Abstract
Septins are conserved GTPases that form heteromultimeric complexes and assemble into filaments that play a critical role in cell division and polarity. Results from budding and fission yeast indicate that septin complexes form around a tetrameric core. However, the molecular structure of the core and its influence on the polarity of septin complexes and filaments is poorly defined. The septin complex of the nematode Caenorhabditis elegans is formed entirely by the core septins UNC-59 and UNC-61. We show that UNC-59 and UNC-61 form a dimer of coiled-coil-mediated heterodimers. By electron microscopy, this heterotetramer appears as a linear arrangement of four densities representing the four septin subunits. Fusion of GFP to the N termini of UNC-59 and UNC-61 and subsequent electron microscopic visualization suggests that the sequence of septin subunits is UNC-59/UNC-61/UNC-61/UNC-59. Visualization of GFP extensions fused to the extremity of the C-terminal coiled coils indicates that these extend laterally from the heterotetrameric core. Together, our study establishes that the septin core complex is symmetric, and suggests that septins form nonpolar filaments.
Keywords: cell polarity, coiled coil, cytoskeleton, electron microscopy, septin
Introduction
The septins form a conserved family of GTPases found in fungi and animal cells (Kinoshita, 2003). During cell division, septins localize to sites of cytokinesis and are essential for this process in budding yeast, Drosophila embryos and mammalian tissue culture cells (Faty et al, 2002). In budding yeast, septins form a ring at the mother-bud neck composed of parallel arrays of filaments (Byers and Goetsch, 1976). This septin ring is closely associated with the plasma membrane and forms a scaffold to recruit myosin II and other cytokinetic factors to the future site of cleavage (Longtine and Bi, 2003). In addition, the septin ring contributes to the formation of a lateral diffusion barrier on the plasma membrane, which helps to maintain polarity factors in the bud (Barral et al, 2000; Takizawa et al, 2000). In metazoans, septins are also thought to be required for compartmentalization of the cell cortex (Schmidt and Nichols, 2004; Joo et al, 2005) and have been implicated in myriad cellular processes, including spindle assembly and orientation (Kusch et al, 2002; Spiliotis et al, 2005), exocytosis and vesicle trafficking (Hsu et al, 1998; Beites et al, 1999), cell migration (Finger et al, 2003), and apoptosis (Larisch et al, 2000; Gottfried et al, 2004). However, despite their important roles, the molecular properties of septins are poorly understood.
The septins share a common core consisting of a short N-terminal polybasic motif involved in phospholipid binding (Zhang et al, 1999), a central GTPase domain, a septin unique element/domain (SUE/SUD) of unknown function (Versele et al, 2004; Russell and Hall, 2005), and, in nearly all cases, a predicted coiled-coil domain at the C terminus (Versele et al, 2004). These proteins form hetero-oligomeric complexes, which are believed to be the basic repeating unit of the septin filament (Versele and Thorner, 2004). However, although the pairwise interactions between septin subunits have been identified (An et al, 2004; Versele et al, 2004; Low and Macara, 2006), the organization of monomers within the septin complex remains unclear. In particular, it is unclear whether the molecular arrangement within the core septin complex results in polar or nonpolar higher-order structures.
Data from budding yeast suggest that septins may form polar complexes. For example, the protein phosphatase regulator Bni4 localizes predominantly to the mother side of the septin ring (DeMarini et al, 1997; Kozubowski et al, 2003), whereas the septin-dependant kinase Hsl1 is found mainly on the daughter side (Barral et al, 1999). These observations suggest that the two faces of the septin ring offer different binding sites, and that septin filaments are therefore intrinsically polar. Alternatively, the asymmetric localization of these factors might reflect a septin-independent mechanism of polarity at the mother-bud neck.
The number of septins varies between organisms and cell types. For example, there are 7 septin genes in Saccharomyces cerevisiae (two of which function specifically in meiosis), 4 in S. pombe, and 13 in humans (Kinoshita et al, 2002; Sheffield et al, 2003; An et al, 2004). One possibility is that the increased number of septins observed in mammalian cells reflects the increased functional variability of septin complexes in these organisms. Interestingly, despite the high degree of conservation between septins, they do not assume equivalent roles within the complex. In each case, a pair of septins forms a tetrameric core for the complex (Versele and Thorner, 2005). For example, only two of the five mitotic yeast septins, Cdc3 and Cdc12, are essential for vegetative growth, suggesting that they are crucial for septin complex assembly. Binding studies using recombinant proteins have shown that these subunits form the tetrameric core of the yeast septin complex (Versele et al, 2004; Farkasovsky et al, 2005). The closest S. pombe homologues of Cdc3 and Cdc12, Spn1, and Spn4, also form the heterotetrameric core of the fission yeast septin subunit (An et al, 2004). Similarly, the mammalian septins Sep2, Sep6, and Sep7 form a hexamer (Kinoshita et al, 2002; Sheffield et al, 2003) in which Sep6 and Sep7 are orthologous to Cdc3 and Cdc12. Thus, despite the variation observed among various septin complexes, the molecular organization of their core is a conserved heterotetramer.
The Caenorhabditis elegans genome encodes two septins, unc-59 and unc-61. Both UNC-59 and UNC-61 have a predicted molecular weight (MW) of 52.9 kDa and share a common domain structure with low complexity regions predicted at the C terminus of UNC-59 and the N terminus of UNC-61 (Figure 1A; Versele and Thorner, 2005). The polybasic region, GTPase domain, and SUD domain of UNC-59 and UNC-61 have ∼40% identity and ∼60% similarity to each other. In vivo, both proteins are present at cleavage furrows and are essential for postembryonic cytokinesis and axon guidance (Nguyen et al, 2000; Finger et al, 2003). Furthermore, the presence of both molecules is required for their functions (Nguyen et al, 2000; Finger et al, 2003), suggesting that they act as an obligate heteromultimer. Thus, C. elegans septins, which are orthologous to Cdc3 and Cdc12, represent a minimal septin subunit and offer a powerful system to study the organization of core septin complexes.
Figure 1.
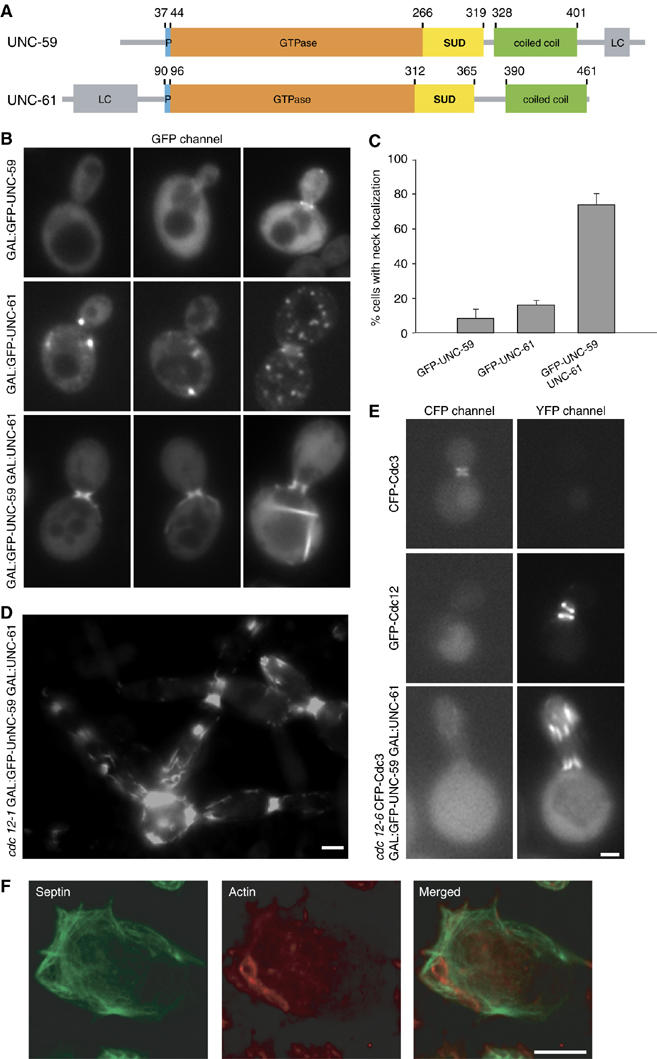
Localization of C. elegans septins expressed in yeast and insect cells. (A) Schematic representation of the domain organization of the C. elegans septins UNC-59 and UNC-61. Both septins contain a polybasic region (P; blue), a GTPase domain (orange), a septin specific domain (SUD; yellow) and a predicted coiled-coil domain (green). The N- and the C-termini of UNC-61 and UNC-59, respectively, contain low complexity regions (LC; gray). Corresponding domain boundaries are indicated by residue positions. (B–E) Localization of C. elegans septins in S. cerevisiae as visualized by fluorescence microscopy. (B) WT yeast cells expressing the indicated combinations of GFP-tagged or untagged UNC-59 and UNC-61. (C) Quantification of budded cells showing septin localization at the bud neck where the indicated septin is expressed. (D) Typical localization of GFP-UNC-59 coexpressed with UNC-61 in the cdc12–1 septin mutant upon overnight incubation at restrictive temperature. (E) Localization of CFP-Cdc3 and GFP-UNC-59 coexpressed with UNC-61 in cdc12–6 cells after growth at 25°C overnight, followed by a 1 h shift to 37°C. As controls, WT yeast expressing either CFP-Cdc3 or GFP-Cdc12 was visualized in both YFP and CFP channels. (F) Visualization of C. elegans septins in SF21 insect cells by indirect immunofluorescence. SF21 cells expressing 6 × His-UNC-59 and UNC-61 were stained with anti-penta-His antibodies (green) and filamentous actin was stained with rhodamin phalloidin (red). The scale bars represent 2 μm in (B, D, E) and 10 μm in (F).
