Abstract
Background
To test whether epithelial sodium channel (ENaC) genes' variants contribute to salt sensitive hypertension in Dahl rats, we screened ENaC α, β, and γ genes entire coding regions, intron-exon junctions, and the 3' and 5' flanking regions in Dahl S, R and Wistar rats using both Denaturing High Performance Liquid Chromatography (DHPLC) and sequencing.
Results
Our analysis revealed no sequence variability in the three genes encoding ENaC in Dahl S versus R rats. One homozygous sequence variation predicted to result in a D75E substitution was identified in Dahl and Wistar rat ENaC α compared to Brown Norway. Six and two previously reported polymorphic sites in Brown Norway sequences were lost in Dahl and Wistar rats, respectively. In the 5' flanking regions, we found a deletion of 5GCTs in Dahl and Wistar rat ENaC α gene, five new polymorphic sites in ENaC β and γ genes, one homozygous sequence variation in Dahl and Wistar rat ENaC γ gene, as well as one Dahl rat specific homozygous insertion of -1118CCCCCA in ENaC γ gene. This insertion created additional binding sites for Sp1 and Oct-1. Five and three Brown Norway polymorphic sites were lost in Dahl and Wistar rats, respectively. No sequence variability in ENaC 3' flanking regions was identified in Dahl compared to Brown Norway rats.
Conclusion
The first comprehensive sequence analysis of ENaC genes did not reveal any differences between Dahl S and R rats that were isogenic in the regions screened. Mutations in ENaC genes intronic sequence or in ENaC-regulatory genes might possibly account for increased ENaC activity in Dahl S versus R rats.
Background
The epithelial sodium channel (ENaC) is made up of three homologous subunits named α, β, and γ that assemble together to form a highly Na+- selective channel [1]. The structure of these subunits is characterized by the presence of two transmembrane domains separated by a large extracellular loop. Identification of mutations in ENaC subunits causing salt-sensitive hypertension and hypotension in humans (Liddle's syndrome and pseudohypoaldosteronism type 1) highlighted the impact of these genes on salt homeostasis and control of blood pressure (BP) [2,3].
In rats, the three α, β and γ ENaC (rENaC) subunits are encoded by three distinct genes Scnn1a, Scnn1b, and Scnn1g respectively, located on chromosomes 4q42, 1q36-41 and 1q36-41. The three subunits share similar structures and show 33–37% amino acid sequence homology in human [4]. rENaC α gene is composed of 12 coding exons [5]. rENaC β and γ genes are composed of 13 exons, the translation initiation codon is present within the second exon for both genes [6,7].
Dahl rats represent a robust animal model of genetically determined salt-sensitive hypertension. High salt intake increases BP in Dahl salt-sensitive (Dahl S), but not in Dahl salt-resistant (Dahl R) rats. Blockade of ENaC in the brain by benzamil prevents the increase in BP in Dahl S rats on high salt diet [8].
So far, only the coding sequences of genes encoding the three subunits have been partially screened in Dahl S and R rats. Analysis of near full length (base 22 till the end, all numbering starts at the A nucleotide of the primary initiation codon) of the Scnn1b cDNAs derived from kidneys of Dahl S and R rats failed to reveal any coding sequence mutations that could affect the predicted peptide sequence of Scnn1b [9]. Sequencing of nucleotides 21667 to 22054, 31172 to 31492, and 29142 to 29522 in the carboxy termini of ENaC α, β, and γ genes respectively revealed no differences in Dahl S versus R rats [10]. The sequence of the entire coding regions, intron-exon junctions, as well as the 3' and 5' flanking regions of ENaC three genes has not yet been reported in Dahl rats. In order to identify any variation, each sequence was analyzed using a combination of two screening methods, Denaturing High Performance Liquid Chromatography (DHPLC), offering 95–100% sensitivity and 100% specificity and automatic sequencing, offering 99.7–100% sensitivity and 100% specificity [11,12]. Dahl S and R rats, as well as Wistar rats, used as a control, were screened and the obtained sequences were compared to Brown Norway sequences retrieved from the rat genome database.
