Abstract
In mammals, perception of pheromones is based on the expression in each vomeronasal sensory neuron of a limited set of receptor genes, chosen among a large repertoire. Here, we report an extremely tight control of the monogenic and monoallelic transcription of the V1rb2 receptor gene. Combining genetic and electrophysiological approaches, we show that the transcription of a non-functional V1r allele leads to the coexpression of another, functional V1r gene. The choice of this coexpressed gene surprisingly includes genes located on the cluster homologous to the one from which the mutant allele is transcribed. However, V1r genes located in cis relative to the transcribed mutant allele are excluded from the coexpression choice. Our observations strongly suggest a monogenic regulatory mechanism acting (a) at a general level, via the expression of the V1r receptor itself, and (b) at a more local level, defined by the V1r gene cluster.
Keywords: gene cluster, gene regulation, monoallelic expression, olfactory receptor, pheromone receptor
Introduction
In most mammalian species, inter-individual interactions related to reproduction are largely dependent on pheromonal communication. In the mouse, the vomeronasal organ (VNO), a chemosensory structure located in the nasal cavity (Figure 1A), represents a major tool for such exchanges (Halpern and Martinez-Marcos, 2003).
Figure 1.
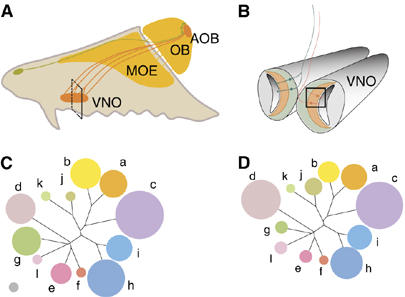
Size and expression variability of the V1r families in the VNO. (A) A schematic representation of a hemisection through the nasal cavity of a mouse. The main olfactory epithelium (MOE) and its projections to the olfactory bulb (OB) are shown in dark yellow. The Gruneberg organ and its projections are indicated in green. The VNO and its axonal projections to the accessory olfactory bulb (AOB) are shown in orange. (B) Coronal section through a mouse VNO, corresponding to the plane indicated in panel A. Basal sensory neurons expressing V2r receptors are shown in green, while apical, V1r- and Gαi2-expressing VSNs are in orange. (C) A tree depicting the percentage of V1rs recognized by our set of in situ probes. The area occupied by the gray disk on the lower left corresponds to 1%. (D) A representation of the proportion of VSNs, which express V1rs from a given family.
Vomeronasal sensory neurons (VSNs) are in contact with the outside environment, and express on their dendrites seven transmembrane receptors encoded by two different superfamilies, V2rs (Herrada and Dulac, 1997; Matsunami and Buck, 1997; Ryba and Tirindelli, 1997) and V1rs (Dulac and Axel, 1995) (Figure 1B). These latter, expressed by VSNs located on the apical portion of the vomeronasal neuroepithelium, have been shown to encode pheromone receptors (Boschat et al, 2002; Del Punta et al, 2002). The mouse genome contains over 150 potentially functional V1r genes (and an equal number of pseudogenes), which are classified into 12 families each containing between one and 30 members (Rodriguez et al, 2002; Zhang et al, 2004). To this already remarkable diversity, which is at least partly the result of positive Darwinian selection, one can add numerous polymorphic allelic variants.
V1r genes are present on most chromosomes, and members of a given family are usually found clustered in the genome. A single V1r cluster of 600 kb located on chromosome 6 has been analyzed in some detail, and contains all members (but one) of the V1ra and V1rb families, intermingled with no apparent specific organization (Del Punta et al, 2000; Lane et al, 2002). It is not known if the organization of V1rs in clusters plays a role in the regulation of their expression, or if each V1r gene behaves as a single independent unit.
A mouse possesses many VSN subpopulations, each composed of only a few hundred neurons expressing monoallelically a given V1r receptor or set of receptors (Rodriguez et al, 1999). Members of each subpopulation project their axons toward the brain, in a specific area of the olfactory bulb, termed the accessory olfactory bulb. There, fibers coalesce into multiple neuropil-rich spherical structures called glomeruli, which are usually exclusively innervated by a functionally identical subpopulation of VSNs (Belluscio et al, 1999; Rodriguez et al, 1999; Wagner et al, 2006). A surprising role is played by V1r receptors at this level: they are involved in the elaboration of the axonal projection map (Belluscio et al, 1999; Rodriguez et al, 1999). V1r receptors are thus both involved in chemodetection and axonal guidance processes.
As for many monoallelically expressed or imprinted genes, including those coding for odorant receptors or interleukins, it has been reported that V1r genes are characterized by asynchronous DNA replication (Singh et al, 2003). This peculiar characteristic is thought to reflect an early tag, of unknown nature, but related to the future expression or non-expression of one of the parental alleles.
We here report that the transcription of a non-functional V1r allele allows the expression of another, functional V1r gene. Unexpectedly, none of the V1r genes located in the V1r cluster containing the expressed non-functional allele is chosen for coexpression, while V1r genes present on the homologous cluster of the other parental chromosome can be chosen. These data strongly suggest that the organization of V1r genes in clusters does not simply reflect evolutionary proximity, but underlies mechanisms regulating monogenic V1r expression.