In this paper, we have used a combination of cell biology, biochemistry, and electron microscopy (EM) to gain insight into the molecular organization of the UNC-59/UNC-61 septin complex. We show that UNC-59 and UNC-61 form a heterotetrameric complex capable of associating into higher-order structures in cells and filaments in vitro. In addition, we find that the C-terminal domains of UNC-59 and UNC-61 form parallel two-stranded coiled coils and that the heterotetrameric septin complex is assembled from UNC-59/UNC-61 heterodimers. EM studies show that the UNC-59/UNC-61 heterotetramer is an elongated, rod-like structure composed of two central UNC-61 subunits flanked by two UNC-59 subunits and that the UNC-59/UNC-61 coiled coils extend laterally from the heterotetramer. Together, our results delineate the molecular arrangement of monomers within the septin complex and suggest that septin complexes assemble into filaments and higher-order structures in a nonpolar fashion.
Results
C. elegans septins form higher-order structures in vivo
Although UNC-59 and UNC-61 have been shown to localize to the cleavage furrow in early embryos, it is not clear whether they assemble into filaments in vivo (Nguyen et al, 2000; Finger et al, 2003). To examine their ability to form filaments in cells, we investigated whether they polymerize when expressed in yeast and insect cells. Both septins localized diffusely throughout the cytoplasm and appeared at the bud neck in a small percentage of cells (8% of budded cells; Figure 1B and C). GFP-UNC-61 displayed similar localization and was present with slightly higher frequency at the bud neck (Figure 1B and C). In some cases, punctate structures were observed, which presumably corresponded to septin aggregates. When GFP-UNC-59 and UNC-61 were coexpressed, the number of yeast cells with a GFP-labeled bud neck increased to 73% (Figure 1B and C). In addition, filamentous structures stretching from the bud neck into the mother were frequently observed (Figure 1B). Together, these data indicate that both C. elegans septins incorporate into higher-order structures in cells, and that they form filamentous structures efficiently only when coexpressed. This is in agreement with the observation that UNC-59 and UNC-61 require each other for localization in the worm (Nguyen et al, 2000).
To determine whether UNC-59 and UNC-61 form complexes on their own or require endogenous septins, we investigated whether disruption of yeast septin complexes affected localization of the worm septins. The septin alleles cdc12-1 and cdc12-6 disrupt the assembly and stability, respectively, of yeast septin filaments at the restrictive temperature of 37°C (Dobbelaere et al, 2003). Cells carrying either mutation and expressing both GFP-UNC-59 and UNC-61 were shifted to the 37°C before visualizing GFP-UNC-59 by fluorescence microscopy (Figure 1D and data not shown). A strong GFP signal was still observed at the necks of these cells and in filamentous structures along the cell cortex. Thus, disruption of endogenous septin complexes did not affect the ability of UNC-59 and UNC-61 to assemble into filamentous structures. However, growth and morphological analyses indicated that coexpression of C. elegans septins was unable to complement the lack of endogenous septin structures. In addition, analysis of CFP-Cdc3 localization in cdc12-6 cells coexpressing GFP-UNC-59 and UNC-61 at the restrictive temperature indicated that Cdc3 localization was not restored in these cells (Figure 1E). Together, these results suggest that coexpressed C. elegans septins assemble into filamentous structures that localize to the bud neck of yeast cells independently of endogenous septins.
Similar results were obtained when the two C. elegans septins were coexpressed in insect cells using baculovirus. Again, expression of UNC-59 or UNC-61 alone did not lead to filamentous structure formation (data not shown), whereas coexpression of both proteins did (Figure 1F). Together, these findings establish that UNC-59 and UNC-61 are necessary and sufficient to form higher-order structures in cells.
C. elegans septins form filaments in vitro
To understand complex formation by the C. elegans septins, we next expressed and purified UNC-59 and UNC-61 from Escherichia coli or insect cells. Neither UNC-61 nor UNC-59 was soluble when expressed alone (data not shown). In contrast, coexpression of UNC-59 and UNC-61 resulted in the production of a soluble complex from both bacteria and insect cells. 6 × His-UNC-59 and UNC-61 expressed in either system co-eluted from a Ni-Sepharose column in a 1:1 ratio, indicating that they form a stochiometric complex (data not shown). These results are consistent with both C. elegans septins being required for proper localization (Figure 1B and C) and function (Nguyen et al, 2000).
Dialysis of purified yeast septins from high to low salt leads to assembly of long filaments (Frazier et al, 1998). We therefore tested the ability of bacterially produced UNC-59 and UNC-61 to form filaments under similar conditions. Affinity-purified septin complexes were subjected to size-exclusion chromatography and the main peak fraction was isolated (see below). This fraction was dialyzed from 500 to 20 mM NaCl and filament formation was assessed by high-speed centrifugation and EM. Under these conditions, a few long and rigid filaments of up to 500 nm in length and 8–9 nm in width were observed (Figure 2A). Thus, bacterially expressed septins are able to polymerize in vitro, although with low efficiency.
Figure 2.
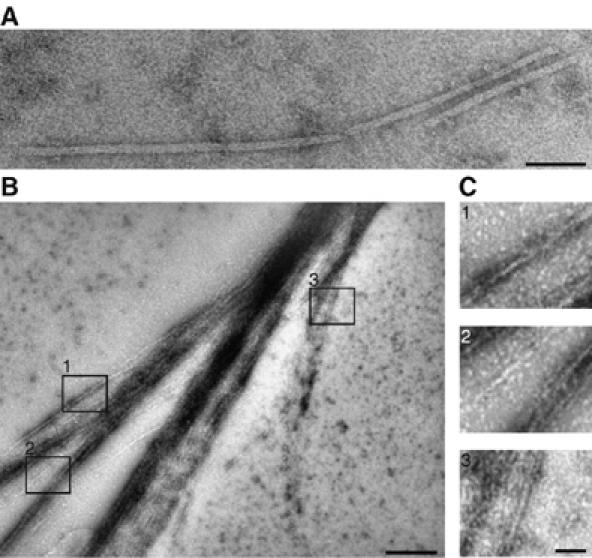
C. elegans septins form filaments in vitro. Electron micrographs of negatively stained septin filaments produced in E. coli (A) and insect cells (B, C) after dialysis from 500 mM NaCl into 20 mM NaCl buffer. (B) Overview of a septin filament sheet. (C) Enlargement of the indicated areas 1, 2, 3 from (B). The scale bar represents 50 nm in (A, C) and 250 nm in (B).