Results
Screening of ENaC α, β, and γ Genes Coding Regions for Variations between Dahl R and S rats
No sequence variability was identified in the entire coding regions, as well as in exon-intron junctions of ENaC α, β, and γ genes in Dahl S versus R rats (Table 1). One homozygous sequence variation G225T in exon 1 of ENaC α, predicted to result in a D75E substitution, was identified in Dahl S and R and Wistar rats compared to the published Brown Norway sequence (Table 1). Previously published Wistar rat sequences did not identify this variation [5]. It is therefore possible that Wistar rats are heterozygous at this position. One reported Brown Norway polymorphic site of ENaC α was lost in both Dahl and Wistar rats (Table 1). No sequence variability was identified in Dahl and Wistar rat ENaCβ gene compared to the Brown Norway sequence available in the public domain. No sequence variation in ENaC γ gene was identified in Dahl and Wistar rats compared to the published Brown Norway sequence. However, five and one previously reported polymorphic sites in Brown Norway ENaC γ sequence were lost in Dahl and Wistar rats, respectively (Table 1). The Wistar rat alleles studied followed a Mendelian independent assortment.
Table 1.
Allelic variants in the coding sequence of ENaC subunits in Dahl S, R, Wistar, and Brown Norway rats
| Subunit | Nucleotide position/exon | Dahl S and R | Wistar | Brown Norway | AA change |
| ENaC α | +225/1 | TT | TT* | GG | D75E |
| +10716/3 | AA | AA | GG, GA, AA | R290R | |
| ENaC γ | +4008/4 | GG | GG | AA, AG, GG | E255K |
| +22549/7 | CC | TT, TC, CC | TT, TC, CC | D376D | |
| +24672/8 | CC | TT, TC, CC | TT, TC, CC | C410C | |
| +29195/13 | TT | CC, CT, TT | CC, CT, TT | C542C | |
| +29291/13 | TT | GG, GT, TT | GG, GT, TT | C573W |
Numbering starts at the A nucleotide of the primary initiation codon [15].
AA: amino acid
* GG genotype was previously reported [5].
Screening of ENaC α, β, and γ Genes 3' and 5' Flanking Regions for Variations between Dahl R and S rats
We first defined the 5' flanking regions to be screened for each ENaC subunit. The predicted putative binding sites on Brown Norway rat ENaC sequences were identical using TRANSFAC® and TFSEARCH®. Because of the presence of potential kidney and brain transcription factor binding sites, we screened 1.8 kbp, 1.5 kbp, and 4.4 kbp of the 5' flanking regions of ENaC α, β, and γ respectively from the transcription start site (Figures 1, 2, and 3). Compared to previously reported transcription factors binding sites [5-7,13], the present analysis determined five, one, and ten new potential transcription factor binding-sites sequences on ENaC α, β, and γ, respectively (Figures 1, 2, and 3).
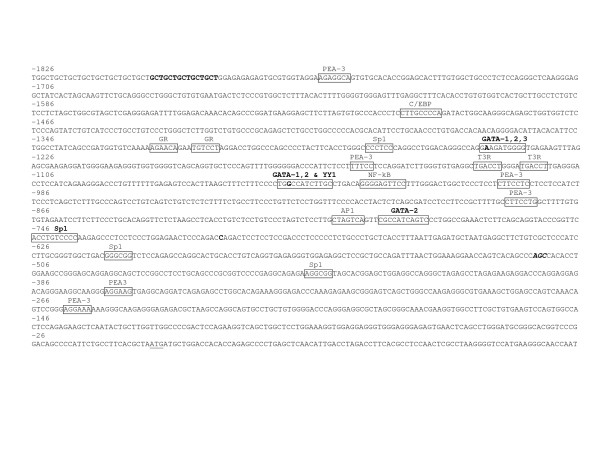
Figure 1.
Location of the variants identified in the 5' flanking region of Dahl S, R, and Wistar rats ENaC α gene on the Brown Norway rat genomic sequence [15]. Position of the variants identified in the current study is highlighted in bold. Boxes represent the putative transcription factor-binding sequences; the putative binding sequences found during the present sequence analysis are labeled in bold; the factor names are written above the boxes. The first three bases for the major kidney and brain transcription start sites are italicized and bold. The translation initiation codon (+1) is underlined. TFSEARCH® scores for the newly assigned putative binding sequences are 93.1, 89.7, and 89.7 for GATA 1, 2, 3 respectively; 89.0 and 88.5 for GATA 1, 2 respectively and 85.8 for YY1; 88.5 for GATA 2, and 87.7 for Sp1.
Figure 2.