Results
Expression of V1r families in the VNO
About 150 putative functional V1r genes are present in the mouse genome, and are therefore potentially expressed by VSNs. The transcribed fraction of these 150 genes is unknown.
To obtain a global view of the transcribed V1r repertoire, we opted for an in situ approach. Since V1r sequences are highly divergent between V1r families (Rodriguez et al, 2002), a situation which prevents a single cRNA probe to recognize all V1r transcripts, we selected one or two V1r genes for each of the 12 V1r families, based on intrafamily homologies. This allowed us to design probes that exclusively recognized members of the corresponding family, without binding to V1r genes outside the family (Supplementary Table 1). Sequences corresponding to the V1ra3, V1ra9, V1rb2, V1rb9, V1rc32, V1rd16, V1re4, V1rf4, V1rg1, V1rh5, V1ri1, V1ri8, V1rj3, V1rk1 and V1rl1 genes were used. Since V1r genes pertaining to families V1ra and V1rb are relatively closely related (up to 75% sequence homology), we tested the family specificity of the probes corresponding to these two families by hybridizing V1rb2-expressing VSNs with V1ra3/a9 probes. No cross-hybridization was observed (4000 VSNs analyzed). We also evaluated the potential cross-hybridization between V1ra3/a9 versus V1rg1/e4/f4 probes, and V1ra3/a9 versus V1ri1/i8 probes. Over 4500 VSNs were scored and none reacted with two probes. Control hybridizations using multiple probes at once showed a labeling of over 75% of the apically located, V1r- and Gαi2-expressing VSNs (data not shown).
With these tools in hand, we investigated the potential correlation between the size of a given V1r gene family and the fraction of VSNs expressing its members. We performed in situ hybridizations using family-specific probes on VNO sections, counted the mean number of VSNs recognized by the probe(s) and calculated, for each family, the fraction of the expressed V1r repertoire it represented (Figure 1D and Supplementary Table 1). We then compared this diagram with the V1r repertoire potentially recognized by our probe sets (i.e., V1r genes sharing over 80% of the probe sequences; Figure 1C and Supplementary Table 1).
This approach showed a remarkably good correlation between the size of the V1r gene families and the number of VSNs expressing their members, except for members of families V1re and V1rg, which appear to be rarely chosen for transcription, and members of family V1rj, which are on the contrary often transcribed, relative to the fraction of the repertoire they represent (Supplementary Table 1).
Our probes are able to recognize several members of a given V1r family. But do the in situ signals reflect expression of different V1r genes pertaining to a given family or the frequent choice of a single or a very limited number of genes? To answer this question, we evaluated the transcriptional status of all V1r genes from the V1ra and V1rb families. Taking advantage of the high V1r intrafamily sequence identity, we designed PCR primer pairs specific and exclusive for each of these two V1r families. We then amplified the corresponding cDNAs from adult vomeronasal neuroepithelium and subcloned the amplicons. The sequencing of 35 random clones indicated that the V1ra6, V1ra11, V1ra3, V1ra4, V1ra1, V1ra7, V1rb4, V1rb8, V1rb2 and V1rb1 genes were transcribed, which corresponds to over 60% of the potential V1ra/b repertoire. The relative representation of each of the V1r sequences varied from 1 to 4 (the potential bias of the primers for any given V1r was tested on genomic DNA and was found negligible, data not shown). No single or small group of V1r gene transcripts were drastically overrepresented, indicating that a majority of the potential V1ra and V1rb genes was transcribed by VSNs.
Expression of a single V1r gene per sensory neuron
Monogenic expression of odorant and vomeronasal receptor genes is thought to be the rule in the olfactory system. However, a systematic investigation of this dogma is still lacking, and reports suggest that olfactory receptors may express multiple chemosensory receptor genes. For example, previous data indicate that probes recognizing about 35% of the complete V1r repertoire (families V1ra, b and d) react with most of the Gαi2-expressing VSNs, suggesting a possible coexpression of different V1rs in single sensory neurons (Pantages and Dulac, 2000).
We investigated the monogenic expression of V1rb2, a V1r gene that we studied previously, and for which we generated gene-targeted animals bearing genetic tags (Supplementary Figure S1) (Rodriguez et al, 1999). We used an in situ approach, taking advantage of these tags, which permit the discrimination between neurons expressing V1rb2 and highly related V1rb genes. We tested the possible coexpression of V1rb2 with any member of the V1r superfamily, excluding the family to which V1rb2 belongs (families a, c–l) (Figures 2A–D). A total of 29 240 VSNs hybridizing with the non-V1rb family probes and 6220 reacting with the V1rb2 probe were analyzed, but none exhibited hybridization for both probe sets (Figure 2E). This analysis included 2241 VSNs expressing V1ra genes, genes that are part of the large V1r cluster containing V1rb2.
Figure 2.
