA2 or BisB5, which mainly appear as a single band of the expected size, irrespective the redox conditions (Figure 3A, lanes e–h; data not shown for BisB5). The redox-dependent oligomerization state of achromo-RpBphP4 in solution was confirmed by size-exclusion chromatography (Figure 3D). Oxidized achromo-RpBphP4 eluted with the void volume of the column (Superdex 200), which corresponds to a molecular mass greater than 500 kDa. However achromo-RpBphP4 in the presence of DTT produced a major elution volume peak corresponding to a molecular mass of 200 kDa. Taking into account the predicted molecular mass of the His-tagged RpBphP4 monomer, we conclude that the reduced protein is folded in solution as a homodimer (Figure 3D). When the native chromo-RpBphP4 from HaA2 was separated by gel filtration, the elution volume obtained did not vary with different redox conditions and corresponded to the dimeric form of the protein (Figure 3C).
Figure 3.
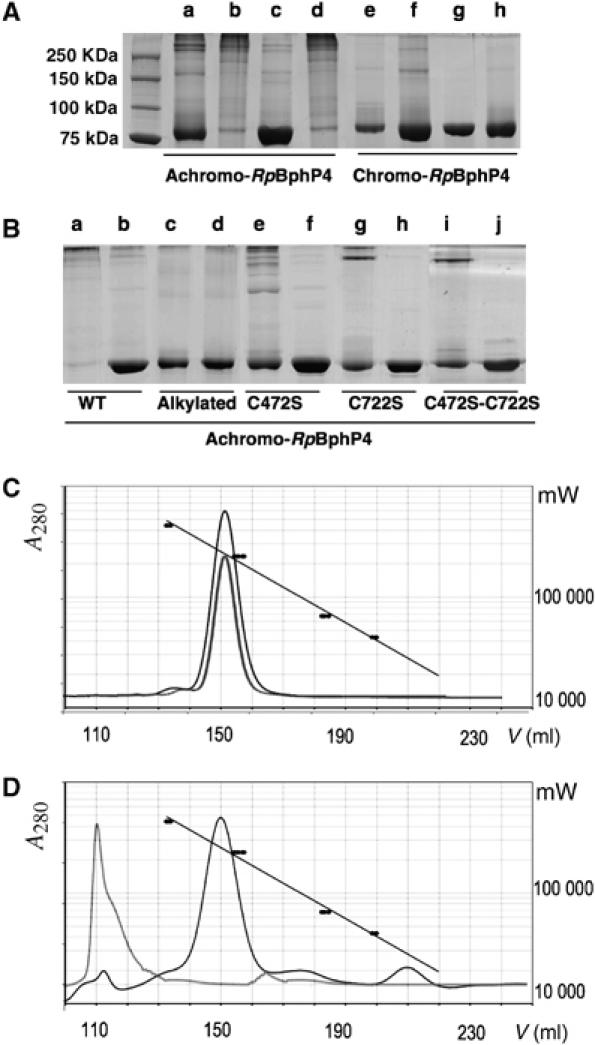
Redox dependence of achromo- and chromo-RpBphP4 proteins. SDS–PAGE analysis and gel filtration of RpBphP4 from CEA001 (achromo) or from HaA2 (chromo) after various treatments. (A) Lanes a and e show, respectively, untreated achromo- and chromo-RpBphP4 obtained after nickel affinity chromatography; lanes b and f show the effect of ultrafiltration (Amicon 30 K NMWL); lanes c and g show the effect of 1 mM DTT; and lanes d and h show the effect of 10 mM ferricyanide after reduction by 1 mM DTT. (B) Disulfide bond formation in achromo-RpBphP4 (WT), alkylated achromo-RpBphP4, RpBphP4-C472S, RpBphP4-C722S and RpBphP4-C472S-C722S, after oxidation with 1 mM ferricyanide (lanes a, c, e, g and i) or reduction with 1 mM DTT (lanes b, d, f, h and j). (C, D) Elution profiles of gel filtration of chromo-RpBphP4 (C) or achromo-RpBphP4 (D) in 50 mM NaCl, 20 mM Tris–HCl pH 8.0 in the absence (gray curves), or in the presence (black curves) of 10 mM DTT. A standard curve is superimposed using the known molecular masses of four protein standards (black circles).
Analyzing spectra and AA sequences has not revealed any signature of a putative redox center such as those for hemes or flavins, so the redox sensitivity of achromo-RpBphP4 may be determined by one or more Cys residues, as has been described for many thiol-based redox sensors (Paget and Buttner, 2003). To investigate this, achromo-RpBphP4 pre-reduced with DTT was incubated with 10 mM iodoacetamide to fully thiol-alkylate the protein. This modified sample was analyzed by SDS–PAGE in reducing and non-reducing conditions. As shown in Figure 3B, lanes c and d, thiol-alkylated achromo-RpBphP4 was found to be redox independent, confirming that at least one Cys is involved in redox sensing. The AA sequences of achromo-RpBphP4 proteins contain seven Cys residues, two of which (C472 and C722) are conserved in all the achromo-RpBphP4s. C472 is in the PHY domain, whereas C722 is in the His kinase module. To investigate the role of these two Cys residues in the redox sensitivity of achromo-RpBphP4, the single mutants, C472S and C722S, and the double mutant C472S-C722S were produced. SDS–PAGE analysis (Figure 3B) and gel filtration (Supplementary Figure 1) showed that all three mutated proteins form fewer aggregates under oxidizing conditions, indicating that Cys472 and Cys722 are key residues in the redox-dependent oligomerization changes of achromo-RpBphP4.
