Abstract
Background
The vertebrate adult axial skeleton, trunk and limb skeletal muscles and dermis of the back all arise from early embryonic structures called somites. Somites are symmetrically positioned flanking the embryo axial structures (neural tube and notochord) and are periodically formed in a anterior-posterior direction from the presomitic mesoderm. The time required to form a somite pair is constant and species-specific. This extraordinary periodicity is proposed to depend on an underlying somitogenesis molecular clock, firstly evidenced by the cyclic expression of the chick hairy1 gene in the unsegmented presomitic mesoderm with a 90 min periodicity, corresponding to the time required to form a somite pair in the chick embryo. The number of hairy1 oscillations at any given moment is proposed to provide the cell with both temporal and positional information along the embryo's anterior-posterior axis. Nevertheless, how this is accomplished and what biological processes are involved is still unknown. Aiming at understanding the molecular events triggered by the somitogenesis clock Hairy1 protein, we have employed the yeast two-hybrid system to identify Hairy1 interaction partners.
Results
Sap18, an adaptor molecule of the Sin3/HDAC transcriptional repressor complex, was found to interact with the C-terminal portion of the Hairy1 protein in a yeast two-hybrid assay and the Hairy1/Sap18 interaction was independently confirmed by co-immunoprecipitation experiments. We have characterized the expression patterns of both sap18 and sin3a genes during chick embryo development, using in situ hybridization experiments. We found that both sap18 and sin3a expression patterns co-localize in vivo with hairy1 expression domains in chick rostral presomitic mesoderm and caudal region of somites.
Conclusion
Hairy1 belongs to the hairy-enhancer-of-split family of transcriptional repressor proteins. Our results indicate that during chick somitogenesis Hairy1 may mediate gene transcriptional repression by recruiting the Sin3/HDAC complex, through a direct interaction with the Sap18 adaptor molecule. Moreover, since sap18 and sin3a are not expressed in the PSM territory where hairy1 presents cyclic expression, our study strongly points to different roles for Hairy1 throughout the PSM and in the prospective somite and caudal region of already formed somites.
Background
The adult vertebrate body is composed of various segmented structures, such as the vertebrae, ribs, intervertebral disks, peripheral nerves and skeletal muscles. One of the earliest manifestations of segmental patterning during vertebrate embryogenesis is the generation of transient, serially repeated structures known as somites. Somites are epithelial blocks of paraxial mesoderm cells that are laid symmetrically on either side of the neural tube from the rostral extremity of the presomitic mesoderm (PSM) in an anterior-posterior (AP) manner. Ongoing gastrulation at the posterior part of the PSM ensures embryo growth as new somite pairs arise from the anterior PSM. These structures are periodically formed and the time required to form a somite pair, as well as the total number of somite pairs generated, is extraordinarily constant and species-specific. In the chick embryo a new pair of somites is formed every 90 min, to a total of 52 somite pairs [1,2].
In 1997, Palmeirim and collaborators disclosed a molecular clock underlying vertebrate embryo somitogenesis when analysing the expression pattern of the hairy1 gene in chick embryos [3]. This gene is cyclically expressed in PSM cells every 90 min, which corresponds to the time required for a new somite pair to be generated [3]. The number of hairy1 oscillations at any given moment is proposed to provide cells with temporal and positional information along both anterior-posterior and medial-lateral PSM axes [4]. Further work performed in our lab showed that a molecular clock is also operating during limb bud development [5], suggesting that the segmentation clock is not an exclusive property of PSM tissue, but may be a more general way to count time during vertebrate development, providing positional information to different types of cells.
Besides participating in the somitogenesis clock in PSM cells, hairy1 is continuously expressed in the caudal region of formed somites. Newly formed somites present two distinct compartments, which are the basis for the resegmentation process that later leads to vertebrae formation [6]. Somite AP polarity is generated in the anterior PSM by Mesp2 and Delta/Notch signalling [7] and the maintenance of rostral and caudal identities in newly formed somites is tightly dependent on tbx18 and uncx4.1 gene expression, respectively [8-10]. The regionalized, persistent hairy1 expression in the posterior somite region suggests that Hairy1 could participate in somite polarity establishment and/or maintenance, although this has not yet been addressed experimentally.
Hairy1 belongs to the hairy-enhancer-of-split (Hairy E(Spl)/HES) family of transcriptional repressor proteins [11]. All HES members share conserved functional motifs: the basic domain (b) required for DNA-binding, a helix-loop-helix dimerization domain (HLH), an orange domain (OR) which confers specificity among family members and the C-terminal WRPW tetrapeptide required for co-factor binding [12,13]. The evidence to date suggests that HES proteins can be involved in an array of repression mechanisms via the recruitment of different protein partners, forming complexes with different specificities [14]. HES-mediated transcriptional repression has been shown to be an important feature of many embryonic developmental processes [15-17]. Data obtained from mouse and zebrafish embryos have shown that Hairy-related proteins act as transcriptional repressors in somitogenesis: hes1 participates in auto-regulatory negative feedback loops [18], probably mediated by direct protein interaction with the Notch signalling component RBP-Jκ[19]; hes7 represses transcription from N box- and E box-containing promoters [20]; hes7 represses its own transcription and also Lfng expression [21,22]; Her13.2 interacts with Her1 and represses both her1 and her7 [23].