Location of the variants identified in the 5' flanking region of Dahl S, R, and Wistar rats ENaC β gene on the Brown Norway rat genomic sequence [15]. Position of the variants identified in the current study is highlighted in bold. Boxes represent the putative transcription factor-binding sequences; the putative binding sequences found during the present sequence analysis are labeled in bold; the factor names are written above the boxes. The first three bases for the major kidney and brain transcription start sites are italicized and bold. TFSEARCH® score for the newly assigned putative binding sequence for STATX is 92.3.
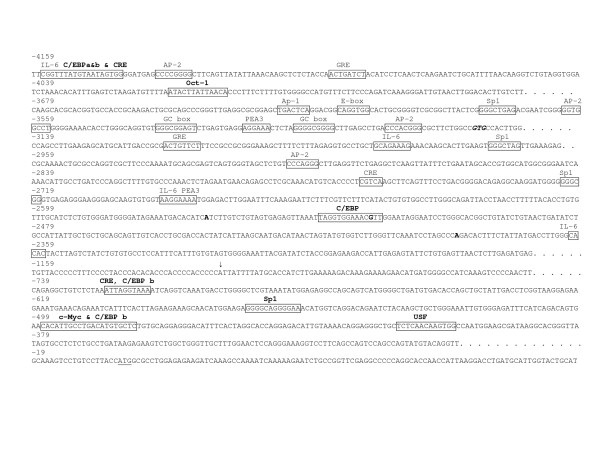
Figure 3.
Location of the variants identified in the 5' flanking region of Dahl S, R, and Wistar rats ENaC γ gene on the Brown Norway rat genomic sequence [15]. Position of the variants identified in the current study is highlighted in bold. Boxes represent the putative transcription factor-binding sequences; the putative binding sequences found during the present sequence analysis are labeled in bold; the factor names are written above the boxes. The first three bases for the major kidney and brain transcription start sites are italicized and bold. The translation initiation codon (+1) is underlined. TFSEARCH® scores for the newly assigned putative binding sequences are 89.2, 87.4, and 89.0 for C/EBP a & b and CRE, respectively; 85.8 for Oct-1; 89.3 for C/EBP, 87.9 and 85.5 for CRE and C/EBPb respectively; 87.7 for Sp1, 91.0 and 87.4 for c-Myc and C/EBPb respectively; and 85.9 for USF.
No sequence variability was identified in both the 3' and 5' flanking regions of ENaC α, β, and γ genes in Dahl S versus R rats (Table 2).
Table 2.
Allelic variants in the 5' flanking regions of ENaC subunits in Dahl S, R, Wistar, and Brown Norway rats.
| Subunit | Nucleotide Position | Dahl S and R | Wistar | Brown Norway |
| ENaC α | -705 | TT | TT | CC, CT, TT |
| -1050 | AA | AA | GG, GA, AA | |
| -1247 | TT | TT | AA, AT, TT | |
| -1788 | Deletion of 5GCTs | Deletion of 5GCTs | ||
| ENaC β | -34381 | AA | GG, GA, AA | GG, GA, AA |
| -34648 | AA | GG, GA, AA | GG | |
| ENaC γ | -1118 | Insertion of CCCCCA | ||
| -1588 | CC | AA, AC, CC | AA | |
| -2054 | TT | TT, CT, CC | CC, CT, TT | |
| -2386 | GG | AA* | AA | |
| -2525 | AA | GG, GA, AA | GG | |
| -2561 | GG | AA, AG, GG | AA | |
| -3313 | CC | CC | TT |
Numbering starts at the A nucleotide of the primary initiation codon [15].
* GG genotype was previously reported [7].
Four homozygous sequence differences were found in both Dahl and Wistar rat ENaC α gene compared to Brown Norway: one homozygous 15 bp deletion (-1788 → -1803 bp), and the loss of three Brown Norway polymorphic sites (Table 2). In addition to previously described transcription factor-binding sites [5,13], computer analyses suggest that the regions from -1248 to -1239, from -1052 to -1047, from -780 to -790, and from -736 to -746 represent putative binding sites for GATA-1, -2, and -3, GATA-1 and 2 and YY1, GATA-2, and Sp1 respectively (Figure 1). The putative GATA-1, -2, and -3 binding site (-1248 to -1239) found in presence of the -1247A allele loses the binding site for GATA-2 in the presence of the T allele. The putative binding site for YY1 (-1052 to -1047) found in presence of the -1050G allele and overlapping GATA-1 and 2 sites, is lost in the presence of the A allele; while GATA-1 and 2 transcription binding sites remained unaltered. Both Dahl and Wistar rats were homozygous TT and AA for respectively the T-1247A and the G-1050A polymorphisms.