The chromo-RpBphP4 protein has unusual photochemical properties
Figure 2B shows the absorption spectra of chromo-RpBphP4 from strain HaA2 after red or far-red illumination and different periods of dark adaptation. Under far-red illumination, chromo-RpBphP4 absorbs maximally at around 708 nm (Figure 2B, spectrum a). This is typical of the Pr form of BphPs. This state is relatively stable in the dark. Only a small increase in absorption is recorded at around 750 nm after dark adaptation (Figure 2B, spectrum b). Starting from this state, illumination with red light induces strong bleaching of the 708 nm band, along with a small increase in absorption around 750 nm (Figure 2B, spectrum c). This state, which has strong similarities with the meta-R states of Agp1 BphP (Borucki et al, 2005) and phyA (Eilfeld and Rüdiger, 1985), is relatively unstable and slowly transforms (with a half-life of about 2 min) to a new state in the dark. This new state, characterized by the greater absorption of light at around 750 nm typical of the Pfr form (Figure 2B, spectrum d), is relatively stable in the dark and only partially reverts to the Pr state over several hours (not shown). Conversely, illumination of this state with 770 nm light induces rapid formation of the Pr form (Figure 2B, spectrum a). The light-induced formation of Pr and ‘meta-R' states starting from the Pfr state is illustrated in Figure 2B (inset). Very similar results were obtained with chromo-RpBphP4 from strain BisB5 (data not shown). Steady-state excitation and emission fluorescence measurements show that only the Pr state of chromo-RpBphP4 is fluorescent (Supplementary Figure 2).
Chromo- and achromo-RpBphP4s use the same signal transduction pathway
Both chromo- and achromo-RpBphP4s have a C-terminal histidine kinase module (Figure 1B). Regulation of the in vitro kinase activity was studied by incubating the proteins in the presence of γ-32P-ATP. The redox conditions significantly affected the autophosphorylation of achromo-RpBphP4 of strain CEA001, which only occurred in its reduced form, that is, the dimeric form (Figure 4A, lanes a–c). Such an effect was not observed for the thiol-alkylated achromo-RpBphP4 (Figure 4A, lanes d–f) or for chromo-RpBphP4 from strain HaA2 (Figure 4B, lanes a–c). In contrast, the phosphorylation state of chromo-RpBphP4 was dependent on the light transition (Figure 4B). Indeed, chromo-RpBphP4 from HaA2 appeared to be maximally phosphorylated in its Pfr form (lane d) or following a 705 nm illumination (Figure 4B, lane f), while transition to the Pr state induced a 60% decrease in the level of phosphorylation (lane e).
Figure 4.
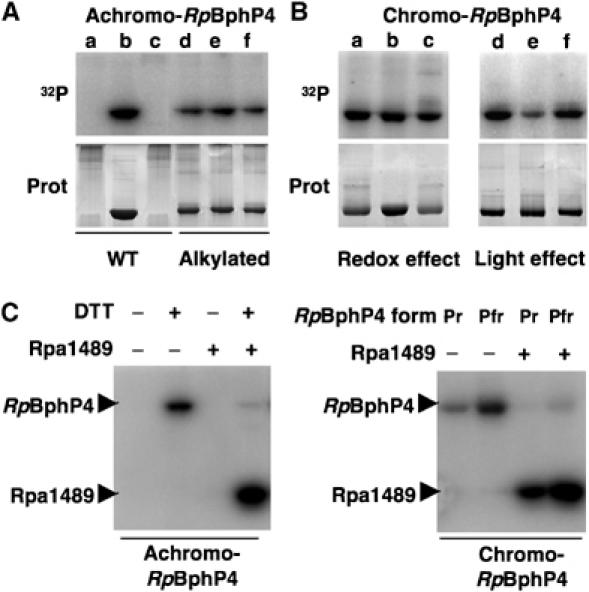
Achromo- and chromo-RpBphP4 proteins act as redox or light-regulated histidine kinases, respectively. (A) Effect of redox conditions on the kinase activity of achromo-RpBphP4 (WT) and alkylated achromo-RpBphP4 (alkylated). Lanes a and d, untreated; lanes b and e, reduced with 1 mM DTT; and lanes c and f, oxidized with 1 mM ferricyanide. Proteins were incubated with γ-32P-ATP for 15 min. The reaction products were separated by SDS–PAGE and the gels were autoradiographed (top) or stained with Coomassie blue (bottom). (B) Effect of redox or light conditions on the kinase activity of chromo-RpBphP4. For the redox effect, the sample was subjected to 15 min of pre-illumination with 705 nm light followed by 15 min of dark adaptation, and was left untreated (lane a), or treated with 1 mM DTT (lane b) or 1 mM ferricyanide (lane c). For the light effect, chromo-RpBphP4 was converted to its Pfr (lane d), Pr (lane e) or ‘meta R' (lane f) forms; the Pfr form was obtained by 15 min of pre-illumination at 705 nm followed by 15 min of dark adaptation, the Pr form by illumination at 770 nm and the ‘meta R' form by illumination at 705 nm. (C) Phosphotransfer between RpBphP4 and the RR Rpa1489. Transfer of the phosphoryl group between the achromo-RpBphP4 and Rpa1489 relative to the redox conditions (left panel). Phosphotransfer between chromo-RpBphP4 and Rpa1489 relative to the light conditions (right panel).