Although the expression of chick hairy1 and its homologues has been demonstrated during somitogenesis in many animal models, it is not clear what the outputs of hairy1 expression are, both in the molecular clock and during somite formation and differentiation. Chick Hairy1 is only known to form homo-dimers and hetero-dimers with two other HES family members, Hey 1 and Hey2, through their bHLH-OR protein domains [24]. Furthermore, the mechanisms of Hairy1-mediated transcriptional repression have not yet been addressed in the chick embryo. Aiming at understanding the molecular events triggered by chick Hairy1, we have employed the yeast two-hybrid system to identify Hairy1 protein interaction partners. The yeast two-hybrid system [25] is a well established technique used to study protein-protein interactions, which are critical in virtually all cellular processes. Sap18 (Sin3-associated polypeptide, 18 kDa), a component of the Sin3/histone deacetylase (HDAC) transcriptional repressor complex [26], was found to interact with the C-terminal portion of the Hairy1 protein. Hairy1/Sap18 interaction was independently confirmed by in vitro co-immunoprecipitation assays. Whole mount in situ hybridization experiments showed that sap18, sin3a and hairy1 mRNA co-localize in the rostral-most part of the chick embryo PSM and in the caudal compartment of already formed somites. Our results indicate that in these tissues chick Hairy1 may mediate gene transcriptional repression by recruiting the Sin3/HDAC complex through a direct interaction with the Sap18 adaptor molecule. Moreover, since sap18 and sin3a are not expressed in the PSM territory where hairy1 presents cyclic expression, our study strongly points to different roles for Hairy1 throughout the PSM and in the prospective somite and caudal region of already formed somites.
Results
Interaction of Hairy1 with Sap18 revealed by yeast two-hybrid analysis
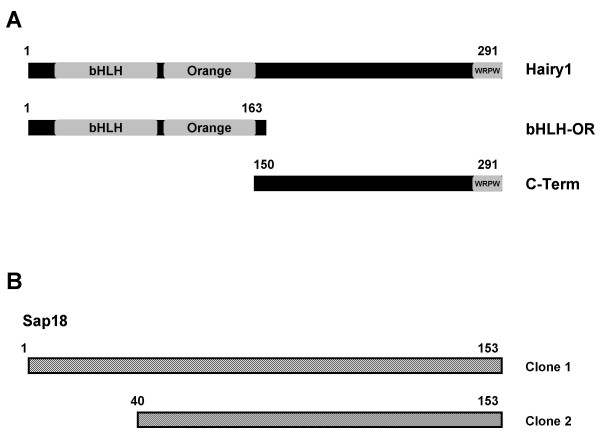
The yeast two-hybrid system was used to screen proteins that associate with the C-terminal portion of the chick Hairy1 protein (Fig. 1A; C-Term, amino acids 150–291). mRNA was extracted from stage HH20 – HH22 chick embryos and a cDNA library was constructed in the pGADT7 plasmid. The C-Term fragment of the Hairy1 protein was cloned in the pGBKT7 vector and the resulting bait construct was used to screen the constructed chick embryo cDNA library. Two different plasmids were isolated from positive-interaction yeast colonies which coded for the entire chick Sap18 protein (Fig. 1B; clone 1) or a truncated form, lacking the first 39 amino acids (Fig. 1B; clone 2).
Figure 1.
Schematic representation of Hairy1 protein domains assayed for interaction and Sap18 clones obtained from the yeast two-hybrid screen. (A) Hairy1 protein domains used as bait in the yeast two-hybrid assays; (B) Yeast two-hybrid positive interaction clones encoding for Sap18. Numbers correspond to amino acids in (A) and (B).
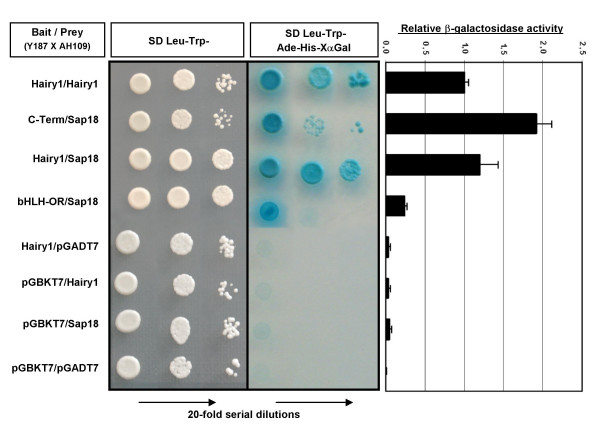
Hairy1 protein interaction with Sap18 was confirmed by performing additional yeast two-hybrid experiments using the whole Hairy1 protein, or its N-terminus (Fig.1A; bHLH-OR, amino acids 1–163) as bait. The documented interaction between Hairy1 proteins in a homodimer [24] was used as a positive control (Fig. 2). Bait and prey constructs were transformed into the opposite mating-type yeast strains Y187 and AH109, respectively, and the transformed strains were mated and spread on appropriate selective media. Positive interactions were observed for Hairy1/Hairy1, C-Term/Sap18 and Hairy1/Sap18, while no interaction was detected between bHLH-OR/Sap18 or in the negative control reactions, as expected (Fig. 2).