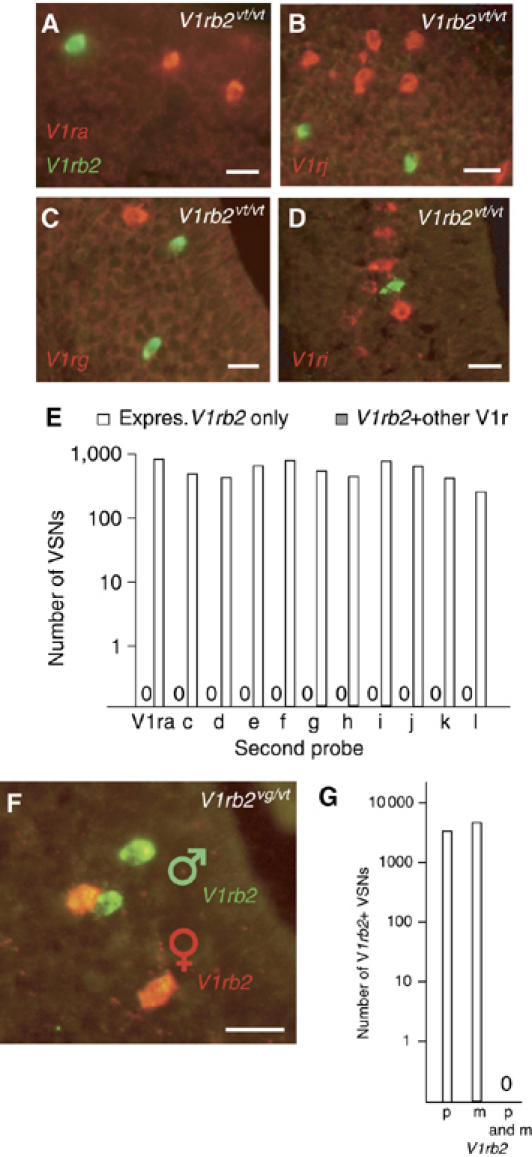
Monogenic expression of V1r genes. (A–D) Representative two-color in situ hybridizations using a probe specific for V1rb2 (green), and probes specific for a given V1r family (red). (E) Empty bars represent the number of VSNs reacting with the V1rb2-specific probe, and potentially with the V1ra-l probes. Dark bars (invisible since no coexpression was found) indicate the number of these V1rb2-expressing VSNs cotranscribing a member of any of the V1r families. (F, G) Monoallelic expression of V1rb2. (F) An in situ hybridization with allele-specific probes for V1rb2 (green and red represent paternal and maternal alleles, respectively) (G) Graph showing the number of VSNs reacting with allele-specific probes for V1rb2 (empty bars), and those coexpressing both alleles (dark bar). Genotypes are shown, in white, in the upper right corner. Scale bar, 30 μm.
We performed a similar analysis with a V1rb2-specific probe and a probe recognizing multiple members of the V1rb family (V1rb9, which also recognizes V1rb2), and found multiple VSNs hybridized exclusively with the V1rb9 probe (data not shown), showing that not all members of a given family are expressed in a given VSN.
The monoallelic expression of the V1rb2 gene was previously reported (Rodriguez et al, 1999). This study however only analyzed the translated products of two tagged V1rb2 alleles (V1rb2vt, Tau::LacZ and V1rb2vg, Tau::GFP; Supplementary Figure S1), and not the transcription of the corresponding mRNAs, leaving post-transcriptional allele-specific silencing as an alternative explanation for the observation. To address this issue, we performed in situ hybridizations with probes specific for either of the two V1rb2 parental alleles on vomeronasal sections from compound heterozygotes, bearing different tags (Tau::LacZ and Tau::GFP) on each of the V1rb2 alleles. We observed 1251 and 1498 VSNs expressing respectively the paternal or the maternal V1rb2 allele, and none expressing both alleles (n=15, seven males and eight females analyzed), confirming the monoallelic transcription of V1rb2 (Figures 2F and G).
Our data thus strongly support a remarkably strict monoallelic and monogenic expression of V1rs in the VNO.
Expression of a null V1r allele allows the coexpression of another V1r gene
VSNs expressing the same V1r gene coalesce into 10–30 glomeruli in the accessory olfactory bulb (Figure 3A) (Belluscio et al, 1999; Rodriguez et al, 1999; Wagner et al, 2006). It was reported that in two mouse lines, carrying non-functional alleles of the V1rb2 and of the V1ra1 receptor genes, the transcription of either of the two nonfunctional genes in a given VSN affected the position of their axonal projection targets, and resulted in a non-convergence of axons from VSNs expressing the deletion alleles (Belluscio et al, 1999; Rodriguez et al, 1999). These observations were suggested to be the result of a lack of targeting information for the misrouted VSNs (information carried by the expressed receptor itself). We analyzed in detail the targeting defect of the V1rb2 deletion line (V1rb2dv), in which the endogenous V1rb2 CDS is replaced by the GFP CDS, leaving all V1rb2 non-coding sequences intact (including introns) (Rodriguez et al, 1999). In this mutant line, we observed that multiple axonal projections emanating from neurons expressing the V1rb2dv allele did enter and arborize into apparently random glomeruli (Figure 3B), similar to what fibers pertaining to VSNs expressing other V1rs would do. This observation and parallel data related to the regulation of odorant receptors suggest a potential coexpression of the mutant V1rb2 allele and of another, possibly functional V1r gene in the same VSN.