The genes for chromo- and achromo-RpBphP4 proteins are both found close to a gene encoding a putative transcription factor (Rpa1489), which consists of an N-terminal RR domain and a C-terminal helix–turn–helix DNA binding domain (Figure 1B). Alignment of the sequences of these putative transcription factors from the CEA001 and HaA2 strains shows they have 92% identity. Efficient phosphotransfer from both types of RpBphP4 to Rpa1489 was observed (Figure 4C). As expected, the level of phosphorylation of Rpa1489 can be correlated to the redox or light conditions depending on the RpBphP4 type. Therefore, both types of RpBphP4 are involved in the same signal transduction pathway that is probably a two-component regulatory system, where the Rpa1489 homologue is the last element.
The transcription factor Rpa1489 binds to pucBA promoters
The position of RpBphP4/rpa1489 genes in the vicinity of the pucBA.e operon suggests that the putative transcription factor Rpa1489 might bind to the pucBA.e promoter. DNA binding of recombinant Rpa1489 was first tested by gel mobility shift assays, using the promoter regions of the five pucBA operons identified in strain CGA009. The results shown in Figure 5A indicate that Rpa1489 binds specifically to the pucBA.b and pucBA.e promoters in vitro. This specific interaction depends on the phosphorylation state of Rpa1489 (Figure 5B). The affinity of unphosphorylated Rpa1489 for the pucBA.b promoter is seven-fold lower than that of phosphorylated Rpa1489. Similar results were obtained for the pucBA.e promoter (data not shown).
Figure 5.
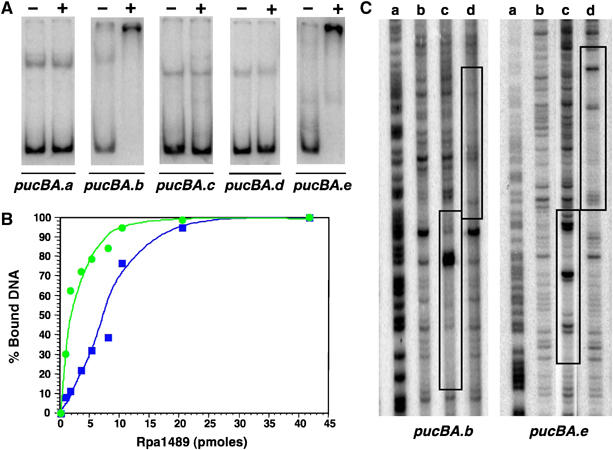
The transcription factor Rpa1489 binds the pucBA.b/e promoters. (A) Gel mobility shift assays with purified Rpa1489. The left lane (−) of each gel contained only 32P-labeled probe, while the right lane (+) contained in addition 0.5 μM Rpa1489. PucBA from a to e represent the five pucBA promoter regions identified in Rps. palustris CGA009 strain (Larimer et al, 2004). (B) Binding affinity of unphosphorylated (blue curve) and phosphorylated (green curve) Rpa1489 to the pucBA.b promoter. Phosphorylated and unphosphorylated forms of Rpa1489 were obtained by incubating Rpa1489 with achromo-RpBphP4 for 30 min under reducing conditions in the presence or absence of ATP, respectively. (C) DNase I footprints of Rpa1489 and PpsR2 binding to the pucBA.b/e promoter regions. G+A ladder (lane a), control without protein (lane b), 1 μM Rpa1489 (lane c), 1 μM PpsR2 (lane d) (for color figure see online version).
In addition, we investigated the Rpa1489 DNA binding sites on the pucBA.b/e promoters by DNase I protection (footprint). The DNase I digestion patterns (Figure 5C) confirmed that Rpa1489 protects both pucBA.b and pucBA.e promoters. The sequences of the protected regions are very similar and both contain the palindromic motif TGTCCGN8CGGACA. Interestingly, the pucBA.b/e promoters contain a TGTN12ACA palindrome downstream and juxtaposed to the Rpa1489 DNA binding site. This sequence corresponds to the DNA binding site of PpsR, a key regulator of photosynthesis gene expression in purple bacteria (Elsen et al, 2005). Rps. palustris is unusual in having two distinct PpsRs (encoded by ppsR1 and ppsR2) like the photosynthetic Bradyrhizobium sp. ORS278 strain (Jaubert et al, 2004). We tested whether purified PpsR2 (Giraud et al, 2004) binds to the promoters of the pucBA.b/e operons. As shown in Figure 5B, PpsR2 binds these promoter regions downstream of the Rpa1489 binding site, strongly suggesting that these two pucBA operons are under the control of Rpa1489, PpsR2 and most probably PpsR1.