Figure 2.
Hairy1 and Sap18 protein interaction evaluated by the yeast two-hybrid assay. Baits were expressed as Gal4 DNA-BD fusion proteins in the pGBKT7 plasmid and transformed into the Y187 yeast strain; preys were expressed as Gal4 AD fusion proteins in pGADT7 vector and transformed into the AH109 strain. Bait- and prey-transformed strains were mated and the resulting diploids were cultured in SD-Leu/-Trp medium, selecting for the presence of both plasmids and also in SD-Leu/-Trp/-His/-Ade/X-α-gal medium, were only the diploids presenting protein interactions are capable of growth. 20-fold serial dilutions were performed prior to colony plating to ensure growth on the selective medium is dependent solely on HIS3, ADE2 and MEL1 reporter gene expression. β-galactosidase activity was assayed for each strain and is presented relative to that obtained for the Hairy1/Hairy1 interaction. In all assays, Hairy1/Hairy1 protein interaction was used as a positive control and the empty vectors were employed in the negative controls. (DNA-BD: DNA binding domain; AD: activation domain).
The relative strength of Hairy1/Sap18 protein interaction was determined by scoring for reporter β-galactosidase activity (Fig. 2), using the Hairy1/Hairy1 protein interaction as a reference [24]. We found that Hairy1 has a similar affinity for Sap18 as it has for Hairy1 itself, and that the association between Hairy1 C-terminal portion and Sap18 is almost twice as strong as that for the whole protein (Fig.2). This is most probably due to the absence of the DNA-binding bHLH domain, leading to a relief of reporter gene repression. Altogether, our yeast two-hybrid results indicate that Hairy1 associates with Sap18 and that Hairy1's C-terminal domain mediates this interaction.
Co-immunoprecipitation of Hairy1 with Sap18
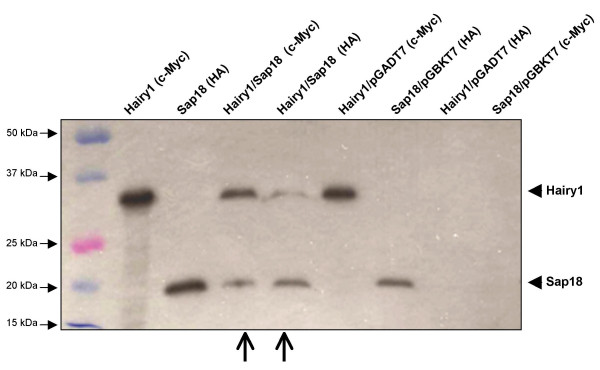
To independently validate the interaction between chick Hairy1 and Sap18 proteins, in vitro co-immunoprecipitation assays were performed. Chick hairy1 and sap18 genes cloned in pGBKT7 and pGADT7 vectors, respectively, were used to generate 35S-labeled c-Myc-Hairy1 and HA-Sap18 through in vitro transcription/translation reactions. Upon incubation of both proteins, Hairy1 immunoprecipitated with Sap18 when either c-Myc or HA antibodies were employed (Fig. 3). This was not the case when in vitro transcription/translation reaction products of the empty vectors were used. This experiment confirms the Hairy1/Sap18 protein association identified in the yeast two-hybrid assay.
Figure 3.
In vitro co-immunoprecipitation of Hairy1 and Sap18. The proteins tested are indicated on top of each lane, as well as the antibody used for immunoprecipitation in each assay (in brackets). Proteins produced form the pGADT7 and pGBKT7 empty plasmids were used in negative controls. Open arrows indicate the gel lanes where both Hairy1 and Sap18 proteins are observed, which is indicative of a protein interaction among them.
Characterization of chick sap18 mRNA expression pattern
Having established that chick proteins Hairy1 and Sap18 can interact, we set out to determine if this protein interaction could be of biological relevance in our system, namely, during the process of somitogenesis. If that were so, sap18 gene expression should co-localize with hairy1 mRNA distribution in the chick embryo.