Figure 3.

Expression of a deletion V1rb2 allele allows for coexpression of another V1r. (A) Two-photon imaging of a wt glomerulus innervated by V1rb2-expressing axons. (B) Similar imaging of fibers emanating from VSNs expressing the V1rb2dv allele, which enter the accessory olfactory bulb glomerular layer, and apparently innervate random glomeruli. (C–E) In situ hybridizations of VSNs coexpressing the V1rb2dv allele (green) and a member of the V1ra family (red). (F) The number of VSNs coexpressing the V1rb2dv allele together with a member of another receptor family (dark bars). Genotypes are shown, in white, in the upper right corner. Scale bar, 20 μm.
To test this hypothesis, we performed in situ hybridizations on vomeronasal sections from V1rb2dv/dv animals, with probes specific for most of the V1r families (families a–j) (Figures 3C–E). VSNs expressing the V1rb2 deletion allele were visualized (2567 VSNs), and hybridized with probes representing most V1r families. We found VSNs coexpressing the V1rb2 mutant allele and members of families V1ra, V1rb, V1rc, V1rd, V1rg, V1rh, V1ri and V1rj (Figure 3F). The percentage of neurons cotranscribing the V1rb2 mutant allele and another V1r ranged from 0 to 7.4% (V1ra: 1.1%, V1rb: 0.4%, V1rc: 7.4%, V1rd: 1.6%, V1rg: 0.9%, V1rh: 5.3%, V1ri: 1.5%, V1rj: 1.8%, V1re/f: 0%), showing no strong bias for a V1r family to be more likely chosen for coexpression with the V1rb2 mutant allele (after normalization of the frequencies with the number of VSNs recognized by the probes).
We did not detect V1r coexpression in about 80% of the V1rb2dv-expressing VSNs, possibly pointing to a limited time window during which sensory neurons can select a V1r gene, or alternatively to the cotranscription of non-V1r chemoreceptor genes. We tested this latter possibility by investigating the potential coexpression of members of the V2r family, a receptor family also expressed in the vomeronasal neuroepithelium, but in the basal, Gαo-expressing zone. We took advantage of the fact that all V2r-expressing VSNs coexpress the V2r2 gene (Martini et al, 2001), and used a corresponding probe to visualize potential VSNs coexpressing the V1rb2dv allele together with a member of the V2r family. In addition, and independently, we used a V2r probe specific for the V2rb1 receptor gene. A total of 276 VSNs expressing the V1rb2dv allele were scored, and none was found to coexpress V2r2 or V2rb1 (Figure 3F).
We then investigated the possibility that odorant receptors could be expressed subsequently to the transcription of the V1rb2dv allele. First, a probe recognizing the MOR204-6 odorant receptor gene (which is part of a subfamily of 28 members) was used. A total of 52 VSNs expressing the deletion allele were scored, none of them coexpressed MOR204-6 or related odorant receptor genes (Figure 3F). Second, we tested the potential coexpression of the odorant receptor gene MOR18-2, a receptor gene known to be transcribed by VSNs (Levai et al, 2006). A total of 1727 MOR18-2 and 713 V1rb2dv-expressing VSNs were scored, none coexpressing the deletion allele and the odorant receptor gene.
Taken together, our data show that expression of the V1rb2dv allele leads to the expression of other V1rs, and strongly support a role played by the V1r protein as a main player in the monogenic/monoallelic V1r regulation.
Functional chemoreceptors are coexpressed with the deletion allele
Do the newly chosen V1r genes modify the chemosensory capacities of VSNs expressing them? We previously showed that VSNs transcribing a functional V1rb2 gene respond to 2-heptanone, while those expressing a V1rb2 non-functional allele (V1rb2dv) are insensitive to this pheromone (Boschat et al, 2002). We applied multiple chemical compounds (known to activate VSNs) to neurons expressing the V1rb2dv allele, and measured their electrical responses using whole-cell patch-clamp recordings. We analyzed 12 VSNs expressing the non-functional allele, among which one responded with reversible inward currents to α and β-farnesenes (26 pA; holding potential −60 mV) and one to pentyl acetate (72 pA; holding potential −60 mV) (Figures 4B and C). The 10 remaining VSNs did not respond to the potential agonists (Figure 4A). Among 15 VSNs that expressed the wild-type V1rb2 allele, none responded to the compounds, except as expected, to 2-heptanone (Figure 4D), which triggered a response in all VSNs tested.
Figure 4.

Coexpressed V1rs are functional receptors. (A–C) Representative whole-cell patch-clamp recordings of VSNs expressing the V1rb2dv allele to 5″ pulses of 2-heptanone, α and β-farnesenes, 2,5-dimethylpyrazine or pentyl-acetate (10−6 M; holding potential, −60 mV). VSNs did not respond to these agonists (A), or responded to α and β-farnesenes (B), or to pentyl-acetate (C). (D) Only 2-heptanone (10−8 M) induced an inward current in VSNs expressing V1rb2 (V1rb2vg line). Cell viability was assessed by KCl (100 mM) pulses before and after the perfusion with agonists.