Chromo- and achromo-RpBphP4 control LH2 synthesis
The above biochemical data are strong indications that RpBphP4, sensing either a light or a redox signal, may control the synthesis of LH2 complexes through the regulation of some pucBA operons using a cognate RR (Rpa1489). The next step was to confirm this role in vivo. The photosystem composition of three Rps. palustris strains, harboring either achromo- (CEA001) or chromo-RpBphP4 (BisB5 and HaA2), and their corresponding RpBphP4 deletion mutants were analyzed after growth under different light and redox conditions. The absorption spectra of intact cells were recorded at 77 K. At this low temperature, absorption in the near infrared range by LH2 and LH1 complexes can be clearly distinguished, making it more straightforward to calculate the relative contribution made by each type of complex. For BisB5 (chromo-RpBphP4-containing) cells grown in the dark at O2 tensions of between 1 and 8%, a small amount of photosynthetic apparatus was observed only at 1% (Figure 6A). In contrast, large amounts of photosynthetic apparatus were synthesized when cells were illuminated with 700 or 770 nm light, irrespective of the O2 tension (Figure 6A, left panel). This effect is related to the activating effect of RpBphP1 (Giraud et al, 2002, 2004). Illumination with each of these wavelengths however has a different effect on the synthesis of LH1 and LH2 complexes. The LH2/LH1 ratios are 1 and 0.7 for illumination with 700 or 770 nm light, respectively (Figure 6A). On the other hand, no differential effect of illumination with 700 or 770 nm light on LH2 synthesis was observed for the BisB5ΔRpBphP4 mutant (Figure 6A, right panel). A similar result was observed at 3 or 8% O2 tension for both the BisB5 and HaA2 strains (not shown). These experiments indicate that 700 nm light absorbed by chromo-RpBphP4 activates the synthesis of LH2 complexes.
Figure 6.
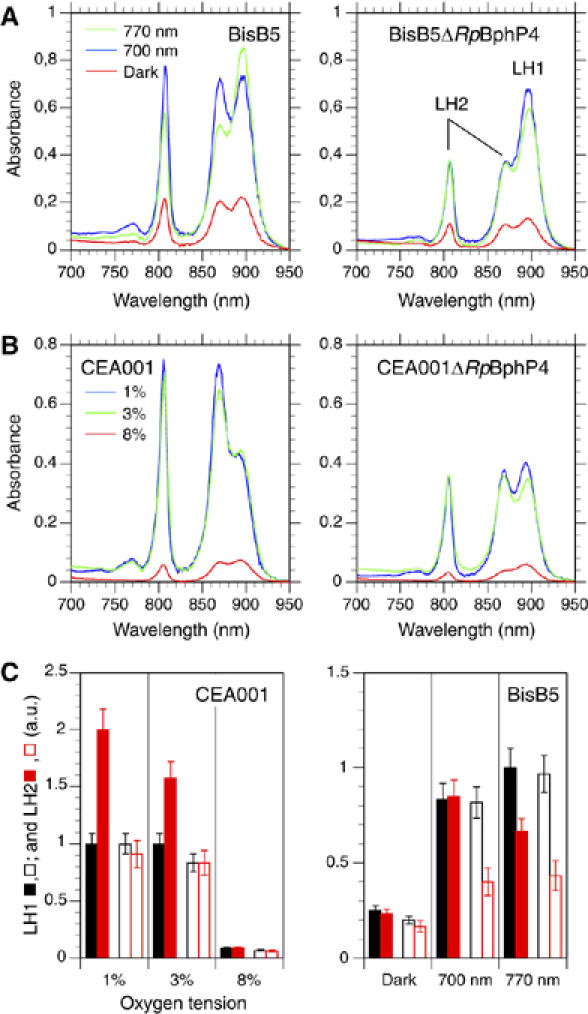
Effect of light and oxygen tension on the synthesis of the photosynthetic apparatus in WT and RpBphP4 deletion mutants. (A) Left panel, absorption spectra recorded at 77 K of intact BisB5 cells grown at 1% oxygen tension in the dark (red spectrum), or with 700 nm (blue spectrum), or 770 nm (green spectrum) light. Right panel, as for the left panel, but for the BisB5ΔRpBphP4 mutant. (B) Left panel, absorption spectra recorded at 77 K of intact CEA001 cells grown in the dark at 1% (blue spectrum), 3% (green spectrum) and 8% (red spectrum) oxygen tension. Right panel, as for the left panel, but for the CEA001ΔRpBphP4 mutant. (C) Left panel, amounts in arbitrary units of LH1 (black bars) and LH2 (red bars) synthesized in the dark in strain CEA001 (filled bars) and mutant CEA001ΔRpBphP4 (open bars) as a function of the oxygen tension. Right panel, amounts of LH1 (black bars) and LH2 (red bars) synthesized in the dark or under 700 or 770 nm illumination in strain BisB5 (filled bars) and mutant BisB5ΔRpBphP4 (open bars). All cell cultures were at 1% oxygen tension.