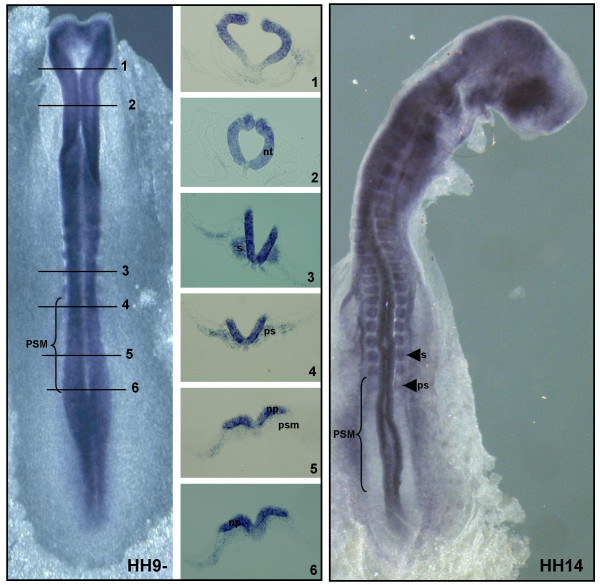
A sap18 antisense mRNA probe was generated and whole-mount in situ hybridization experiments were performed with the purpose of characterizing sap18 expression pattern in the chick embryo. At early stages, sap18 is expressed at the level of the primitive streak in prospective neural cells (Fig. 4, section 6), is absent from the neural tube facing the PSM (Fig. 4, section 5) and is upregulated throughout all the neural tube positioned rostrally to the PSM territory (Fig.4, sections 1–3). In older embryos, sap18 is expressed along the entire neural tube AP axis (Fig.4, HH12+ and HH14). In all embryos, sap18 expression is absent from most of the PSM tissue, being upregulated only in the rostral part of the PSM, the region that will give rise to the prospective somites, and in already formed somites. Chick hairy1 is also present in the prospective somites and in the caudal region of formed somites [3], indicating that the Hairy1/Sap18 protein interaction might play a functional role in these tissues.
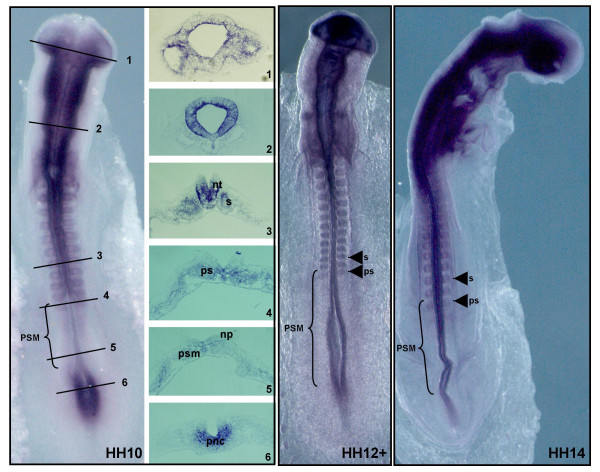
Figure 4.
sap18 mRNA expression pattern in chick embryos. Chick embryos staged HH10, HH12+ and HH14 were processed for in situ hybridization using a probe for chick sap18. Transverse sections at the indicated AP levels on stage HH10 embryo are shown. At early stages, sap18 is expressed at the level of the primitive streak in prospective neural cells (section 6, pnc), is absent from the neural plate (np) facing the PSM (section 5, psm) and is upregulated throughout all the neural tube positioned rostrally to the PSM territory (sections 1–3, nt). In older embryos, sap18 is expressed along the entire neural tube AP axis (stages HH12+ and HH14). In all embryos, sap18 expression is absent from most of the PSM tissue (brackets) being upregulated only in the rostral part of the PSM, the region that will give rise to the prospective somites (whole embryos and section 4, ps) and in already formed somites (whole embryos and section 3, s).
Co-localization of hairy1, sap18 and sin3a mRNA expression patterns
Sap18 has been shown to interact directly with Sin3a and act as an adaptor molecule bridging the Sin3/HDAC complex to sequence-specific DNA-binding proteins [26]. Since Sap18 associates with Hairy1, it could be recruiting the Sin3a complex for Hairy1-mediated transcriptional repression during chick embryo development. For this to be true, both sap18 and sin3a mRNAs should co-localize with each other and with hairy1 gene expression domains.
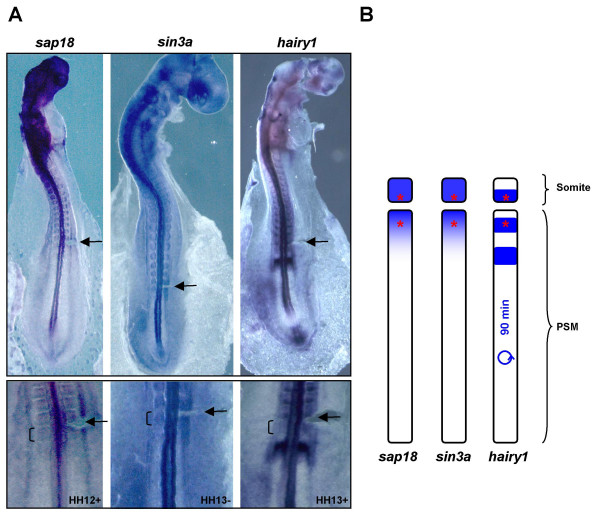
A sin3a antisense mRNA probe was generated and whole-mount in situ hybridization experiments revealed that many sin3a expression domains (Fig. 5) coincide with those of sap18 in the chick embryo, namely in the prospective somite region and in somites (Fig. 6). This is in accordance to their reported protein interaction and biological role in transcriptional regulation. Moreover, both sap18 and sin3a expression patterns co-localize with hairy1 mRNA in the caudal region of somites and in the rostral part of the PSM (Fig. 6). These results suggest that in these tissues Hairy1/Sap18 interaction could be mediating the recruitment of the Sin3/HDAC repressor complex to Hairy1-target gene promoters.