The G-34648A ENaC β gene variant, not reported in Brown Norway rats, was present at the homozygous state in Dahl rats and at the heterozygous state in Wistar rats (Table 2). One Brown Norway ENaCβ gene polymorphic site was lost in Dahl rats, but not Wistar rats (Table 2). In addition to previously described transcription factor-binding sites [6] the regions from -34650 to -34635 represent a putative binding site for STAT proteins respectively (Figure 2). However, the presence of the A-34648G polymorphism within this STATX potential site is not predicted to alter STAT protein binding. Dahl rats were homozygous AA for the G-34648A polymorphisms, while Wistar rats were heterozygous.
Four ENaC γ gene sequence variations (A-1588C, G-2525A, A-2561G and T-3313C) were identified in both Dahl and Wistar rats compared to Brown Norway rats, these variations were all homozygous in Dahl rats; in Wistar rats T-3313C was found at the homozygous state, while A-1588C, G-2525A, A-2561G were heterozygous (Table 2). A 6 bp deletion at -1118 was only found in Dahl rat ENaC γ gene (Table 2). The A-2386G ENaC γ gene variant, not reported in Brown Norway rats, was present at the homozygous state in Dahl rats. Our screening of the Wistar ENaC γ gene only found the homozygous AA allele, however, the presence of the G allele was previously reported [7] (Table 2). One previously reported Brown Norway polymorphic site ENaC γ was lost in Dahl rats, but not Wistar rats (Table 2). In addition to previously described transcription factor-binding sites [7], the regions from -4102 to -4120, from -4010 to -4022, from -2483 to -2497, from -696 to -706, from -607 to -619, from -442 to -463, and from -400 to -413 represent putative binding sites for C/EBP a & b and CRE, Oct-1, C/EBP, CRE and C/EBP b, Sp1, c-Myc and C/EBP b, and USF respectively (Figure 3). One polymorphism, the G-2525A polymorphism, out of the seven found in the 5' flanking region of ENaC γ gene, is located within a potential consensus binding sites for C/EBP (Figure 3). The C/EBP binding site is present when the -2525G allele is present and the binding site is lost in the presence of the A allele. Dahl rats were homozygous AA for the G-2525A polymorphism, while Wistar rats were heterozygous and Brown Norway rats homozygous for the G allele. Finally, the 6 bp homozygous insertion at position -1118, which is only present in Dahl rats, is predicted to create Sp1 and Oct-1 sites (CCCCACCCCATT) (TFSEARCH® scores 87.7 and 85.8, respectively).
Discussion
The results of the current study show that Dahl S and R inbred rats are isogenic in the entire coding regions, exon-intron junctions, 3' and 5' flanking studied regions of ENaC α, β, and γ genes. These results are in agreement with the initial partial screenings of ENaC subunits in Dahl rats [9,10]. Nine homozygous sequence variations were identified in ENaC genes in Dahl rats compared to the Brown Norway sequence available in the public domain and eight of these nine sequence variations (5 polymorphic and 3 homozygous) were also identified in Wistar rats. However, the 6 bp deletion at -1118 which in the γ ENaC 5' flanking region, was specific for Dahl rats and not found in Wistar or Brown Norway rats. Eleven and five Brown Norway polymorphic sites were lost in Dahl and Wistar rats respectively. Among the identified variations in Dahl and Wistar rats, three are non synonymous variations. Four variants in the 5' flanking regions of ENaC α, β, and γ genes are present within putative DNA consensus regulatory elements and two putative DNA consensus sites were introduced with the Dahl rat specific 6 bp insertion at -1118 in ENaC γ.
Of the three non synonymous variants identified in the present study (αD75E, γE255K, and γC573W), only γC573W, located in the transmembrane domain M2, was previously functionally assessed and was found not to modify ENaC activity in Xenopus oocytes [10]. The αD75E substitution is present in the N terminus of the channel, prior to the transmembrane domain M1 and close to the channel pore, a critical region for kinetics properties of the channel predicted to participate in channel gating [10,14]. γE255K located in the extracellular loop might alter the amiloride sensitivity since residues essential for the formation of the high affinity amiloride-binding sites reside within this domain [1]. The silent polymorphism C542C was previously documented in ENaC γ carboxy terminus within and between rat strains [10].