When purified from insect cells, septin complexes assembled into filamentous sheets and bundles in 20 mM NaCl (Figure 2B and C). The number and thickness of these bundles increased, when dialysis was carried out at room temperature (Figure 2B and data not shown). Septin sheets and bundles often terminated in split endings. At these sites, the width of septin filaments measured either 8–9 or 16 nm (Figure 2C). These dimensions are similar to those of individual (7–9 nm) or pairs (14–18 nm) of yeast septin filaments (Frazier et al, 1998; Versele et al, 2004).
The C. elegans septin subunit is a dimer of UNC-59/UNC-61 heterodimers
To obtain a more detailed picture of the C. elegans septin subunit, we assessed nucleotide content and characterized the size and composition of recombinant UNC-59/UNC-61 complexes. In complexes purified from both E. coli and insect cells, low nucleotide content was observed; the ratio of GDP per septin monomer was <0.1 and no GTP was detected in any of the preparations (data not shown), suggesting that recombinant C. elegans septins were substoichiometrically loaded with nucleotide.
Next, we examined the size of the UNC-59/UNC-61 complex. Septins from both bacteria and insect cells eluted in three peaks over gel filtration (Figure 3A). The first peak corresponded to the void volume and contained aggregated septins. Based on the analysis of different fractions on protein gels and subsequent Coomassie staining, this aggregated material represented approximately 5% of septin prepared from E. coli (Figure 3B, lane 1). The second peak contained the largest fraction of the preparation (Figure 3B, lanes 2–4) and was further characterized by sedimentation velocity and equilibrium analytical ultracentrifugation (AUC). The average molecular mass of this fraction was 203 kDa with a sedimentation coefficient of 4S (data not shown), which corresponds to an elongated tetramer of UNC-59 and UNC-61. Based on static light scattering (SLS) analysis, the third peak in the size-exclusion chromatogram corresponds to a dimer (Figure 3B, lanes 5–7) with a molecular weight of 113 kDa. As UNC-59 and UNC-61 co-eluted from affinity- and size-exclusion chromatography at a 1:1 ratio, these results indicate that UNC-59 and UNC-61 contribute an equal number of subunits in both dimeric and tetrameric complexes.
Figure 3.
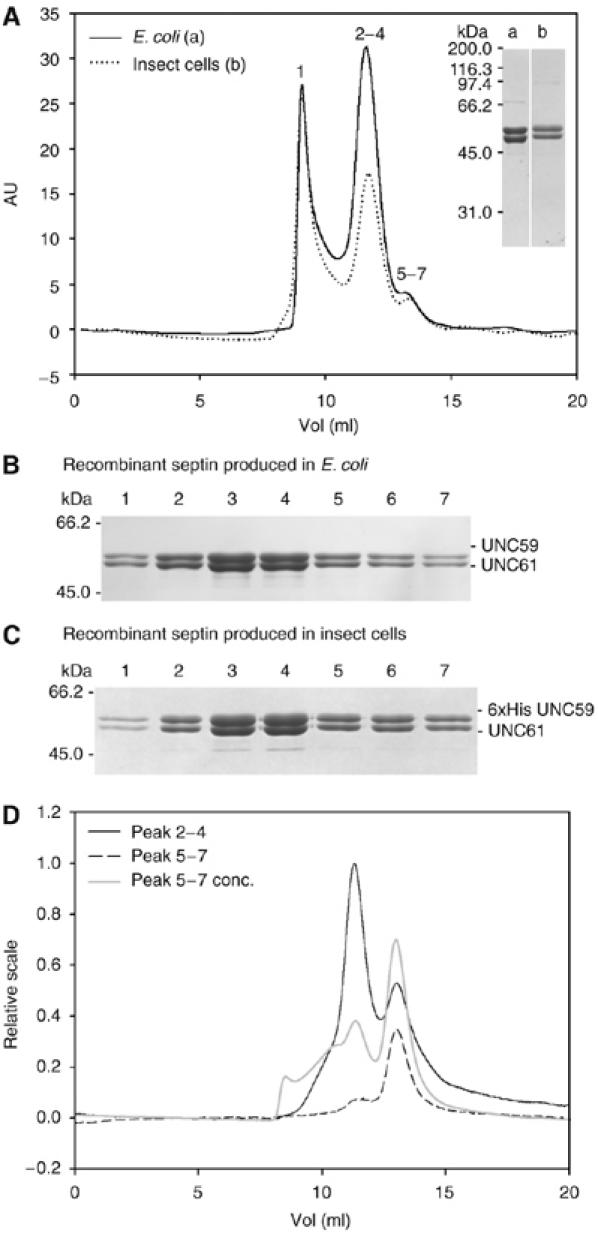
Analysis of the UNC-59/UNC-61 complex by size-exclusion chromatography. (A) Affinity-purified septin complex expressed in E. coli (solid line; 100 μl of 3 mg/ml injected) or in insect cells (dotted line; 100 μl of 1 mg/ml injected) was analyzed on a Superdex 200 column. Numbers on top of the peaks correspond to the lanes in the SDS–PAGE analysis (Coomassie stained) of septins produced in E. coli (B) or in insect cells (C). For SDS–PAGE comparison of septins (6 × His-UNC-59/UNC-61) produced in bacteria (a) or insect cells (b) see inset in (A). (D) Analysis of tetramer and dimer fractions. The tetramer fractions (2–4) (black line; 100 μl of 0.5 mg/ml injected), the dimer fractions (5–7) (black dashed line; 100 μl of 0.05 mg/ml injected) and a fourfold concentrated dimer fraction (gray line; 100 μl of 0.2 mg/ml injected) were rechromatographed on a Superdex 200 column.
To test the stability of the tetramer and dimer complexes, we analyzed these fractions by size-exclusion chromatography. The reapplied tetramer fraction remained primarily tetrameric, although a fraction dissociated into dimers (Figure 3D, black line). Molecular weights were determined by SLS to be 236 and 113 kDa, respectively. Similarly, the reapplied dimer fraction showed some tendency to associate into tetramers (Figure 3D, dashed black line). This tendency was enhanced upon four-fold concentration of the dimer fraction (Figure 3D, gray line).
C. elegans septins coexpressed in insect cells were also analyzed. 6 × His-UNC-59 and UNC-61 co-eluted from a Ni-Sepharose column in a 1:1 ratio. Analysis of this complex by size-exclusion chromatography led to a similar chromatogram (Figure 3A, dotted line) with similar peak distribution and complex stability (Figure 3A and C). Together, these data indicate that septins produced in insect cells and bacteria behave similarly. In both cases, the heterotetrameric complex, which resulted from dimerization of UNC-59/UNC-61 heterodimers, formed the main protein fraction. As the heterotetrameric complex is competent for polymerization (see above and Figure 2), it likely represents the basic building block of septin filaments.
C. elegans septin coiled coils form parallel heterodimers
To delineate the protein–protein interactions within the heterotetrameric septin complex, we next analyzed the role of the UNC-59 and UNC-61 coiled-coil domains. Both septins lacking their coiled-coil domains were insoluble when expressed alone and exhibited a strong tendency to precipitate when coexpressed (data not shown), suggesting that removal of these domains destabilize the proteins. We next investigated the homo- and heterotypic interactions of the coiled-coil domains of UNC-59 (residues 328-401, ccUNC−59) and UNC-61 (residues 390-451, ccUNC−61). At 5°C, ccUNC−59 (9.0 kDa) and ccUNC−61 (8.8 kDa) displayed far-ultraviolet (UV) CD spectra with minima at 205 and 222 nm (Figure 4A), characteristic of proteins with a helical content of 30–35%. Thermal unfolding experiments revealed broad transitions, indicating that the coiled-coil domains are largely denatured at 30°C (Figure 4A, inset). Sedimentation equilibrium experiments of ccUNC−59 and ccUNC−61 at 4°C yielded average molecular masses of 14 kDa in both cases, indicating an equilibrium between monomers and homodimers.