Figure 5.
sin3a mRNA expression pattern in chick embryos. Chick embryos staged HH9- and HH14 were processed for in situ hybridization using a probe for chick sin3a. Transverse sections at the indicated AP levels on stage HH9- embryo are shown. In all embryos, sin3a is expressed in the neural plate (np) and along the entire neural tube AP axis (nt), is absent from the PSM (section 5, psm; brackets in whole embryos) and is upregulated in the rostral part of the PSM, the region that will give rise to the prospective somites (whole embryos and section 4, ps) and in already formed somites (whole embryos and section 3, s).
Figure 6.
Co-localization of hairy1, sap18and sin3a mRNA expression in the prospective somitic territory and somites. (A) Whole mount in situ hybridization with the probe indicated on top was performed in chick embryos in somitogenesis developmental stages (upper panel). The lower panel is an amplified view of the rostral PSM and somites. Prior to in situ hybridization processing, a slit was made on the right side of the embryo (arrow), in the boundary between the last formed somite and the rostral PSM, evidencing the prospective somitic territory (bracket). (B) Schematic representation of the genes' expression domains, indicating (with an asterisk) the prospective somite and caudal region of already formed somites, where hairy1, sap18 and sin3a mRNA expression co-localize. Cyclic expression of hairy1 with a 90 minute periodicity is indicated.
Although hairy1 expression can be observed throughout the whole chick PSM, both sap18 and sin3a mRNAs are absent, except in the rostral-most region. This implies that Hairy1 is not capable of exerting gene transcriptional repression through Sap18-mediated Sin3/HDAC complex recruitment in the PSM regions where it is cyclically expressed.
Discussion
Members of the hairy-enhancer-of-split (Hairy E(Spl)/HES) family of proteins are reported to be transcriptional repressors. HES proteins can form homo- or hetero-dimers and repress gene expression either through direct binding to cis regulatory elements in the promoter or by sequestering, and thereby inactivating, transcriptional activators of the target gene. When bound to promoter DNA, HES proteins can recruit transcriptional regulatory protein complexes through their C-terminal domain and therefore exert repressor activity (reviewed by [13]).
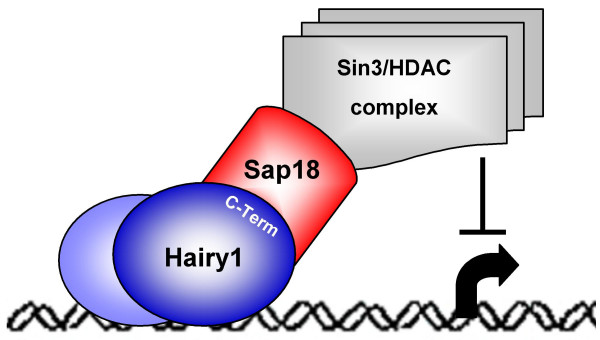
This study reports a protein interaction screen performed with the C-terminal portion of the chick hairy-enhancer-of-split protein Hairy1, using yeast two-hybrid technology. We have established that Sap18, the 18-kDa Sin3-associated protein, is a Hairy1-interacting protein, using both yeast two-hybrid experiments and in vitro co-immunoprecipitation assays. Chick embryo sap18 and sin3a expression patterns were characterized for the first time and were found to co-localize with each other and with hairy1 mRNA in the prospective somitic territory and in the caudal region of already formed somites. Our results provide novel insights into Hairy1-mediated transcriptional repression during chick embryo development, suggesting that Hairy1 may repress target gene transcription through a direct interaction with Sap18 protein, recruiting the Sin3/HDAC transcriptional repressor protein complex (Fig. 7).
Figure 7.
Proposed model for Hairy1-mediated transcriptional repression in the prospective somite and caudal region of somites. In the prospective somite and caudal region of already formed somites, where sap18, sin3a and hairy1 expression co-localize, a homo- or hetero-dimer containing Hairy1, bound to specific promoter regulatory sequences, may interact with Sap18 through the Hairy1 C-terminal domain and recruit the Sin3/HDAC complex to the transcriptional site, thus repressing target gene expression.