A different situation was observed for strain CEA001 (an achromo-RpBphP4 containing strain). A significant amount of photosynthetic apparatus was synthesized in the dark at O2 tensions lower than 8% (Figure 6B, left panel). In addition, a clear increase in the LH2/LH1 ratio was observed when the O2 tension decreased from 8 to 1% (Figure 6B, left panel). The deletion of RpBphP4 in strain CEA001 resulted in an approximately two-fold lower LH2/LH1 ratio, but did not affect the amount of LH1 at the various O2 tensions tested (Figure 6B, right panel). Contrary to the effect observed in BisB5 or HaA2, illumination with 700 nm light induced a decrease in LH2 synthesis, as previously reported (Giraud et al, 2005).
Another difference in the behavior of strains CEA001 and BisB5/HaA2 was observed for cells grown under anaerobic conditions under white light. CEA001 cells synthesized many more LH2 complexes than the BisB5 and HaA2 strains (data not shown). On the one hand, this is consistent with the activation of CEA001 achromo-RpBphP4 under reducing conditions, and on the other, with the inability to synthesize the chromophore of BisB5/HaA2 chromo-RpBphP4 in the absence of O2.
Altogether these data, summarized in Figure 6C, indicate that both achromo- and chromo-RpBphP4s play an activating role in the synthesis of LH2 at low O2 tension or under specific light wavelengths, respectively. The conditions of this induction are consistent with their respective kinase activities described above.
Discussion
Evolution of RpBphP4 from a light to a redox sensor
Rps. palustris is known as one of the most energetically versatile microorganisms, as it is able to grow under different environmental conditions by using photosynthesis, aerobic or anaerobic respiration, or fermentation. To rapidly adapt its metabolism to changes in oxygen or light availability, Rps. palustris uses a set of sensors like PpsRs and BphPs.
In this study, we show that there is an additional level of regulation of LH2 complex synthesis by the sensor RpBphP4. Depending on the strain considered, RpBphP4 responds to either redox or light signals to activate synthesis of LH2 complexes. Light- and redox-sensing RpBphP4 proteins, chromo- and achromo-RpBphP4, respectively, correspond to two versions of the same protein, found in the same synthon of different Rps. palustris strains. They form two sister clades phylogenetically related to the phytochrome family, suggesting that a BphP ancestor could have evolved from a light sensor to a redox sensor in some of the Rps. palustris strains studied. To our knowledge this is the first description of evolution in the nature of signal perception by a histidine kinase sensor. The loss and acquisition of specific Cys residues would thus be decisive events in modifying the sensory properties of RpBphP4; the canonical N-terminal Cys used as the chromophore binding site in all BphPs is specific to chromo-RpBphP4 while Cys422 and Cys722 are specific to the redox-sensing achromo-RpBphP4.
Among the 10 strains of Rps. palustris, which have RpBphP4, only two have a chromo-RpBphP4. Interestingly, all the achromo-RpBphP4s were found in collection strains, while the two chromo-RpBphP4 are from strains recently isolated from the environment. However, given the very high similarity between the different achromo-RpBphP4 sequences, it is very unlikely that the same AA modifications have occurred in different laboratories. As already mentioned, Rps. palustris strains with achromo-RpBphP4 contain more LH2 complexes than those containing chromo-RpBphP4, when grown under anaerobic conditions in the light, the standard conditions for selecting anoxygenic photosynthetic bacteria. These strains are highly pigmented due to the large quantity of LH2 complexes. When isolating photosynthetic bacteria, selection may be biased toward the more strongly colored colonies, and this may explain the large number of achromo-RpBphP4 strains found in laboratory collections.
Achromo-RpBphP4, a redox sensor
The in vivo phenotypes of deletion mutants indicate that achromo-RpBphP4 is a redox-dependent activator of LH2 synthesis. In vitro analyses suggest that this redox sensing is mediated by two conserved Cys residues, Cys422 and Cys722, located in the PHY and in the His kinase domains, respectively. These two Cys are involved in the reversible redox-dependent formation of aggregates via intermolecular disulfide bonds. This change of oligomerization state can be correlated to the kinase activity of the protein, which is only active in its dimeric form. This is in agreement with previous studies, which showed that histidine kinases of two-component systems always act as dimers where the subunits trans-phosphorylate each other (Yang and Inouye, 1991; Ninfa et al, 1993).
In the absence of a complete structural description of a classical BphP, including both the PHY and histidine kinase domains, it is not possible to fully explain the role of these cysteines in the oligomerization of the protein. However, the involvement of redox-active cysteines in modulating the activity of regulators of photosystem synthesis has already been proven in purple bacteria. For example, an intramolecular disulfide bond, mediated by oxygen, enhances the DNA binding activity of CrtJ, the aerobic repressor of photosynthetic apparatus synthesis in Rhodobacter capsulatus (Masuda et al, 2002). In contrast, PpsR1, the counterpart transcription factor in Bradyrhizobium has only one Cys residue and can form an intermolecular disulfide bond. This leads to a switch from a tetrameric to an octameric form when the redox potential increases (Jaubert et al, 2004). Regulation by intermolecular disulfide bonds has also been demonstrated for the global regulator RegB. RegB and its cognate RR RegA control a large number of oxygen response processes (Elsen et al, 2004). Thanks to the essential role of a metal cofactor, the formation of an intermolecular disulfide bond modulates the oligomerization state from a tetramer under oxidizing conditions to a dimer under reducing conditions. This intermolecular bond also affects RegB autophosphorylation, which can only autophosphorylate in its dimeric form (Swem et al, 2003). The changes in oligomerization state observed in vitro for RpBphP4 are probably related to the mechanisms described above.