Figure 4.
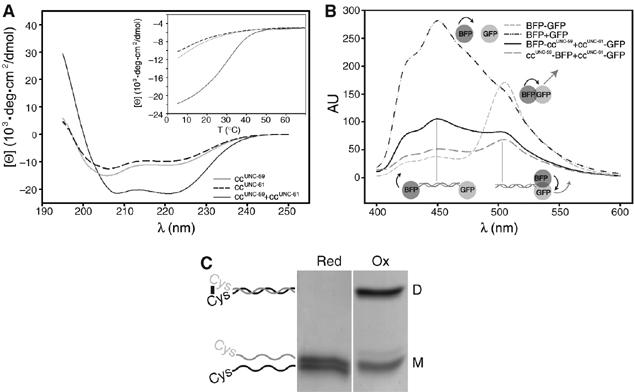
Analysis of the coiled-coil domains of UNC-59 and UNC-61. (A) Far-UV CD spectra of ccUNC−59 (gray line), ccUNC−61 (dashed line), and ccUNC−59 and ccUNC−61 mixed in equimolar ratio (solid line). (B, inset) Thermal unfolding profile recorded by CD at 222 nm. (B, C) Analysis of septin coiled-coil orientation. (B) FRET efficiency of soluble fractions of cells expressing ccUNC−59BFP and ccUNC−61GFP (solid line) or BFPccUNC−59 and ccUNC−61GFP (large dashed line), respectively. Positive control: BFP-GFP (small dashed line); negative control: BFP and GFP (dashed/dotted line). (C) Coomassie blue stained SDS–PAGE of CGGccUNC−59 mixed in equimolar ratio with CGGccUNC−61 under reducing (red) or oxidizing (ox) conditions. The migration positions of monomers (M) and dimers (D) are indicated schematically.
In contrast, the far-UV CD spectrum of a 1:1 mixture of ccUNC−59 and ccUNC−61 suggested a helical content of ∼65% (Figure 4A). Thermal unfolding revealed a sigmoidal shaped melting profile with a midpoint of the transition, Tm, centered at 31°C (Figure 4A, inset). Both the CD spectrum and the melting profile are characteristic for moderately stable α-helical coiled-coil structures (Steinmetz et al, 1998). Sedimentation equilibrium AUC revealed an average molecular mass of 17 kDa, corresponding to a dimer of ccUNC−59 and ccUNC−61 (data not shown). Thus, ccUNC−59 and ccUNC−61 preferentially assemble into heterodimeric coiled-coil structures.
The relative orientation of ccUNC−59 and ccUNC−61 within the heterodimeric coiled coil was determined by fluorescence resonance energy transfer (FRET) and crosslinking experiments. For FRET experiments, ccUNC−59 was labeled at either the N or C terminus with BFP and tested for its ability to act as a FRET donor for ccUNC−61-GFP. Combining ccUNC−59-BFP and ccUNC−61-GFP, but not BFP-ccUNC−59 and ccUNC−61-GFP, resulted in energy transfer (Figure 4B), indicating that the septin coiled-coil domains engage in a parallel fashion. This interpretation was confirmed by cysteine crosslinking experiments (Harbury et al, 1993) in which the ccUNC−59 and ccUNC−61 constructs were engineered to contain an N-terminal cysteine, followed by two flexible glycines (CGGccUNC−59 and CGGccUNC−61). These proteins were mixed in a 1:1 ratio and analyzed on reducing and nonreducing SDS–PAGE gels to separate monomers and disulfide bonded dimers (Figure 4C). Consistent with Cys-Cys disulfide-mediated crosslinking, dimers observed under nonreducing conditions were resolved into monomers under reducing conditions. In contrast, nearly no crosslinking was observed using ccUNC−59GGC and CGGccUNC−61 (data not shown). Together, these data show that ccUNC−59 and ccUNC−61 assemble into heterodimeric, parallel coiled coils.
The septin heterotetramer is an elongated UNC-59/UNC-61/UNC-61/UNC-59 complex
To understand its architecture, we visualized the UNC-59/UNC-61 heterotetramer using EM. Electron micrographs of negatively stained 6 × His-UNC-59/UNC-61 complexes derived from the tetrameric fraction (Figure 3A) showed elongated particles (Figure 5A). After image classification and averaging, these particles appeared as four linearly arranged globular densities, each representing a subunit within the tetramer (Figure 5A, first eight panels). These particles are approximately 17–19 nm long and 5–7 nm wide, similar to the width of 7–9 nm measured for nematode (Figure 2), yeast and mammalian septin filaments (Field et al, 1996; Frazier et al, 1998; Kinoshita et al, 2002; Versele and Thorner, 2004). Panel 9 of Figure 5A shows a particle with only two densities, which most likely corresponds to the dimer observed by size-exclusion chromatography (Figure 3A and D). Together, these findings are consistent with the C. elegans septins forming a linear and stable UNC-59/UNC-61 heterotetrameric subunit.
Figure 5.
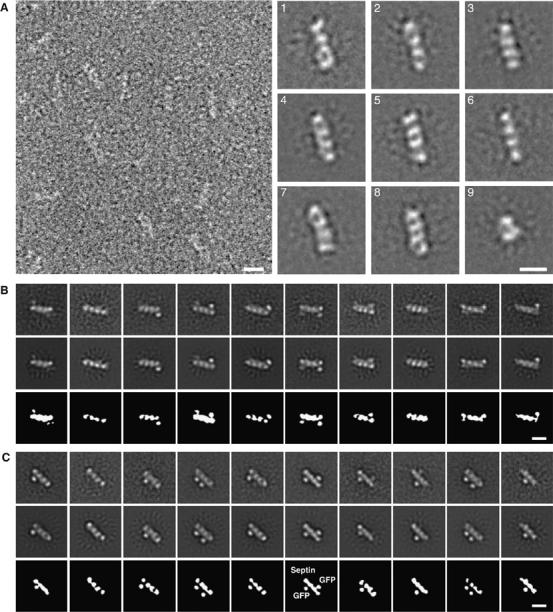
EM of septin complexes. (A) Electron micrograph and projection averages of negatively stained 6 × His-UNC-59/UNC-61 heterotetramers expressed in E. coli. The septin complexes stain poorly, making it difficult to see the linear septin molecules. The first eight panels show averages obtained from representative classes of septin heterotetramers containing four linearly arranged densities. The last panel is an average of heterodimers, which are composed of only two densities. (B) Projection averages of negatively stained GFP-UNC-59 (23–459)/6 × His-UNC-61 heterotetramers. The top panels show ten representative class averages from the multi-reference alignment using randomly selected raw images as initial references. These averages were used as input references for the second round of multi-reference alignment, whose outputs are shown in the middle row. Schematic diagrams of the averages from the second alignment are shown in the bottom row. (C) Projection averages of negatively stained 6 × His-UNC-59/GFP-UNC-61 (77–461) heterotetramers. The top panels show ten representative class averages from the multireference alignment using random references. These averages were used as input references for the second round of multireference alignment, whose outputs are shown in the middle row. Schematic diagrams of the averages from the second alignment are shown in the bottom row. All scale bars represent 10 nm.
To determine the positions of UNC-59 in the tetramer, an N-terminal GFP tag was fused to residues 23–459 of UNC-59. Affinity purified GFP-UNC-59 (23−459)/6 × HIS-UNC-61 behaved similarly to UNC-59/UNC-61, co-eluting during size-exclusion chromatography as a tetramer (data not shown). The GFP-labeled complexes were analyzed by EM (Figure 5B). Class averages revealed densities representing either or both of the GFP domains in addition to the four linear densities of the septin tetramer. Although several classes failed to resolve either of the GFP domains, in many of the class averages the position of at least one GFP was clearly discernable. To improve the signal-to-noise ratio of the averages, we selected 10 representative classes and used them as inputs for a second multireference alignment (Figure 5B, top row). The resulting class averages are very similar to the input images, but with clearer features (Figure 5B, middle row). In these averages, the GFP domain is localized to the terminal densities of the heterotetramer, suggesting that UNC-59 is located at the extremities of the septin complex.