The Sin3/HDAC complex is highly conserved from yeast to human and contains HDAC activity, causing transcriptional repression mainly by deacetylating histones (reviewed by [27]). Sap18 is an integral member of the Sin3/HDAC transcriptional repressor complex and it functions as an adaptor polypeptide, tethering the protein complex to promoters by interacting with sequence-specific DNA binding proteins, since members of the Sin3/HDAC complex lack intrinsic DNA binding activity [26,28]. To date, the mechanisms of Hairy1-mediated transcriptional repression have not been addressed in the chick embryo. Our work suggests that upon binding to specific DNA promoter elements, chick Hairy1 is capable of interacting with Sap18, which in turn recruits the Sin3/HDAC repressor complex, repressing Hairy1-target gene transcription (Fig. 7). In fact, the recruitment of the Sin3/HDAC complex through interaction with Sap18 has already been shown for the Drosophila GAGA factor [29] and Bicoid genes [30], as well as for mouse suppressor of fused protein, mSu(fu) [28] and DNA polymerase, DPE2 [31].
The gene encoding Hairy1 presents cyclic expression in the chick embryo PSM, while in somites hairy1 expression ceases to cycle and is persistent in the posterior somitic region [3]. In their recent review on the HES gene family, Kageyama and collaborators [13] point out that low levels of Hes1 down-regulate cell cycle inhibitors, promoting cellular proliferation. On the other hand, continuous and high levels of Hes1 expression inhibit the cell cycle. This highlights another, as yet, unaddressed issue in somitogenesis: Is Hairy1 involved in the same biological functions in the PSM, where it is cyclically expressed in undifferentiated mesenchymal cells, and in the caudal somitic region, where its expression is persistent?
Our study strongly points to different roles for Hairy1 in the PSM cyclic expression domain and in the rostral-most PSM and caudal region of somites. The expression of both sap18 and sin3a genes is absent from PSM cells and is up-regulated only in prospective and already-formed somites. This clearly indicates that Hairy1 is only capable of interacting with Sap18, recruiting the Sin3/HDAC complex for transcriptional repression in these tissues and not along the entire PSM, suggesting that this mechanism is not involved in the somitogenesis clock machinery. Instead, we can hypothesize that Hairy1 transcriptional repression through Sap18-mediated Sin3/HDAC complex recruitment may be required for somite formation and/or polarization.
Conclusion
We have identified an interaction between the C-terminal domain of the chick protein Hairy1 and the Sin3/HDAC repressor complex adaptor molecule, Sap18. The expression patterns of chick sap18 and sin3a genes were characterized for the first time and were shown to co-localize with hairy1 mRNA in the rostral PSM and caudal region of formed somites. Our results indicate that Hairy1 may be mediating transcriptional repression in these tissues by interacting with Sap18 and recruiting the Sin3/HDAC transcriptional repressor complex to the promoters of its target genes (Fig. 7). Moreover, since sap18 and sin3a are not expressed in the PSM territory where hairy1 presents cyclic expression, our study strongly points to different roles for Hairy1 throughout the PSM and in the prospective somite and caudal region of already formed somites.
Methods
Eggs and embryos
Fertilised chick (Gallus gallus) eggs obtained from commercial sources were incubated at 37.2°C in a 49% humidified atmosphere and staged according to the Hamburger and Hamilton (HH) classification [32].
Yeast two-hybrid assays
Chick embryo cDNA library construction and yeast two-hybrid screening were performed using the Matchmaker Library Construction & Screening Kit (Clontech, USA). A cDNA library in the pGADT7-Rec (prey) vector was constructed using RNA extracted from the forelimbs of chick embryos in stages HH 20 – HH22. PCR-generated hairy1 fragments were cloned in the pGBKT7 plasmid for preparation of the bait vectors. The primers used were: Hairy1 – forward: 5'ATGCCCGCCGACACGGGCATG3', reverse: 5'CTACCACGGCCGCCAGACG3'; C-Term – forward: 5'CAGATCGTGGCCATGAACTACCTGC3', reverse: 5'CTACCACGGCCGCCAGACG3'; bHLH-OR – forward: 5'ATGCCCGCCGACACGGGCATG3', reverse: 5'GCCAGCAGGAGGGGGTGGCAG3'. Plasmid construction was confirmed upon sequencing.
The haploid yeast strain Y187 was transformed with the bait vectors and tested for toxicity. The haploid AH109 strain was transformed with the generated pGADT7-chick embryo cDNA library. Positive and negative protein interaction controls supplied with the kit were performed with the expected outcomes. Yeast screening for potential interacting proteins with the C-terminal domain of Hairy1 protein was performed by mating Y187 transformed with pGBKT7+C-Term and AH109 transformed with pGADT7-cDNA library. Diploids were plated on SD-Leu/-Trp/-His medium and growing colonies, indicative of a potentially positive interaction between the bait (C-Term) and the prey (library protein), were re-streaked several times on SD-Leu/-Trp/-His/-Ade plates. Finally, the positive clones were replica-plated on maximally selective SD-Leu/-Trp/-His/-Ade/X-α-gal medium to ensure that colonies contained the correct phenotype. Colony PCR using primers flanking the multiple cloning site of the pGADT7 plasmid was performed on the colonies that successfully passed the reporter tests. Clones presenting an insert over 300 bp were harvested for plasmid isolation and sequencing.