Neurons expressing the V1rb2dv allele are therefore able to acquire chemosensory capacities not shared by VSNs, which express a functional V1RB2 receptor, a situation, which very likely reflects the acquisition of novel V1r receptors.
Cluster lock after gene choice and coexpression of genes on homologous clusters
We showed an apparent random coexpression of a V1r gene together with the V1rb2 deletion allele, including V1rs from the V1ra or V1rb families. But are the parental chromosomes containing the newly transcribed V1rs randomly chosen? Previous reports related to the regulation of monoallelically expressed genes suggest that (a) a particular chromatin state surrounding an expressed odorant receptor gene may favor the choice of another gene located closeby, in cis, on the same chromosome (Lewcock and Reed, 2004), and that (b) a transcriptionally active gene is, relative to its homologue located on the other parental chromosome, marked early during development, marking that reflects its future expression or non-expression (Simon et al, 1999; Singh et al, 2003).
A priori, based on the publications mentioned earlier, our results showing the coexpression of the V1rb2dv allele and another V1ra or V1rb gene likely reflect the coactivation of V1r genes located in cis relative to the active V1rb2dv allele. To test this apparently logical conclusion, we opted for the combination of an in situ and a genetic approach, allowing the visualization of V1ra and V1rb genes transcribed from a known parental chromosome.
The entire functional V1ra and V1rb repertoires include nine V1ra and eight V1rb genes, located in a 600 kb V1r cluster on chromosome 6, except for one V1rb member, V1rb9, which is present on the X chromosome. We took advantage of a previously generated mouse line (Supplementary Figure S1, V1rabΔ) which lacks the entire V1ra/b cluster (Del Punta et al, 2002). To identify the parental origin of the V1ra/V1rb genes coexpressed with the V1rb2dv allele, we crossed the V1rabΔ with the V1rb2dv line, generating V1rabΔ;V1rb2dv compound heterozygotes (Figure 5A). To perform hybridizations revealing exclusively V1ra and V1rb transcripts produced by genes surrounding the active V1rb2dv allele, we had to test the following caveats: (a) a potential transcription of the X-linked V1rb9 gene. This turned out not to represent a problem since hybridization of V1rabΔ/Δ VNO sections with a V1rb9/V1rb2 probe set showed no V1rb9 transcripts in mice whose age matched the one chosen for the present study (Figure 5D); (b) a potential general trans-acting V1r control element lying inside the V1ra/b cluster and therefore absent in mice carrying the V1rabΔ deletion; its absence could thus affect the expression of all V1rs. We tested this hypothesis by hybridizing V1rabΔ/Δ VNOs with V1ri1/V1ri8 probes, and observed an unaltered expression profile of this gene family, thus rejecting this hypothesis (Figure 5E); (c) a similar unknown trans-acting control element specific for V1ra and V1rb genes and lying inside the V1rabΔ deletion could, if removed, prevent expression of all members of the V1ra/b cluster present on the other parental chromosome. This was tested by hybridizing V1rabΔ/+ VNOs with V1rb2/V1rb9 probes. V1rb expression was observed, rejecting the hypothesis (Figure 5C). We thus disposed of compound V1rabΔ;V1rb2dv heterozygous mice that lacked a complete set of parental V1ra/V1rb genes, and possessed a functional V1ra/b repertoire limited to the V1ra/V1rb genes surrounding the V1rb2dv allele.
Figure 5.
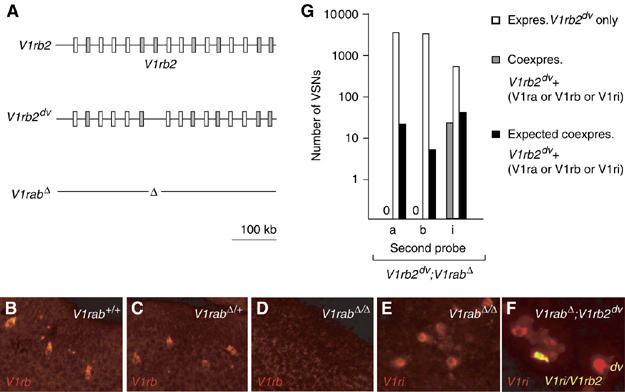
Cluster lock associated with V1r gene expression. (A) A schematic representation of the V1ra/V1rb alleles used. (B–D) In situ hybridizations of V1rab+/+, V1rabΔ/+ and V1rabΔ/Δ VNO sections using V1rb9-V1rb2 probes. V1rb transcripts were identified in V1rab+/+ and V1rabΔ/+ animals, while no expression of V1rb genes was observed in V1rabΔ/Δ mice. (E) in situ hybridization of V1rabΔ/Δ VNO sections using the V1ri1-V1ri8 probe set; expression of V1ri genes was observed in V1rabΔ/Δ mice. (F) V1rabΔ /V1rb2dv VNO sections hybridized with V1ri1-V1ri8 probes. A single VSN (yellow) coexpresses the V1rb2dv allele and a V1ri gene. (G) Graph corresponding to in situ hybridizations, indicating an absence of coexpression between the V1rb2dv allele and V1ra/V1rb genes surrounding it. Expected values of coexpressing VSNs were based on the observed frequencies, for each family, of coexpressing VSNs in V1rb2dv/dv mice. Genotypes are shown, in white, in the upper right corner.