To independently confirm this result, we decorated 6 × His-UNC-59/UNC-61 with Fab fragments directed against the 6 × His-tag at the N terminus of UNC-59 (Supplementary Figure 1). In decorated molecules, the Fab fragment was localized to one or both ends of the septin tetramer, consistent with the GFP-labeling experiment, indicating that UNC-59 localizes to the extremities of the heterotetramers.
We also identified the position of UNC-61 within the heterotetramer using GFP-labeling. To do this, we generated a construct containing an N-terminal GFP tag fused to UNC-61 residues 77–461. 6 × HIS-UNC-59/GFP-UNC-61 (77–461) co-eluted as a tetramer over gel filtration, suggesting that the predicted disordered region of UNC-61 is dispensable for tetramer formation. Purified 6 × HIS-UNC-59/GFP-UNC-61 (77–461) complexes were imaged by EM and particles were aligned using two rounds of multireference alignment as was performed for GFP-UNC-59 (23–459)/6 × HIS-UNC-61. The class averages that were used as the input references for a second multireference alignment are shown in Figure 5C, top row, and the resulting class averages are shown below (middle row). The GFP domain appears to be somewhat flexible. However, it is clearly localized to the central densities, placing the UNC-61 subunits at the center of the heterotetramer.
Taken together, our EM data strongly suggest that the heterotetramer corresponds to the tail-to-tail assembly of heterodimers into an elongated UNC-59/UNC-61/UNC-61/UNC-59 complex and thus is symmetric along its longitudinal axis.
The coiled coils extend laterally from the tetrameric septin core
To address the position of the coiled-coil domains in the heterotetrameric septin complex, we fused GFP to the C terminus of UNC-61, which corresponds to the extremity of the coiled-coil domain (Figure 1A). The GFP was introduced at the terminus of the coiled coil, because a two-stranded coiled coil does not have sufficient mass to be visualized by negative stain EM. Affinity-purified 6 × His-UNC-59/UNC-61-GFP behaved similarly to UNC-59/UNC-61 during size-exclusion chromatography, eluting primarily as a tetramer with a molecular weight of 256 kDa, as determined by SLS (data not shown). Visualization of this complex by EM revealed that the GFP density lies at a distance from the heterotetramer (Figure 6A). Due to this inherent structural variability, most averages resolved only one of the two GFPs, whereas the other one was averaged out. This implies that the two heterodimeric coiled coils extend laterally from the heterotetramer. Reminiscent to the ‘thin strands' previously visualized by rotary metal shadowing EM (Hsu et al, 1998), the coiled-coil domains appear to radiate out from the septin heterotetramer at a variety of angles, resulting in the GFP moiety appearing in various positions relative to the heterotetramer core.
Figure 6.
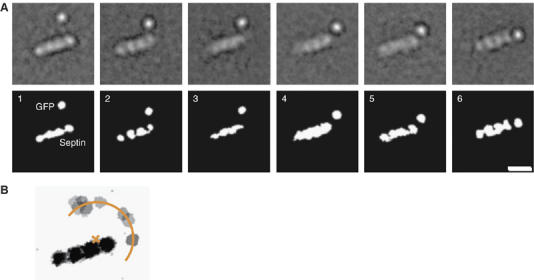
Flexibility of the coiled-coil domain. (A) Six representative class averages of UNC-59/UNC-61-GFP complexes showing one resolved GFP. The GFP is located at various positions relative to the heterotetramer core, demonstrating the flexibility of the coiled-coil/septin core junction. The scale bars represent 10 nm. (B) Schematic representation obtained by overlaying nine class averages of negatively stained 6 × His-UNC-59/UNC-61-GFP. Averages with the one resolved GFP moiety at the maximum distance from the four aligned globular domains were chosen. The coiled-coil domains occupy a semicircle around a heterodimer. The center lies at the interface between the UNC-59 and UNC-61 globular domains. The radius is about 10 nm.
When we considered the averages in which the GFP moiety was located at the maximum distance from the four aligned globular densities, these GFPs formed an arc with a constant radius with respect to the center of either heterodimer core (Figure 6B). The radius of the GFP semicircle is 10 nm, which corresponds to the predicted length of the UNC-59/UNC-61 coiled coil. Thus, these data indicate that the heterodimeric coiled coil emanates from a point between the globular domains of the UNC-59/UNC-61 heterodimer and that the junction between the coiled-coil and globular domains is flexible.
Discussion
Critical aspects of the function and mechanism of a cellular cytoskeletal filament is directly related to the molecular organization of its core components. To date, the precise organization of the septin core structure has not been elucidated. To address this question, we investigated the structural organization of the C. elegans septins UNC-59 and UNC-61. Coexpression of UNC-59 and UNC-61 in S. cerevisae, insect cells or E. coli revealed that these two septins are necessary and sufficient for the formation of higher-order structures in cells and filaments in vitro, and are functionally similar to other, more complex septin assemblies (Field et al, 1996; Frazier et al, 1998; Kinoshita et al, 2002; Mendoza et al, 2002; Sheffield et al, 2003; Versele and Thorner, 2004). In vivo, these heterologous filaments formed and localized independently of endogenous septins. Thus, the C. elegans septins UNC-59 and UNC-61 represent an attractive minimal system to investigate the molecular structure of the septin core.
Our biochemical analysis revealed that the recombinantly expressed UNC-59 and UNC-61 are substoichiometrically loaded with guanine nucleotide (<0.1) and have no detectable GTP. This result is in contrast with studies of the S. cerevisiae septins showing that both endogenous and recombinant complexes are fully complexed with nucleotide and have a GDP:GTP ratio of 2.3:1 (Vrabioiu et al, 2004; Farkasovsky et al, 2005). This difference may be due to a limitation of our expression and purification methods or a species-specific difference between nematode and yeast septins. More specifically, because C. elegans has only two septin genes, it is possible that additional factors are required for full nucleotide binding by UNC-59 and UNC-61. In addition, although the role of GTP binding and hydrolysis remains unclear, it is likely to regulate some aspect of higher-order complex assembly. Thus, the lack of an additional factor may also explain the inefficient filament formation by UNC-59 and UNC-61 we observed (Figure 2).
However, our analysis establishes that UNC-59 and UNC-61 assemble into an obligate heterodimer that, in turn, assembles into a stable heterotetramer (Figure 3). Formation of the heterodimer required interactions between the parallel two-stranded coiled-coil domains of UNC-59 and UNC-61 (Figures 4, 7A and B). These data are consistent with the finding that the hSep6/hSep7 and Cdc3/Cdc12 septin pairs, orthologs of the UNC-61/UNC-59 pair, interact via their coiled coils (Sheffield et al, 2003; Versele et al, 2004). UNC-61/UNC-59 heterodimers dimerize to form an elongated heterotetramer. Along the long axis of the core tetramer, subunit arrangements could result in a symmetric or asymmetric organization of heterodimers.
Figure 7.