To confirm Hairy1 interaction with Sap18, yeast Y187 was transformed with bait vectors while AH109 was transformed with prey plasmids. After mating, the diploid strains were cultured in SD-Leu/-Trp medium and in SD-Leu/-Trp/-His/-Ade/X-α-gal medium. Diploids were suspended in sterile water and three 20-fold serial dilutions were performed, after which a drop of each suspension was placed on each medium. This procedure ensures that the growth observed is not due to feeding on underlying cells, but can clearly be attributed to reporter gene expression. Hairy1/Hairy1 protein interaction was used as a positive control and the empty vectors were employed in the negative controls.
Quantification of β-galactosidase activity
Quantification of reporter β-galactosidase activity was performed using the Yeast β-Galactosidase Assay Kit (Pierce, USA). Four co-transformed colonies of each combination were grown in liquid SD medium lacking leucine and tryptophan, assayed in triplicate and the mean value was calculated. Hairy1/Hairy1 protein interaction was used as a positive control and the empty vectors were employed in the negative controls. Mean activity values were normalized to those obtained for the Hairy1/Hairy1 protein interaction.
In vitro co-immunoprecipitation assays
Chick hairy1 and sap18 genes cloned in pGBKT7 and pGADT7 vectors, respectively, were used to generate 35S-labeled Hairy1 fused to the c-Myc epitope and Sap18 fused to the HA epitope using the TnT Quick Coupled Transcription/Translation System (Promega, USA). In vitro co-immunoprecipitation assays were performed using the Matchmaker Co-IP Kit (Clontech, USA) following the manufacturers instructions. Samples were run on 12 % SDS-PAGE gel and the gel was dried at 80°C under constant vacuum and exposed to an X-ray film for autoradiography. Empty vectors were employed in the negative controls.
In situ hybridisation probes
In situ hybridisation probes for sap18 and sin3a were generated. Reverse transcription and polymerase chain reactions (RT-PCR) were used to isolate chick sap18 (Genbank: NM_204312) and portions of sin3a (Genbank: XM_413695) using the sense oligos 5'-CCGGAATTCATGGCGGTGGAGTCGCG-3', 5'-TGAAGACTCCACTTTTGTGAGC-3', and antisense oligos 5'-CCGCTCGAGTCAGTATGGCCTCATGCGGC-3', 5'-TGTACCAGTTGCTGTTCACG-3', respectively. The DNA fragments generated were cloned into the pCR®II-TOPO® vector (Invitrogen, USA) and plasmid DNA was isolated. The constructs were confirmed upon sequencing. Digoxigenin-labelled RNA probes were obtained from linearised plasmids, according to standard procedures.hairy1 probe generation was performed as previously described [3].
Embryo whole mount in situ hybridisation and cryosectioning
Embryos were fixed overnight at 4°C in 4% formaldehyde with 2 mM ethylene glycol-bis (β-amino-ethyl ether) tetraacetic acid (EGTA) in PBS, rinsed in PBT (PBS, 0,1% Tween 20), dehydrated in methanol and stored at -20°C. Whole mount in situ hybridisation was performed according to the previously described procedure [33].
Some in situ hybridized embryos were processed for cryosectioning as previously described [34]. 12 μm cryostat (Leica CM1900) sections were collected on Superfrost Plus Slides (Menzel-Glasse). Slides were mounted in Aquatex (Merck, Germany) and photographed.
Imaging
Embryos processed for in situ hybridization were photographed in PBT/0.1% azide, using a Leica DFC320 digital camera coupled to a Leica MZFLIII stereomicroscope equipped with Leica IM50 image manager (Leica Microsystems, Germany). Cryosectioned embryo sections were photographed using an Olympus DP70 camera coupled to an Olympus BX61 microscope.
Authors' contributions
CJS prepared the sin3a probe plasmid, synthesized the probes, staged and collected the chick embryos, performed in situ hybridizations, performed the yeast two-hybrid confirmations, β-galactosidase and co-immunoprecipitation experiments and assisted in manuscript writing; IP contributed to experimental design and interpretation of results, was involved in the conception and coordination of this project and assisted in manuscript preparation; RPA prepared all yeast two-hybrid plasmids plus the sap18 probe vector, constructed the chick embryo cDNA library in pGADT7, performed the yeast-two hybrid screen, contributed to experimental design and interpretation of results, was involved in the conception and coordination of this project and wrote the manuscript. All authors approved the final manuscript.
Acknowledgments
Acknowledgements
The authors wish to thank Fernanda Bajanca for informatics technical support and for critical reading of the manuscript. This work was supported by the Fundação para a Ciência e Tecnologia, Portugal (project POCTI/BIA-BCM/62688/2004) and by EU/FP6 – Network of Excellence – Cells into Organs [35]. RPA and CJS were supported by grants n° SFRH/BPD/9432/2002 and UMINHO/POCI-62688/BI/1/06, respectively.
Contributor Information
Caroline J Sheeba, Email: sheeba@ecsaude.uminho.pt.
Isabel Palmeirim, Email: ipalmeirim@ecsaude.uminho.pt.