In situ hybridizations on these compound V1rabΔ;V1rb2dv heterozygous mice were performed with V1ri1/V1ri8, V1ra3/V1ra9 or V1rb2/V1rb9 probes. First, using the V1ri probes we confirmed that the non-functional V1rb2 allele in the V1rabΔ;V1rb2dv configuration could be coexpressed with members of the family I (Figure 5F and G). We then analyzed 2513 V1rb2dv-expressing VSNs hybridized with the V1ra3/V1ra9 probes and 2542 V1rb2dv-expressing VSNs hybridized with the V1rb2/V1rb9 probes (Figure 5G). Unexpectedly, we did not find a single VSN coexpressing the V1rb2dv allele and any of the V1ra/V1rb genes surrounding this latter. A comparison of these results with the corresponding hybridizations on V1rb2dv/dv sections (6/637 for V1ra3/V1ra9 and 3/733 for V1rb2/V1rb9, Figure 3F), showed a clear unequal propensity (Fisher exact test, P<0.001), of the V1r genes physically associated with the deletion allele to be co-opted for expression (Figure 5G).
These surprising results show that (a) the co-opted V1r genes can include those located on the homologous cluster present on the other parental chromosome and that (b) V1r genes located in the cluster from which the deletion allele is expressed are not available for coexpression.
Discussion
Tight control of pheromone receptor gene expression
Our results strongly support the fact that a single functional V1r gene and allele is transcribed in each VSN, and show that the expression of a non-functional V1r allele allows neurons to choose another, functional V1r gene. Why express in each VSN a single V1r receptor gene, moreover from a single allele? Monogenic, but also monoallelic expression, are tools to achieve the expression of a single V1r chemodetector ‘morph', since allelic polymorphic variants are common in V1r genes (Rodriguez et al, 1999). In terms of information coding, the use of multiple and narrowly tuned parallel coding lines allows the vomeronasal system to avoid a too complex decoding strategy. But the monogenic expression does not only affect peripheral chemodetection: V1r receptors are indeed involved in axon guidance mechanisms (although their role in this function remains elusive). The coexpression of multiple V1r receptor genes, even highly related, may thus simply be incompatible with the formation of functional topographical maps in the accessory olfactory bulb. This explanation is supported by the fact that, in the main olfactory system, very minor odorant receptor sequence modifications lead to different axonal responses to guidance cues in the olfactory bulb (Feinstein and Mombaerts, 2004). A dual pressure may therefore act on the mechanisms leading to the expression of a single V1r ‘morph' per VSN: on one side to avoid a too complex decoding expense, and on the other side, to facilitate axonal wiring processes.
In the main olfactory system, cotranscription of a pseudo- and a functional odorant receptor gene is compatible in a single odorant sensory neuron, while the coexpression of more than one functional odorant receptor gene is found, although rarely (Rawson et al, 2000). The mechanisms responsible for a single receptor gene per neuron are unknown, but are thought to be based on negative feedback mediated by the expressed chemoreceptor, and/or on negative selection allowing only sensory neurons expressing a single chemoreceptor morph to survive (Serizawa et al, 2003; Feinstein et al, 2004; Lewcock and Reed, 2004; Shykind et al, 2004).
Despite the remarkable tightness of V1r regulation we report here (at least of the V1rb2 gene), which somehow contrasts with an apparently looser regulation of some odorant receptor genes (Rawson et al, 2000), and despite a complete lack of sequence homology between V1rs and odorant receptors, parallels between the two olfactory subsystems are striking. It is therefore likely that a common mechanism, in which the receptor itself is an essential regulator of the process leading to the expression of a single monomorphic chemoreceptor gene per neuron, is at work in the main olfactory and vomeronasal systems.
Coactivation of two homologous V1r gene clusters
We show here that the choice of a non-functional V1r gene accompanies the expression of a functional V1r. Unexpectedly, this latter can originate from V1r genes located in trans, from the cluster homologous to the one from which the nonfunctional gene is transcribed. This observation was surprising for the following reason: asynchronous chromosome replication during S phase is known to characterize pheromone receptor gene clusters (a characteristic also shared by odorant receptor gene clusters and by many imprinted genes) (Mostoslavsky et al, 2001; Singh et al, 2003; Gimelbrant and Chess, 2006). The identity of the asynchronously replicated chromosome is established early during development, apparently randomly, and is clonally inherited. Although the significance of the unequal treatment of these chemosensory gene clusters is not understood, it is thought to reflect a functional difference between different parental chromosomes, and the availability or non-availability for expression of the genes subject to this asynchronous replication. Our data, which indicate that two allelic V1r clusters can be chosen for expression and cotranscribed in a single VSN, thus contradict the idea that a tag labels the parental chromosomes for V1r expression, or non-expression.
Cluster lock after gene choice
To date, no experimental evidence supporting a mechanism involved in V1r regulation is available. An obvious hint to the regulatory option possibly taken by the pheromone receptor gene repertoire may be represented by the very peculiar genomic organization of V1r genes, which are almost invariably found in clusters of variable sizes, usually as an uninterrupted string of genes. Does this arrangement underlie regulatory mechanisms taking place at the cluster level, or does it simply reflect a physical proximity resulting from the multiple duplication events, which led to the emergence of the V1r clusters?