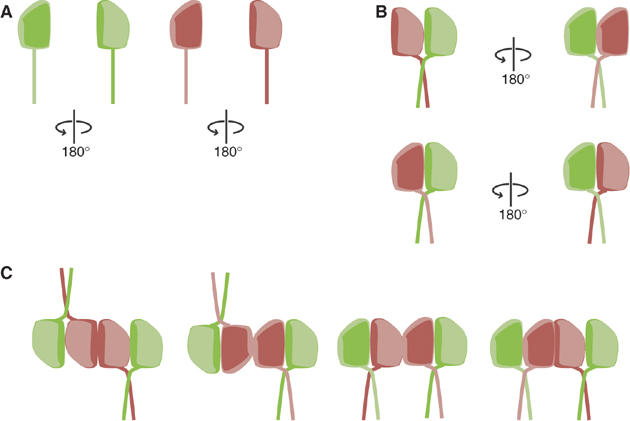
Molecular organization of the heterotetrameric septin core complex. (A) Schematic representations of UNC-59 (green) and UNC-61 (red), showing their GTPase domains and coiled-coil domains (rod). The GTPase domains are not internally symmetric, as represented by the ‘top' face of the protein (arbitrary) shown as pale green or pink, and the ‘bottom' side as dark green or red. (B) The septin heterodimer interacts via its globular domains and its parallel coiled coils. Diagrams show two possible modes of interaction between UNC-59 and UNC-61: ‘top' to ‘top', in which UNC-59 and UNC-61 are in the same orientation (top), and ‘top' to ‘bottom', in which they face opposite directions (bottom). (C) All models for heterotetramer formation require that two of three axes are nonpolar. Two of UNC-59/UNC-61 heterodimers dimerize along their longitudinal axis (X-axis). One two-fold symmetry axis between the two UNC-59/UNC-61 heterodimers is perpendicular to the drawing plane (Z-axis). Thus, the tetrameric septin subunit is nonpolar along X. In the first two models, the tetramer is also symmetric along the Y-axis, but asymmetric along the Z-axis. In the second two models, the tetramer is asymmetric along the y-axis, but symmetric along the Z-axis.
EM analysis of tetramers labeled with GFP on either the UNC-59 or UNC-61 subunits, as well as tetramers decorated with anti-His Fab fragments, indicated that the core septin tetramer is formed by homotypic interactions between UNC-61 subunits along its longitudinal axis, with a two-fold symmetry axis perpendicular to this (Figures 5B, C and 7C and Supplementary Figure 1). The width of the tetrameric subunit (5–7 nm) and the width of a single septin filament (8–9 nm) suggest that the subunits assemble along this longitudinal axis, presumably through UNC-59/UNC-59 interactions. As the two-fold axis of the C. elegans septin subunit is perpendicular to this direction, the resulting filament growth is nonpolar (Figure 7C). Although the coiled-coil domains were not visible in our EM preparations, we were able to visualize the extremity of the coiled coil using a GFP fusion. The resulting images indicated that the coiled coils are flexible around their point of origin and extend laterally from the heterotetramer core (Figure 6). If we assume that filament growth occurs by tetramer addition along the long axis of the core septin, then the coiled coils are available to make lateral contacts with other septin filaments in the context of septin bundles and sheets.
Parallel assemblies of septin filaments form sheets closely associated to the plasma membrane (Byers and Goetsch, 1976). Interestingly, our proposed model (Figure 7) results in an arrangement in which septin sheets have distinct polarities, consistent with data indicating that one face of the septin assembly engages the membrane, whereas the other face is exposed to the cytoplasm. Overall, we propose that, within a septin sheet, all axes within the plane of the sheet are nonpolar, but the axis perpendicular to the sheet is polar.
In conflict with this model, the asymmetric localization of Bni4 and Hsl1 to opposite sides of the septin ring has been proposed to depend on intrinsic septin polarity (Gladfelter et al, 2001). However, we have observed that mutants of Hsl1 that fail to interact with the septin ring localize in a polar manner in the cell and accumulate specifically at the bud cortex (Y. Barral, unpublished result), suggesting that Hsl1 polarity is not provided by intrinsic septin polarity, but rather by a septin-independent mechanism. This conclusion is supported by a recent study of Bni4 and Kcc4 in yeast, suggesting that the actin cytoskeleton is involved in the asymmetric distribution of these proteins (Kozubowski et al, 2005).
Our model of the septin core complex is compatible with genetic and proteomic data reported for the S. cerevisiae septin complex (Versele et al, 2004; Farkasovsky et al, 2005). The two essential septins Cdc3 and Cdc12 form a protease resistant tetrameric core within the nonameric yeast septin complex (Versele et al, 2004; Farkasovsky et al, 2005). However, the entire complex was proposed to be polar (Versele et al, 2004; Farkasovsky et al, 2005). In light of our data, the orthologues of UNC-59 (Cdc12) and UNC-61 (Cdc3) are instead predicted to form a nonpolar tetrameric septin core: Cdc12-Cdc3-Cdc3-Cdc12, flanked by additional septin subunits. It is unclear how these peripheral septins would engage the tetrameric core, but one possibility is that the additional subunits would associate with the core at the extremities of the elongated tetramer. These models are supported by data on the dimensions of the septin complexes; the C. elegans septin core is 5–7 nm wide and 17–19 nm long (Figure 5), whereas the S. cerevisae complex has approximately the same width (7–9 nm), but is longer (32 nm) (Frazier et al, 1998), as would be expected if additional subunits are accommodated at each end.
Our results define the molecular organization of the tetrameric core of the septin complex, suggesting that septin filaments assemble in a nonpolar manner. Together, these findings provide a molecular basis for understanding of the formation and regulation of septin complexes. Importantly, these findings also suggest a mechanism for the formation of other higher-order septin structures containing polar surfaces, including sheets and gauzes.
Materials and methods
Yeast strain construction and fluorescence microscopy
All yeast media and standard genetic techniques were performed as described previously (Ausubel et al, 1987). All yeast strains were in the S288C background. For expression of C. elegans septins in yeast, the sequences encoding full-length unc-59 and unc-61 were cloned into the univector plasmid fusion system (Liu et al, 2000). GFP-unc-59, GFP-unc-61, and loxH-unc-61 were expressed from a centromeric plasmid under the control of the GAL1–10 promoter. Wild type (WT), cdc12–1, and cdc12–6 yeast strains (Dobbelaere et al, 2003) were transformed with pGFP-unc-59 (pFM250), pGFP-unc-61(pFM247), or pGFP-unc-59 and ploxH-unc-61 (pFM296). Fluorescence microscopy was carried out as described previously (Kusch et al, 2002).
For colocalization of endogenous CFP-Cdc3 with GFP-UNC-59 and UNC-61, pGFP-unc-59 and ploxH-unc-61 were transformed into either WT or cdc12–6 strains containing CFP-CDC3 integrated at the URA3 locus (Dobbelaere et al, 2003). Expression of unc-59 and unc-61 was induced by growth on galactose plates overnight at 25°C. CFP- and GFP-labeled proteins were visualized using a BX50 microscope (Olympus) equipped with 425 nm and 504 nm filters and a monochromator (TILLphotonics) as a light source. As a control, WT strains expressing GFP-CDC12 or CFP-CDC3 and cdc12–6 expressing CFP-CDC3 were used. Pictures were taken with an IMAGO cooled CCD camera (TILLphotonics).
Insect cell culture
6 × His-unc-59 and 6 × His-unc-61 were coexpressed in SF21 insect cells using the Multibac baculovirus expression system (Berger et al, 2004). Protein purification, using a metal affinity column and size-exclusion chromatography, was performed exactly as for bacterially expressed protein (see below). The 6 × His-tag was not removed.
Immunofluorescence
At 72 h post-infection, insect cells were seeded on glass coverslips and processed for immunofluorescence essentially as described previously (Reinsch and Karsenti, 1994). Cells were labeled with penta-HIS antibody (dilution 1:150, Qiagen), followed by incubation with Alexa488-labeled secondary antibody (Molecular Probes) to visualize UNC-59 and UNC-61, and with Rhodamine-phalloidin (Molecular Probes) diluted in blocking solution to visualize filamentous actin. Pictures were acquired using confocal microscopy (LSM 510, Zeiss).