Raquel P Andrade, Email: rpandrade@ecsaude.uminho.pt.
References
- Andrade RP, Pascoal S, Palmeirim I. Thinking clockwise. Brain Res Brain Res Rev. 2005;49:114–9. doi: 10.1016/j.brainresrev.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Freitas C, Rodrigues S, Saude L, Palmeirim I. Running after the clock. Int J Dev Biol. 2005;49:317–24. doi: 10.1387/ijdb.041955cf. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/S0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Freitas C, Rodrigues S, Charrier JB, Teillet MA, Palmeirim I. Evidence for medial/lateral specification and positional information within the presomitic mesoderm. Development. 2001;128:5139–47. doi: 10.1242/dev.128.24.5139. [DOI] [PubMed] [Google Scholar]
- Pascoal S, Carvalho CR, Rodriguez-Leon J, Delfini MC, Duprez D, Thorsteinsdottir S, Palmeirim I. A Molecular Clock Operates During Chick Autopod Proximal-distal Outgrowth. J Mol Biol. 2007;368:303–9. doi: 10.1016/j.jmb.2007.01.089. [DOI] [PubMed] [Google Scholar]
- Saga Y, Takeda H. The making of the somite: molecular events in vertebrate segmentation. Nat Rev Genet. 2001;2:835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Koizumi K, Takagi A, Kitajima S, Inoue T, Koseki H, Saga Y. Mesp2 initiates somite segmentation through the Notch signalling pathway. Nat Genet. 2000;25:390–6. doi: 10.1038/78062. [DOI] [PubMed] [Google Scholar]
- Bussen M, Petry M, Schuster-Gossler K, Leitges M, Gossler A, Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18:1209–21. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitges M, Neidhardt L, Haenig B, Herrmann BG, Kispert A. The paired homeobox gene Uncx4.1 specifies pedicles, transverse processes and proximal ribs of the vertebral column. Development. 2000;127:2259–67. doi: 10.1242/dev.127.11.2259. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Voss AK, Thomas T, Yokota Y, Gruss P. Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development. 2000;127:2251–2258. doi: 10.1242/dev.127.11.2251. [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Hogan A, Pinchin SM, Howe KM, Lardelli M, Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. Embo J. 1989;8:3095–103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–57. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–51. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. Embo J. 1998;17:2067–78. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–34. doi: 10.1016/S0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8:2729–42. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Wainwright SM, Ish-Horowicz D. Point mutations in the Drosophila hairy gene demonstrate in vivo requirements for basic, helix-loop-helix, and WRPW domains. Mol Cell Biol. 1992;12:2475–83. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- King IN, Kathiriya IS, Murakami M, Nakagawa M, Gardner KA, Srivastava D, Nakagawa O. Hrt and Hes negatively regulate Notch signaling through interactions with RBP-Jkappa. Biochem Biophys Res Commun. 2006;345:446–52. doi: 10.1016/j.bbrc.2006.04.097. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kang L, Zhang N. Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis. 2005;43:196–204. doi: 10.1002/gene.20171. [DOI] [PubMed] [Google Scholar]
- Kawamura A, Koshida S, Hijikata H, Sakaguchi T, Kondoh H, Takada S. Zebrafish hairy/enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev. 2005;19:1156–1161. doi: 10.1101/gad.1291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C, Dale K, Fischer A, Klamt B, Hrabe de Angelis M, Radtke F, McGrew MJ, Pourquie O, Gessler M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev Biol. 2000;227:91–103. doi: 10.1006/dbio.2000.9884. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–64. doi: 10.1016/S0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99:5442–7. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas ML, Canudas S, Fanti L, Pimpinelli S, Casanova J, Azorin F. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 2000;1:253–9. doi: 10.1093/embo-reports/kvd046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Foehr M, Jaynes JB, Hanes SD. Drosophila SAP18, a member of the Sin3/Rpd3 histone deacetylase complex, interacts with Bicoid and inhibits its activity. Dev Genes Evol. 2001;211:109–17. doi: 10.1007/s004270100135. [DOI] [PubMed] [Google Scholar]
- Wada M, Miyazawa H, Wang RS, Mizuno T, Sato A, Asashima M, Hanaoka F. The second largest subunit of mouse DNA polymerase epsilon, DPE2, interacts with SAP18 and recruits the Sin3 co-repressor protein to DNA. J Biochem (Tokyo) 2002;131:307–11. doi: 10.1093/oxfordjournals.jbchem.a003104. [DOI] [PubMed] [Google Scholar]
- Hamburger , Hamilton Stades du d, veloppement de l'embryon de poulet. J Morph. 1951;88 [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Bajanca F, Luz M, Duxson MJ, Thorsteinsdottir S. Integrins in the mouse myotome: Developmental changes and differences between the epaxial and hypaxial lineage. Dev Dyn. 2004;231:402–415. doi: 10.1002/dvdy.20136. [DOI] [PubMed] [Google Scholar]
- http://www.cellsintoorgans.net