The observation of the cluster lock naturally argues in favor of a global regulatory mechanism. Different explanations can be proposed. We could, for example, consider each V1r gene, irrespective to the chromosome on which it is located, as a single and independent unit, chosen randomly among all other members of the V1r repertoire. The potential repertoire would in this case be composed of twice the number of V1r genes, that is, 300 members. The unavailability of the V1r genes surrounding the expressed allele could in this case represents a hypothetical physical or epigenetic constraint on these latter, negative constraint and side effect mediated by the transcription environment responsible for the expression of the first chosen and expressed allele.
However, data related to the regulation of other seven transmembrane receptor gene clusters, such as the X-linked opsin gene cluster (Pichaud et al, 1999) or those containing odorant receptor genes (Serizawa et al, 2003; Lomvardas et al, 2006), point at least partly to cluster-mediated or non-gene autonomous regulatory mechanisms.
One of the simplest models to explain a cluster-mediated type of regulation for V1r genes, could involve an activator sequence unique to each cluster, and responsible for the transcription of any member of the cluster. This random interaction would naturally be at the expense of the other members of the cluster, and lead to a single V1r gene expressed in the cluster.
We thus favor a model in which the expressed pheromone receptor is an essential and global tool in the establishment of monogenic and monoallelic V1r expression, but is only part of a more complex diagram, involving regulation at the V1r gene cluster level.
Materials and methods
Animals
Animals were housed and handled in accordance with the guidelines and regulations of the institution and of the state of Geneva.
In situ hybridizations
Cryostat slices (14 μm) of adult mouse VNOs were arranged on SuperFrost+ slides (Menzel-Glaser), dried for 40′, fixed for 20′ at 4°C with 4% paraformaldehyde, and hybridized overnight at 65°C in the following buffer: 1 × salt buffer (10 mM NaCl, 5 mM NaH2PO4·H2O, 5 mM Na2HPO4·2H2O, 5 mM EDTA, pH 7.5), 50% formamide, 10% dextran sulfate, 1 μg/μl tRNA, 1 × Denhardt's, with 20 ng/μl cRNA probe(s). Following hybridizations, slides were washed twice at 65°C and once at RT with 1 × SSC, 50% formamide and 0.1% Tween20. Slides were preincubated in 1 × MABT, 2% blocking reagent (Roche) for 30′ followed by 1 h incubation with alkaline phosphatase-anti-digoxigenin antibody (1:500, Roche) or/and peroxidase-anti-fluorescein antibody (1:200, Roche). Peroxydase activity was detected by washing slides three times with TNT (Tris 150 mM, NaCl 150 mM, Tween20 0.05%, pH 7.5), incubation for 30′ with a biotinyl-tyramide solution (PerkinElmer), 3 × washes with TNT and incubation for 30′ with streptavidin-alexa488 (Molecular Probes). Alkaline phosphatase activity was detected by incubating slides with Fast Red substrate (DAKO) for 30′. Sections were mounted in DABCO mounting medium (Sigma). Fluorescein- and digoxigenin-labeled RNA probes were prepared using the DIG RNA labeling kit (Roche) following the manufacturer's instructions. The following sequences were used: V1ra3, nt 249–839; V1ra9, nt 193–750; V1rb2, nt 1–933; V1rb9, nt 196–581; V1rc32, nt 103–467; V1rd16, nt 417–619; V1re4, nt 178–754; V1rf4, nt 397–920; V1rg1, nt 311–833; V1rh5, nt 163–740; V1ri8, nt 496–933; V1ri1, nt 313–750; V1rj3, nt 334–775; V1rk1, nt 332–804; V1rl1, nt 433–876; V2R2, nt 1898–2733; MOR204-6, nt 158–881; MOR18-2, nt 257–937; Gαi2, nt 248–1138; LacZ (1.8 kb HindIII fragment) or EGFP (entire ORF) probes were used to specifically identify VSNs expressing wt or mutant V1rb2 alleles.
Microscopy
For two-photon imaging (Figures 3A and B), GFP was excited at 900 nm with a pulsed Ti-sapphire laser (Spectraphysics Mai Tai). The image was reconstructed from the fluorescence collected by descanned photomultipliers as maximum intensity projections of a z-stack of 20 slices (Olympus Fluoview 3.0).
Electrophysiology
Isolated mouse VSNs were studied at room temperature by whole-cell patch-clamp using an Axopatch 200B amplifier (Axon Instruments, USA). The softwares Pulse (Instrutech, USA) and Igor (Wavemetrics, USA) were used to analyze the signals. Patch pipettes were made from borosilicate glass microcapillaries (TW150F-6, World Precision Instruments, USA) and fire polished with a microforge (MF-830, Narishige, Japan) to have a resistance of 8–10 MΩ. The pipette solution contained (in mM) the following: 120 KCl, 10 Hepes, 1 CaCl2, 2 MgCl2, 11 EGTA and 2 ATP (pH adjusted to 7.3 with KOH). The bath and perfusion solutions contained (in mM) the following: 140 NaCl, 5 KCl, 10 Hepes, 2 CaCl2, 1 MgCl2 and 10 Glucose (pH adjusted to 7.6 with NaOH). Solutions were perfused with a fast perfusion apparatus (SF-70, Warner Instruments, USA). VSNs were exposed to 5″ pulses of α,β-farnesenes, 2,5-dimethylpyrazine, pentyl-acetate and 2-heptanone added to the perfusion solution at 10−6, or otherwise mentioned. The holding potential was maintained at −60 mV. KCl pulses (100 mM) were used as a test of neuronal viability.