Chromo-RpBphP4, an atypical light sensor
We show that chromo-RpBphP4 has unusual spectral properties, with significant bleaching of the Pr form when illuminated with a small concomitant increase in absorption in the near infrared region (Figure 2B). This is probably a consequence of a very slow conversion from the intermediate meta-R state to the stable Pfr state. In fact, this unusual property is consistent with the light regulation of photosynthetic apparatus synthesis in Rps. palustris. Indeed, the synthesis of the photosystem and its associated pigments is activated by the Pr state of RpBphP1, that is, with 770 nm illumination (Giraud et al, 2002, 2004). In contrast, we show in this study that the synthesis of LH2 is activated by 700 nm light, that is, via both the ‘meta-R' and the Pfr states of chromo-RpBphP4. In other words, LH2 synthesis in strains BisB5 or HaA2 requires both 770 and 700 nm light. Such light conditions would yield a mixture of both Pr and Pfr states for a classical BphP. However, due to its poor absorption of near infrared light, chromo-RpBphP4 is preferentially in its activated ‘meta-R' state when excited by both 700 and 770 nm light (Supplementary Figure 3).
RpBphP4, the first element in a two-component system
Our data show that both chromo- and achromo-RpBphP4s have a kinase activity regulated by light or redox conditions, respectively. Both phosphorylate a transcription factor homologous to Rpa1489, increasing its binding affinity for the promoter regions of the pucBA.b/e operons. The enhancement of LH2 synthesis at low oxygen tensions for CEA001 (achromo-RpBphP4) or in 700 nm light for BisB5 and HaA2 (chromo-RpBphP4) implies that the Rpa1489 homologues activate the transcription of both pucBA operons under their phosphorylated form. Such regulation is also suggested by the phenotype of RpBphP4 deletion mutants, which are strongly affected in LH2 synthesis. Altogether, this describes a complete two-component signaling pathway to control LH2 synthesis initiated by a BphP sensing either light or oxygen.
The presence of a PpsR binding motif immediately downstream of the DNA binding site of Rpa1489 indicates, however, that additional regulators (PpsR1 and PpsR2) could be involved in LH2 synthesis (Figure 7). We show that PpsR2 binds to pucBA.b/e promoters (Figure 5C). This transcription factor represses the synthesis of the entire photosynthetic apparatus, including the LH chromophores. This repressive effect is antagonized by far-red light via RpBphP1 (Giraud et al, 2004). Similarly, LH2 synthesis is strongly repressed in a PpsR1 mutant, showing that this transcription factor also acts as a regulator of the expression of pucBA genes (Supplementary Figure 4).
Figure 7.
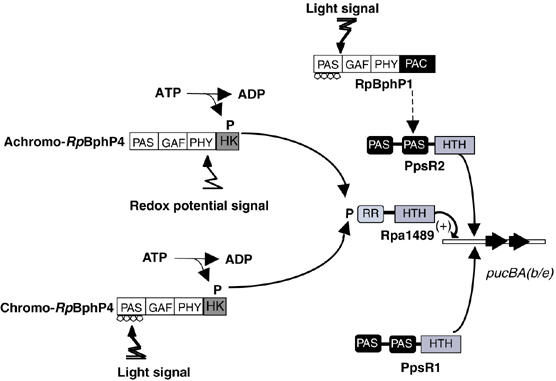
Proposed model for the regulation of LH2 antennae synthesis in Rps. palustris.
In conclusion, the control of photosystem synthesis in Rps. palustris is highly sophisticated and involves a complex network of different light and redox regulatory pathways. This complexity, coupled with the sensor flexibility, enables this species to adapt precisely to different environments and to rapid changes in light and redox conditions.
Materials and methods
Bacterial strains and growth conditions
Ten different strains of Rps. palustris were used: the CEA001 strain (Giraud et al, 2004); the CGA009, HaA2 and BisB5 strains (kindly provided by Dr Harwood, Department of Microbiology, University of Iowa, USA); the DSM126, DSM131, DSM7375 and DSM8283 strains (purchased from the DSMZ Collection) and the LMG4316 and LMG4317 strains (purchased from the BCCM/LMG Bacteria Collection). Cells were grown in liquid medium under different light or oxygen conditions, as previously described (Giraud et al, 2004). Light was provided by light-emitting diodes emitting at 700 or 770 nm (ELD700-524 and ELD 770-524 from Roithner), with an irradiance of 200 and 100 μmol of photon/m2/s, respectively.
Isolation of RpBphP4 and sequencing
The RpBphP4 gene from different Rps. palustris strains was amplified by PCR, using the following primers: pucB.e.f (5′-CAACGACGAGTCGCCTCGCGTTG-3′)/rpa1489.r (5′-AGCGCCTTCATGCCGCTGAGCAC-3′). The sequences of the PCR products have been submitted to the GenBank databases under the following accession numbers (CEA001, EF010855; LMG4317, EF010856; DSM126, EF010857; DSM8283, EF010858; DSM7375, EF010859; LMG4316, EF01860; DSM131, EF010861).