Previous studies analyzed 1.5 kbp of ENaC α, 1.3 kbp of ENaC β, and 4.2 kbp of ENaC γ from the transcription start site in Wistar, Sprague Dawley and Wistar rats respectively as well as the first intron in Wistar rat ENaC γ [5-7,13]. In the present analysis, we determined five, one, and ten new potential transcription factor binding-sites sequences on ENaC α, β, and γ, respectively. Among all variants identified within putative transcription factors binding sites, one of them, the insertion of -1118CCCCCA, located on ENaC γ gene was only present in Dahl rats. To our knowledge, this variant has not been previously reported on Dahl rats or any other rat strains [7,15,16]. It creates binding sites for Sp1 and Oct-1. Sp1 is a DNA-binding protein which interacts with a variety of gene promoters containing GC-box elements. Moreover, the activity of TATA-less promoters is frequently dependent on Sp1 sites in the proximal promoter region. Deletion of one of the two clusters of Sp1 consensus binding sites within the 5' flanking region of the ENaC β gene indicated that the proximal cluster was essential to basal promoter activity in transfected cell lines [6]. Studies of both the human and rat γ -subunits [7,17] also reported the absence of a TATA box in their promoters and the presence of GC boxes and Sp1 consensus sites. In human ENac γ, Sp1 may be part of the transcription complex that binds at the core promoter [17]. Sp1 protein is part of a much larger family of mammalian transcription factors, the Sp/XKLF family [18], it is ubiquitously expressed, and contains three highly conserved C2H2-type zinc fingers in the C-terminal region and have a glutamine-rich activation domain. Sp1 can bind GC boxes, and acts as a transcriptional activator. Therefore the presence of a potential additional Sp1 consensus site in Dahl rats may enhance the transcription of the ENaC γ gene in Dahl rats compared to other rat strains. Similarly, Oct-1 sites have previously been reported to be able to function as repressors or activators of transcription depending on context [19-21]. Overexpression of ENaC γ in collecting duct cells has been shown to enhance Na+ transport [22]. Remarkably, overexpression of marginally detectable amount of ENaC γ was sufficient to produce a full increase in Na+ transport [22]. However, determination of the precise mechanism of all above variants in influencing promoter activity awaits further investigation.
It remains possible that mutations in the intronic regions of the ENaC are involved in the generation of hypertension in Dahl S rats on high salt intake. Commonly occurring ENaC variants, including intronic substitution (i12-17CT) were found associated with an increased urinary potassium excretion rate in relation to the renin levels as well as with hypertension in humans [23]. However, when expressed in Xenopus oocytes, the variants did not show a significant difference in activity compared with ENaC wild-type [23].
One can speculate that mutations in genes encoding a protein interacting with ENaC to regulate its activity might increase ENaC activity leading to hypertension. SGK1, which activates ENaC in tubules, maps to a known BP QTL [24]. Abnormal regulation of SGK1 mRNA and protein level by aldosterone in Dahl S compared to Dahl R rat was observed suggesting that regulation of ENaC via SGK1 signaling pathway may be disturbed in Dahl S rat [25].
Conclusion
To our knowledge, this report presents the first comprehensive screening for variations in the entire coding sequences including intron-exon junctions, and in the 3' and 5' flanking regions of ENaC three genes in the hypertensive Dahl S rats and their normotensive Dahl R control rats together with an additional control group of Wistar rats. We could not link salt induced hypertension in Dahl S to differences in ENaC sequences in Dahl S versus R rats. Further characterization of SNPs across candidate genes contributing to the salt-sensitive hypertension phenotype will be useful in designing genetic mapping panels for association studies. If disordered activity of the epithelial cell sodium channel contributes to the pathogenesis of hypertension in Dahl S rats, it appears to stem from genetic variations in genes encoding proteins that regulate ENaC or in intronic sequences important for the structure or function of the sodium channel.
Methods
Animals
Male Dahl S and R rats (4 rats/group), 4–5 wks of age, were obtained from Harlan Sprague Dawley (Indianapolis, IN) and handled as previously described [8]. To assess the salt sensitivity of Dahl S rats, at 5 wks of age, Dahl S and R rats were placed on a high-salt (1,370 μmol Na/g, Teklad; Madison, WI) diet for 4 wks. After 4 wks, BP was measured invasively by intraarterial catheter and the average mean arterial pressure was estimated to be 156 ± 11 for S, and 131 ± 1 for R rats (P < 0.05). Wistar rats (Charles River Breeding Laboratories Montreal, QC, Canada) were used as control, and were not subjected to high salt diet. The animals were then killed by decapitation and whole blood was collected for DNA isolation. All experiments were carried out in accordance with the guidelines of the University of Ottawa Animal Care Committee for the care and use of laboratory animals.