RT–PCRs
mRNAs were extracted from 6-week-old male mouse VNOs. For the amplification of V1ra cDNAs, the following primers were used: fw GGG AAG TCT TTC TTA TTA GTC TCA TGG TCC TCT C, rev CAA TGC TGT CAA AGA TGG ACA TCA GCA CAA AGA AG; for V1rb: fw TCA CCC TTT GTG CTA CCT GTC TGC TGA AT, rev GTA ACT CAT GGG TAG AAG TGA GCA GGA CT. Amplifications were performed under the following conditions: 5′ at 95°C, followed by 34 cycles of 1′ at 95°C, 1′ at 60°C and 2′ at 72°C, and a final extension of 10′ at 72°C. For V1ra amplicons, 17 cDNA and 18 genomic clones were sequenced. For V1rb amplicons, 18 cDNA and 21 genomic clones were sequenced.
Supplementary Material
Supplementary Figure S1
Supplementary Table 1
Acknowledgments
We thank Véronique Pauli-Jungo and Caroline Mareda for expert technical help. We also thank Pierre Vassalli, Paul Feinstein and members of the laboratory for comments on the manuscript. This work was supported by grants from the Swiss National Science Foundation and the Leenaards Foundation.
References
- Belluscio L, Koentges G, Axel R, Dulac C (1999) A map of pheromone receptor activation in the mammalian brain. Cell 97: 209–220 [DOI] [PubMed] [Google Scholar]
- Boschat C, Pelofi C, Randin O, Roppolo D, Luscher C, Broillet MC, Rodriguez I (2002) Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci 5: 1261–1262 [DOI] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P (2002) Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 419: 70–74 [DOI] [PubMed] [Google Scholar]
- Del Punta K, Rothman A, Rodriguez I, Mombaerts P (2000) Sequence diversity and genomic organization of vomeronasal receptor genes in the mouse. Genome Res 10: 1958–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Axel R (1995) A novel family of genes encoding putative pheromone receptors in mammals. Cell 83: 195–206 [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P (2004) A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell 117: 817–831 [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P (2004) Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell 117: 833–846 [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Chess A (2006) An epigenetic state associated with areas of gene duplication. Genome Res 16: 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M, Martinez-Marcos A (2003) Structure and function of the vomeronasal system: an update. Prog Neurobiol 70: 245–318 [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C (1997) A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell 90: 763–773 [DOI] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Axel R, Hood L, Trask BJ (2002) Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc Natl Acad Sci USA 99: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levai O, Feistel T, Breer H, Strotmann J (2006) Cells in the vomeronasal organ express odorant receptors but project to the accessory olfactory bulb. J Comp Neurol 498: 476–490 [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR (2004) A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA 101: 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R (2001) Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci 21: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck LB (1997) A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 90: 775–784 [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, Bergman Y (2001) Asynchronous replication and allelic exclusion in the immune system. Nature 414: 221–225 [DOI] [PubMed] [Google Scholar]
- Pantages E, Dulac C (2000) A novel family of candidate pheromone receptors in mammals. Neuron 28: 835–845 [DOI] [PubMed] [Google Scholar]
- Pichaud F, Briscoe A, Desplan C (1999) Evolution of color vision. Curr Opin Neurobiol 9: 622–627 [DOI] [PubMed] [Google Scholar]
- Rawson NE, Eberwine J, Dotson R, Jackson J, Ulrich P, Restrepo D (2000) Expression of mRNAs encoding for two different olfactory receptors in a subset of olfactory receptor neurons. J Neurochem 75: 185–195 [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Del Punta K, Rothman A, Ishii T, Mombaerts P (2002) Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat Neurosci 5: 134–140 [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P (1999) Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell 97: 199–208 [DOI] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R (1997) A new multigene family of putative pheromone receptors. Neuron 19: 371–379 [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H (2003) Negative feedback regulation ensures the one receptor–one olfactory neuron rule in mouse. Science 302: 2088–2094 [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G (2004) Gene switching and the stability of odorant receptor gene choice. Cell 117: 801–815 [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H (1999) Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401: 929–932 [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A (2003) Coordination of the random asynchronous replication of autosomal loci. Nat Genet 33: 339–341 [DOI] [PubMed] [Google Scholar]
- Wagner S, Gresser AL, Torello AT, Dulac C (2006) A multireceptor genetic approach uncovers an ordered integration of VNO sensory inputs in the accessory olfactory bulb. Neuron 50: 697–709 [DOI] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S (2004) Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics 83: 802–811 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Table 1