Expression and protein procedures
The RpBphP4 gene was amplified by PCR from Rps. palustris genomic DNA from the CEA001, HaA2 and Bis5 strains and the rpa1489 gene from CEA001. Primers designed to add appropriate restriction sites for expression as His6-tagged versions in the pBAD/HisB expression vector (Invitrogen) were used. In order to reconstitute RpBphP4 holobacteriophytochromes in vivo, the hmuO gene from Bradyrhizobium ORS278 was inserted in the above pBAD∷RpBphP4 constructs (described in Giraud et al, 2004). The recombinant proteins were overexpressed in E. coli LMG194 and purified, as previously described (Giraud et al, 2004). To test the reduction/oxidation of RpBphP4 proteins, reducing conditions were created with 1 mM DTT and oxidizing conditions with 1 mM K3Fe(CN)6. After addition of the oxidizing or reducing reagents, the protein samples were incubated for at least 30 min on ice and then analyzed by non-reducing SDS–PAGE.
Site-directed mutagenesis
Single or double mutations were introduced into the achromo-RpBphP4 protein from CEA001 using the QuickChange(tm) site-directed mutagenesis kit (Stratagene), according to the manufacturer's recommendations. To construct the achromo-RpBphP4 C472S and C722S mutants, the plasmid pBAD∷RpBphP4 from CEA001 was used as the template with the following primer pairs: RpBphP4-C472Ssens (5′-GAACGCCGAAAGGCAGCGCGTCGAGCTGG-3′)/BphP4-C472Santisens (5′-CCAGCTCGACGCGCTGCCTTTCGGCGTTC-3′) and BphP4-C722Ssens (5′-CGAGGATGACGGCTCAGGCTTCGTCACCG-3′)/BphP4-C722Santisens (5′-CGGTGACGAAGCCTGAGCCGTCATCCTCG-3′). To construct the double mutant, the pBAD∷RpBphP4-C472 plasmid was used as the template with the BphP4-C722Ssens/BphP4-C722Santisens primer pair.
Sulfhydryl alkylation
To alkylate protein thiol groups, purified achromo-RpBphP4 was treated with 10 mM DTT for 30 min. DTT was removed from the sample on a desalting column and the reduced achromo-RpBphP4 was then incubated at room temperature in the presence of 10 mM iodoacetamide for 3 h. Finally the alkylated sample was thoroughly dialyzed against 20 mM Tris–HCl (pH 8), 50 mM NaCl.
Gel mobility shift assay and DNase I footprint
Five probes corresponding to the pucBA promoter regions identified in Rps. palustris CGA009 were prepared by PCR using 32P 5′-end-labeled oligonucleotide primers, as previously described (Giraud et al, 2004). For gel mobility shift assays, the purified Rpa1489 protein (0.5 μM) was added to 20 μl of reaction buffer composed of 5 fmol of 32P-labeled DNA probe, 1 μg of polydIdC as competitor, 50 mM Tris–HCl (pH 8), 1 mM DTT, 50 mM potassium acetate, 5 μg of bovine serum albumin and 10% glycerol. The reaction mixture was incubated at room temperature for 30 min and was subjected to non-denaturing 5% Tris–glycine–EDTA-buffered polyacrylamide gel electrophoresis at 4°C for 4 h at 70 V. DNase I footprint experiments were performed as previously described (Giraud et al, 2004).
Protein kinase assays
Protein kinase reactions were performed as previously described (Giraud et al, 2005). For the phosphotransfer experiments, achromo- and chromo-RpBphP4 were phosphorylated for 15 min in the presence of γ-32P-ATP, under reducing conditions or following a 705 nm illumination, respectively. This phosphorylation step was followed by a 25 min incubation in the presence of a stoichiometric amount of Rpa1489. 32P-labeled products were quantified with a Typhoon Phosphor-Imager (Amersham Biosciences).
Construction of RpBphP4 mutants
To create RpBphP4-null mutants for strains HaA2 (RpBphP4.HaA2) and BisB5 (RpBphP4.BisB5), a fragment of each gene was deleted (0.35 kb BamHI for RpBphP4.HaA2 and 50 bp SalI for RpBphP4.BisB5) and replaced by the lacZ-Kmr cassette of pKOK5 (Kokotek and Lotz, 1989). The RpBphP4.CEA001-null mutant was obtained by inserting the lacZ-Kmr cassette directly into the unique BamHI site of the RpBphP4.CEA001 gene. These constructs were introduced into the pJQ200 suicide vector (Quandt and Hynes, 1993) and delivered by conjugation into the corresponding Rps. palustris strains, as described (Giraud et al, 2004). Double recombinants were selected on sucrose and confirmed by PCR.