Construction of expression plasmids
For expression of full-length unc-59 and unc-61, the sequences encoding amino acids 1–459 of unc-59 and 1–462 of unc-61 were PCR amplified from C. elegans cDNA clones yk465c12 or yk109f1, respectively (kind gift from Yuji Kohara, Genome Biology Lab, National Institute of Genetics, Mishima, Japan) and cloned into a pET15b (Novagen) derivative to generate plasmids pCJ37 (6 × His-unc-59), pCJ38 (6 × His-unc-61), and pCJ43 (6 × His-unc-59 + unc-61). Sequences encoding the predicted coiled-coil domains of unc-59 (residues 328–401) and unc-61 (residues 390–451) were PCR amplified and cloned into vector pPEP-2 (Brandenberger et al, 1996), containing a 6 × His tag, fusion protein, and a thrombin cleavage site, to generate plasmids pCJ50 and pCJ51, respectively. The same sequences were inserted into vectors containing GFP and BFP (vectors kindly provided by I. Berger, Berger et al, 2003) to generate plasmids coexpressing ccUNC−59-BFP and ccUNC−61-GFP (pCJ68) and BFPccUNC−59 and ccUNC−61GFP (pCJ67).
To generate plasmid pCW156, the sequences encoding residues 1–461 of unc-61 and GFP fused to residues 23–459 of unc-59 were inserted into vector pDEST-PCA (kind gift of James Berger). Similarly, to generate plasmid pCW211, the sequences encoding residues 1–459 of unc-59 and GFP fused to residues 77–461 of unc-61 were inserted into the same vector.
Protein purification
The E. coli host strain JM109(DE3) (Promega) was used for expression. Bacterial cultures coexpressing either 6 × His-UNC-59/UNC-61 or 6 × His-UNC-59/UNC-61-GFP were induced at A600=0.6 with 1 mM IPTG for 5 h at 25°C. Purification was carried out on Ni2+-Sepharose (GE Healthcare) under native conditions at 4°C as described in the manufacturer's instructions. Where indicated, the 6 × His-tag was removed. Proteins were dialyzed against high salt buffer (HSB: 20 mM HEPES 2 (pH 7.5), 500 mM NaCl, 1 mM DTT). After sedimentation (see below), the supernatant was applied to a Superdex 200 10/300 column (GE Healthcare). Peak fractions were collected, concentrated, and analyzed in downstream applications.
Bacterial cultures expressing the coiled coils were grown to A600=0.6 and induced with 1 mM IPTG for 5 h at 37°C. Metal affinity purification was carried out under denaturing conditions. The N-terminal tag was removed by thrombin cleavage.
For purification of GFP-UNC-59 (23–459)/6 × His-UNC-61 (1–462) and 6 × His-UNC-59 (1–459)/GFP-UNC-61 (77–462) complexes, cells were grown at 37°C to A600 = 0.3, shifted to 20°C, and induced with 0.25 mM IPTG for 12–16 h. After centrifugation, cells were resuspended in lysis buffer (50 mM HEPES (pH 7.5), 800 mM NaCl, 10% glycerol, 10 mM imidazole, 5 mM beta-mercaptomethanol, and 1 mM PMSF) and lysed by sonication. Complexes were purified using Ni-affinity chromatography (GE Healthcare), concentrated, and applied to a Superdex200 (10/300) in HSB.
Biochemical and biophysical characterization
Sedimentation experiments and filament preparation. Septin complexes from E. coli or insect cells (0.5–0.8 mg/ml) were dialyzed into low salt buffer (LSB: HSB with 20 mM NaCl) at 4 or 22°C for 12–16 h, then sedimented at 90 000 r.p.m. for 15 min at 4°C in a Beckman Optima TLX ultracentrifuge equipped with a TLA 100.1 rotor. Pellet and supernatant were analyzed by SDS–PAGE.
Nucleotide analysis. Nucleotide analysis using a C18 reversed-phase column was performed as described previously (Tucker et al, 1986). Alternatively, nucleotide was analyzed after protease treatment (Suter et al, 2006) with the following modifications: protein samples in buffer A (125 mM NaCl, 5 mM HEPES (pH 7.5), 0.25 mM DTT) were heated to 100°C for 5 min or treated with ProteinaseK for 30 min at 50°C before centrifugation using a table top centrifuge (Eppendorf) at 13 200 g for 5 min. GDP and GTP were separated and quantified on an anion exchange column (Nucleosil 4000–7 PEI, %0/4, Macherey-Nagel) with a linear gradient (0–1.5 M NaCl in 10 mM Tris–HCl, pH 8.0) using an HPLC system equipped with two independent UV-visible spectrometers (Shimadzu). Elution of samples was monitored at 259 nm.
Analytical ultracentrifugation. For AUC, UNC-59/UNC-61 was dialyzed into HSB; ccUNC−59 and ccUNC−61 were dialyzed into PBS. Measurements were carried out at a concentration of 0.5 mg/ml. Analysis was performed as described previously (Fitzgerald et al, 2004). Equilibrium experiments were carried out at 9000, 12 000, and 14 000 r.p.m. for UNC-59/UNC-61 and at 40 000, 45 000 and 50 000 r.p.m. for ccUNC−59 and ccUNC−61. Four individual sedimentation velocity experiments of UNC-59/UNC-61 were carried out at 30 000 r.p.m. under identical solution conditions as the equilibrium measurements.
Static light scattering. SLS experiments were performed using a miniDAWN TriStar with Optilab rex refractometer (Wyatt) coupled to a Superdex 200 10/30 gel filtration column on an Agilent 1100 Series HPLC. Molecular masses were calculated using the Wyatt Astra version 4.90.08 software package.
Circular dichroism spectroscopy. For CD spectroscopy, protein samples were in PBS. Far-UV CD spectra and thermal unfolding profiles were recorded on a J-810 spectropolarimeter (Jasco Inc.) as described previously (Honnappa et al, 2005).
Coiled coil orientation. FRET experiments were carried out as described previously (Berger et al, 2003; Philipps et al, 2003). Clarified cell extracts in PBS containing ccUNC−59-BFP and ccUNC−61-GFP or BFP-ccUNC−59 and ccUNC−61-GFP were normalized to similar levels of GFP emission, and analyzed for FRET efficiency. Cysteine crosslinking experiments were carried out as described previously (Harbury et al, 1993). CGG septin coiled-coil combinations (CGG-ccUNC−59 with CGG-ccUNC−61 or ccUNC−59-CGG with CGG-ccUNC−61) in 8 M urea were dialyzed into PBS buffer and analyzed by SDS–PAGE under reducing and nonreducing conditions.
Electron microscopy and image processing. Septin filaments were adsorbed on glow discharged, carbon coated copper grids, and negatively stained with 2% uranyl acetate. Electron micrographs were obtained using a Philips Morgagni Transmission Electron Microscope operated at 80 kV, which was equipped with a Megaview III CCD camera.
Septin tetramers (3 μl, 0.05–0.1 mg/ml) were applied to glow discharged carbon-coated copper grids, washed in two drops of deionized water and stained in two drops of 0.75% uranyl formate (Ohi et al, 2004). Imaging was performed with a FEI Tecnai T12 electron microscope operating at an acceleration voltage of 120 kV. Images were taken at × 52 000 at a defocus of −2 μm using low-dose procedures on Kodak SO-163 film. Films were developed for 12 min with full-strength Kodak D-19 developer at 20°C. Films were digitized using a Zeiss SCAI scanner using a step size of 7 μm, and 3 × 3 pixels were binned to produce a pixel size of 4.04 Å.
For native septins, 12 957 particles were manually selected from 32 images using WEB, the display program associated with the image processing package SPIDER (Frank et al, 1996), which was used for all further image processing steps. Selected particles were windowed into 100 × 100 pixel images. Particles were normalized against the background and then subjected to eight rounds of multireference alignment. Each round of multireference alignment was followed by principal component analysis and K-means classification specifying 100 output classes. The references used for the first round of alignment were randomly chosen from the raw images. Nine representative images of the class averages are shown in Figure 5A.