Genomic DNA isolation and amplification
Genomic DNA was isolated from white blood cells (Qiagen FlexiGene, Qiagen Canada, Mississauga, ON, Canada). Using PRIMER3-based Web application [26], 73 sets of specific oligonucleotide primers (Table 3) where designed based on the November 2004 rat (Rattus norvegicus) genome assembly [15] in order to screen the coding sequence of ENaC three genes, including the exon-intron boundaries, as well as the 5' and 3' flanking regions [GenBank: NM_031548; NM_012648; NM_017046] [GeneID: 25122; 24767;24768]. For large exons (> 350 bp) overlapping primer sets were employed.
Table 3A.
Primers designed to screen the coding sequence of ENaC α, β, and γ genes. Optimum temperatures (°C) for PCR and DHPLC are included in the table.
| Sequence | PCR | DHPLC | ||
| ENaC α | ||||
| Exon 1 | Forward | GGTAGCACGGAGCTGGAG | 64 | 63 |
| Reverse | CCCAGGCTGAGTTCACTCTC | |||
| Forward | AGCTCAATACTGCTTGGTTGG | 61 | 63 | |
| Reverse | GAAGAGCTCCCGGTAGGAG | |||
| Forward | GGGACAAACGTGAAGAGCAG | 61 | 63 | |
| Reverse | CTTGCTTTTGTGCTGCTGAG | |||
| Exon 2 | Forward | CACTGATCCCCTCCGTGTTA | 60 | 64 |
| Reverse | TCCATCAGGCCTCTATCTGAA | |||
| Exon 3 | Forward | GCTCTCTGTCCCTCACCTTG | 63 | 62 |
| Reverse | ACCACCAAGCATTTCCTGAG | |||
| Exon 4 | Forward | CACATTGGAGGTGACAGGAA | 63 | 61 |
| Reverse | CCAAGGTGAGACCCAGAAGA | |||
| Exon 5 | Forward | GCTGGCCTTGTTCTATCAGG | 62 | 62 |
| Reverse | CTCCCTTCAGTCTCCTGCTG | |||
| Exon 6 | Forward | GGTGAAGCCTGAGTCATTCC | 63 | 62 |
| Reverse | CCAACCTACTTCCCCTCCAT | |||
| Exon 7 | Forward | GAGGGATGGAGGGGAAGTAG | 61 | 61 |
| Reverse | AGCCAGCACCTAGGGAAAA | |||
| Exon 8 | Forward | GGCACCATTGAAATGCTCTT | 59 | 61 |
| Reverse | ATCAAAGTGCCCAGTTACGG | |||
| Exon 9 | Forward | GCAGCTGCTTAACCTGGTAGA | 61 | 61 |
| Reverse | ATGTCCACTTGTGCGTGTGT | |||
| Exon 10 | Forward | CCATCCCTGTAAACATGAGG | 61 | 61 |
| Reverse | CCCCAATATCTCCACCAGAA | |||
| Exon 11 | Forward | TGGTGGAGATATTGGGGAGA | 61 | 61 |
| Reverse | CAAACCCTTCTGACCCTTCA | |||
| Exon 12 | Forward | TGACAGGAGGCGCTAGAGT | 62 | 63 |
| Reverse | AGTAGCATAGGCAGGTGGAG | |||
| Forward | CTCCACTCCAGCTTCCTCCT | 62 | 63 | |
| Reverse | ATCGTTAGCCCCTGTCCTCT | |||
| Forward | TCTCACTTCAGCACATCTTCC | 61 | 63 | |
| Reverse | GGCCTACCCTGGTCTGTCTT | |||
| Forward | ACCCAAAAGCCCCCTTGT | 61 | 61 | |
| Reverse | AGTACACTGTGGGGGTGAGG | |||
| Forward | CCAAAGGCACCATTTCTTTT | 59 | 61 | |
| Reverse | ATGTAGGCGGTGCCTCAG | |||