Absorbance and fluorescence measurements
Absorbance and fluorescence spectra of purified chromo-RpBphP4 from HaA2 and BisB5 were recorded as previously described (Giraud et al, 2005). Excitation was provided by light-emitting diodes emitting at 770 or 705 nm (ELD770-524 and ELD 700-524 from Roithner), with an irradiance of 15 μmol of photon/m2/s.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Acknowledgments
We thank Professor C Harwood (University of Iowa, USA) for the generous gift of the CGA009, HaA2 and BisB5 strains. LV is indebted to the French Ministry of National Education, Higher Education and Research for a doctoral grant.
References
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414: 776–779 [DOI] [PubMed] [Google Scholar]
- Borucki B, von Stetten D, Seibeck S, Lamparter T, Michael N, Mroginski MA, Otto H, Murgida DH, Heyn MP, Hildebrandt P (2005) Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J Biol Chem 280: 34358–34364 [DOI] [PubMed] [Google Scholar]
- Butler WL, Lane HC (1965) Dark transformations of phytochromes in vivo. II. Plant Physiol 40: 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilfeld P, Rüdiger W (1985) Absorption spectra of phytochrome intermediates. Z Naturforsch 40C: 109–114 [Google Scholar]
- Elsen S, Jaubert M, Pignol D, Giraud E (2005) PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol Microbiol 57: 17–26 [DOI] [PubMed] [Google Scholar]
- Elsen S, Swem LR, Swem DL, Bauer CE (2004) RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol R 68: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K, Fordham-Skelton AP, Mistry H, Reynolds CD, Lawless AM, Papiz MZ (2005) A bacteriophytochrome regulates the synthesis of LH4 complexes in Rhodopseudomonas palustris. Photosynth Res 85: 169–180 [DOI] [PubMed] [Google Scholar]
- Evans K, Grossmann G, Fordham-Skelton AP, Papiz MZ (2006) Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state. J Mol Biol 364: 655–666 [DOI] [PubMed] [Google Scholar]
- Evans MB, Hawthornthwaite AM, Cogdell RJ (1990) Isolation and characterisation of different B800-850 light-harvesting complexes from low-and high-light grown cells of Rhodopseudomonas palustris, strain 2.1.6. Biochim Biophys Acta 1016: 71–76 [Google Scholar]
- Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Verméglio A (2002) Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417: 202–205 [DOI] [PubMed] [Google Scholar]
- Giraud E, Zappa S, Jaubert M, Hannibal L, Fardoux J, Adriano J-M, Bouyer P, Genty B, Pignol D, Verméglio A (2004) Bacteriophytochrome and regulation of the synthesis of the photosynthetic apparatus in Rhodopseudomonas palustris: pitfalls of using laboratory strains. Photochem Photobiol Sci 3: 587–591 [DOI] [PubMed] [Google Scholar]
- Giraud E, Zappa S, Vuillet L, Adriano J-M, Hannibal L, Fardoux J, Berthomieu C, Bouyer P, Pignol D, Verméglio A (2005) A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J Biol Chem 280: 32389–32397 [DOI] [PubMed] [Google Scholar]
- Hartigan N, Tharia HA, Sweeney F, Lawless AM, Papiz MZ (2002) The 7.5-Å electron density and spectroscopic properties of a novel low-light B800 LH2 from Rhodopseudomonas palustris. Biophys J 82: 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert M, Lavergne J, Fardoux J, Hannibal L, Vuillet L, Adriano J-M, Bouyer P, Pignol D, Giraud E, Verméglio A (2007) A singular bacteriophytchrome acquired by lateral gene transfer. J Biol Chem 282: 7320–7328 [DOI] [PubMed] [Google Scholar]
- Jaubert M, Zappa S, Fardoux J, Adriano J-M, Hannibal L, Elsen S, Lavergne J, Verméglio A, Giraud E, Pignol D (2004) Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J Biol Chem 279: 44407–44416 [DOI] [PubMed] [Google Scholar]
- Jiang ZY, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE (1999) Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285: 406–409 [DOI] [PubMed] [Google Scholar]
- Karniol B, Viestra RD (2003) The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc Natl Acad Sci USA 100: 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Wagner JR, Walker JM, Vierstra RD (2005) Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J 392: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokotek W, Lotz W (1989) Construction of a lacZ-kanamycine–resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84: 467–471 [DOI] [PubMed] [Google Scholar]
- Lamparter T, Michael N, Mittmann F, Esteban B (2002) Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc Natl Acad Sci USA 99: 11628–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS (2004) Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotech 22: 55–61 [DOI] [PubMed] [Google Scholar]
- Masuda S, Dong C, Swem D, Setterdahl AT, Knaff DB, Bauer CE (2002) Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc Natl Acad Sci USA 99: 7078–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa EG, Atkinson MR, Kamberov ES, Ninfa AJ (1993) Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol 175: 7024–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ (2003) Thiol-based regulatory switches. Annu Rev Genet 37: 91–121 [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF (1993) Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127: 15–21 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, Knaff DB, Zaleski JM, Bauer CE (2003) Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J 22: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros MH, Waterkamp K (1989) Multiple copies of the coding regions for the light-harvesting B800-850 alpha- and beta-polypeptides are present in the Rhodopseudomonas palustris genome. EMBO J 8: 1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438: 325–331 [DOI] [PubMed] [Google Scholar]
- Yang Y, Inouye M (1991) Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci USA 88: 11057–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4