For the GFP-UNC-59 (23–459)/6 × HIS-UNC61 (1–461) complex, 8802 particles were manually selected from 56 images. For the 6 × HIS-UNC-59 (1–459)/GFP-UNC61 (77–461) complex, 7883 particles were selected from 112 images. Particles were windowed into 100 × 100 pixel images and classified into 100 classes as described above. An additional multireference alignment step was added. Ten representative class averages from the final round of the initial multireference alignment were chosen and used as initial models for a second iteration of 10 rounds of multireference alignment. Each round of the alignment was followed by principal component analysis and K-means classification into 10 output classes. The input (top row) and output (middle row) classes of the GFP-UNC-59 (23–459)/6 × HIS-UNC61 (1–461) complexes are shown in Figure 5B and of the 6 × HIS-UNC-59 (1–459)/GFP-UNC61 (77–461) complexes in Figure 5C.
For UNC-59 (1–459)/UNC-61 (1–461)-GFP complexes, 11 092 particles were selected from 59 images, windowed into 100 × 100 pixel images and classified into 200 classes as described above. Six representative classes in which one GFP was resolved are presented in Figure 6A.
The α-His Fab fragment was prepared from an α-His antibody (Serotech) using the ImmunoPure® IgG1 Fab preparation kit (Pierce Biotechnology), according to the manufacturer's instructions. Fab-Septin complexes were formed by incubating 6 × His-UNC-59/UNC-61 with Fab fragments at a molar ratio of 1:4 and 1:8 (single Fab-labeled) or 1:20 (double Fab-labeled) overnight at 4°C in 20 mM HEPES (pH 7.5), 500 mM NaCl, 1 mM DTT (Nakagawa et al, 2005). For the Fab-labeled septins, 1189 singly labeled particles were selected from 41 images, windowed into 100 × 100 pixel images, classified into 20 classes to produce the representative class average shown in Supplementary Figure 1 (panel 1). A total of 364 doubly labeled particles were initially selected from 167 images and windowed into 100 × 100 pixel images. Due to the heterogeneity and poor contrast of the raw images, only the very best 42 particles were selected and classified into four classes using the procedure described. The resulting class average of the doubly labeled molecules is shown in Supplementary Figure 1 (panel 4).
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank I Berger, J Dobbelaere, Y Kohara, and J Berger for strains and reagents. We are grateful to S Honnapa for assistance with CD experiments, B Maco, H Gross and P Tittman for assistance with EM, C Norden for assistance with fluorescence microscopy, and JP Erzberger for assistance with construct design, protein purification, and manuscript preparation. The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from NIH grant GM62580 (to TW). This work was supported by the Swiss National Science Foundation, within the framework of the National Center of Competence in Research Structural Biology program, and ETH funding. RKH is supported by NIH training grant 5T32HD007390. CSW is an EMBO fellow. YB is an EMBO young investigator.
References
- An H, Morrell JL, Jennings JL, Link AJ, Gould KL (2004) Requirements of fission yeast septins for complex formation, localization, and function. Mol Biol Cell 15: 5551–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smth JA, Struhl K (eds). (1987) Current Protocols in Molecular Biology. Harvard Medical School, Boston: John Wiley and Sons [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M (2000) Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell 5: 841–851 [DOI] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M (1999) Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev 13: 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble WS (1999) The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci 2: 434–439 [DOI] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Berger P, Schaffitzel C, Berger I, Ban N, Suter U (2003) Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci USA 100: 12177–12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Kammerer RA, Engel J, Chiquet M (1996) Native chick laminin-4 containing the beta 2 chain (s-laminin) promotes motor axon growth. J Cell Biol 135: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L (1976) A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol 69: 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR (1997) A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol 139: 75–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y (2003) Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell 4: 345–357 [DOI] [PubMed] [Google Scholar]
- Farkasovsky M, Herter P, Voss B, Wittinghofer A (2005) Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol Chem 386: 643–656 [DOI] [PubMed] [Google Scholar]
- Faty M, Fink M, Barral Y (2002) Septins: a ring to part mother and daughter. Curr Genet 41: 123–131 [DOI] [PubMed] [Google Scholar]
- Field CM, al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ (1996) A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol 133: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Kopish KR, White JG (2003) A role for septins in cellular and axonal migration in C. elegans. Dev Biol 261: 220–234 [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ (2004) Reaction cycle of the yeast Isw2 chromatin remodeling complex. EMBO J 23: 3836–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field C (1998) Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol 143: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ (2001) The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol 4: 681–689 [DOI] [PubMed] [Google Scholar]
- Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S (2004) The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J 23: 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO (2005) Structural insights into the EB1-APC interaction. EMBO J 24: 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Hazuka CD, Roth R, Foletti DL, Heuser J, Scheller RH (1998) Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron 20: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Joo E, Tsang CW, Trimble WS (2005) Septins: traffic control at the cytokinesis intersection. Traffic 6: 626–634 [DOI] [PubMed] [Google Scholar]
- Kinoshita M (2003) The septins. Genome Biol 4: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Field CM, Coughlin ML, Straight AF, Mitchison TJ (2002) Self- and actin-templated assembly of Mammalian septins. Dev Cell 3: 791–802 [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Larson JR, Tatchell K (2005) Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol Biol Cell 16: 3455–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Panek H, Rosenthal A, Bloecher A, DeMarini DJ, Tatchell K (2003) A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol Biol Cell 14: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, Meyer A, Snyder MP, Barral Y (2002) Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev 16: 1627–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, Book-Melamed N, Timberg R, Duckett CS, Lechleider RJ, Steller H, Orly J, Kim SJ, Roberts AB (2000) A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol 2: 915–921 [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Liu D, Elledge SJ (2000) Rapid construction of recombinant DNA by the univector plasmid-fusion system. Methods Enzymol 328: 530–549 [DOI] [PubMed] [Google Scholar]
- Longtine MS, Bi E (2003) Regulation of septin organization and function in yeast. Trends Cell Biol 13: 403–409 [DOI] [PubMed] [Google Scholar]
- Low C, Macara IG (2006) Structural analysis of septin 2, 6, and 7 complexes. J Biol Chem 281: 30697–30706 [DOI] [PubMed] [Google Scholar]
- Mendoza M, Hyman AA, Glotzer M (2002) GTP binding induces filament assembly of a recombinant septin. Curr Biol 12: 1858–1863 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T (2005) Structure and different conformational states of native AMPA receptor complexes. Nature 433: 545–549 [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG (2000) The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci 113 (Part 21): 3825–3837 [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T (2004) Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps B, Hennecke J, Glockshuber R (2003) FRET-based in vivo screening for protein folding and increased protein stability. J Mol Biol 327: 239–249 [DOI] [PubMed] [Google Scholar]
- Reinsch S, Karsenti E (1994) Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol 126: 1509–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SE, Hall PA (2005) Do septins have a role in cancer? Br J Cancer 93: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Nichols BJ (2004) A barrier to lateral diffusion in the cleavage furrow of dividing mammalian cells. Curr Biol 14: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Sheffield PJ, Oliver CJ, Kremer BE, Sheng S, Shao Z, Macara IG (2003) Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J Biol Chem 278: 3483–3488 [DOI] [PubMed] [Google Scholar]
- Spiliotis ET, Kinoshita M, Nelson WJ (2005) A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science 307: 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz MO, Stock A, Schulthess T, Landwehr R, Lustig A, Faix J, Gerisch G, Aebi U, Kammerer RA (1998) A distinct 14 residue site triggers coiled-coil formation in cortexillin I. EMBO J 17: 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D (2006) Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J Biol Chem 281: 32207–32216 [DOI] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD (2000) Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290: 341–344 [DOI] [PubMed] [Google Scholar]
- Tucker J, Sczakiel G, Feuerstein J, John J, Goody RS, Wittinghofer A (1986) Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J 5: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, Gullbrand B, Shulewitz MJ, Cid VJ, Bahmanyar S, Chen RE, Barth P, Alber T, Thorner J (2004) Protein–protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol Biol Cell 15: 4568–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, Thorner J (2004) Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol 164: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, Thorner J (2005) Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol 15: 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu AM, Gerber SA, Gygi SP, Field CM, Mitchison TJ (2004) The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J Biol Chem 279: 3111–3118 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS (1999) Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol 9: 1458–1467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