| ENaC β | ||||
| Exon 2 | Forward | CTAGTCTCCAGGCCCATGAC | 62 | 64 |
| Reverse | CTACTGGAAGGGGCTGGAAT | |||
| Exon 3 | Forward | CCCCATGTTCCACACTCTTT | 60 | 62 |
| Reverse | AACAAAAATCGATTGCTACCAG | |||
| Exon 4 | Forward | TAATACGGTGCTGCCATTCC | 61 | 63 |
| Reverse | GCATAGATCAGCCTGTGTGC | |||
| Exon 5 | Forward | CTCCAGCAGAGCAGGACAAT | 62 | 61 |
| Reverse | GGTCTTTCCGCCCTGTGT | |||
| Exon 6 | Forward | GGTGATGGCCTCCCTTCTAT | 61 | 61.5 |
| Reverse | AGGCAGCCTGAACACAAGAG | |||
| Exon 7 | Forward | GCTCCATGGGGAGGTACATA | 63 | 63 |
| Reverse | GTCCGGCCCTCATAGGTAAG | |||
| Exon 8 | Forward | AAAGTCTCTGGGCTCCAAGG | 63 | 61.5 |
| Reverse | AGAGGCCCCCTTGCCTAAC | |||
| Exon 9 | Forward | GAGGGAAGACCCCTGGAAG | 62 | 63.5 |
| Reverse | ATACTGGGTGTCGCTGGAAG | |||
| Exon 10 | Forward | TCCTGCAAGTGAGTGTGTCC | 62 | 63 |
| Reverse | AGGGGGAGAAGACCCTCTTT | |||
| Exon 11 | Forward | AGCATGTGTGTGCGTGTGTA | 60 | 63 |
| Reverse | CAGCAAGAGCAGTTTGGACA | |||
| Exon 12 | Forward | CAACTCAGACTGGAGATGAGCA | 62 | 62.5 |
| Reverse | GGAACCATAACCCCCACCTA | |||
| Exon 13 | Forward | CCTAGGTGGGGGTTATGGTT | 62 | 64 |
| Reverse | GCAGCCTCAGGGAGTCATAG | |||
| Forward | CTTCCAGCCTGACACAACC | 62 | 63.5 | |
| Reverse | GCCTGTCTGCTAGGTCAACA | |||
| Forward | CTGAGGGGTTCATAGGGTCA | 62 | 60.5 | |
| Reverse | ATCCTACACCCCAGACATGC | |||
| ENaC γ | ||||
| Exon 2 | Forward | AGTCGCAGGCTCCAGAGAT | 62 | 64 |
| Reverse | GAGGGGCTTTGACATCCAT | |||
| Exon 3 | Forward | GGAAGGCAACATGAAGAAGC | 61 | 60 |
| Reverse | ACTGTGAGCCCACCAGCTC | |||
| Exon 4 | Forward | AAGGACCTACCCTGGCATCT | 63 | 60 |
| Reverse | CTCAAGGCCTACAGGTGAGC | |||
| Exon 5 | Forward | ACGCAATGCTTCCTGACTTC | 60 | 60 |
| Reverse | TGCACTGCTGCTGTCAAGAT | |||
| Exon 6 | Forward | CCCTGGGGACTGCTTTTT | 60 | 60 |
| Reverse | TCCATTAGCAGCACCTCCTT | |||
| Exon 7 | Forward | AGGGGAATCCTCCTATCTGG | 61 | 62 |
| Reverse | CCTTGGCCTAGATATAGCTTCA | |||
| Exon 8 | Forward | AGGGAGTTCCCGGTCTCTAC | 63 | 61 |
| Reverse | GCGTGGCCAAGCTGATTC | |||
| Exon 9 | Forward | AGATGGTGGAGGTTCCACAG | 63 | 60 |
| Reverse | GGGAGAAAGGCACAGAGTGA | |||
| Exon 10 | Forward | ACTGGGGCAGGTAGGACTTT | 62 | 60 |
| Reverse | GCTTTGGCTGTGTTGCTGTA | |||
| Exon 11 | Forward | CCAAAGCCAGAGACAGGTTG | 59 | 61 |
| Reverse | TCGAATGAACGAAAAGGTGA | |||
| Exon 12 | Forward | ACCTGGCAGGAAGCCAAG | 62 | 61 |
| Reverse | CCCTCTGGCAGCAAAACTAC | |||
| Exon 13 | Forward | CCCTGAGTGCAGGATTTATCA | 61 | 64 |
| Reverse | GTATCTGGGAGGTGGTGTGC | |||
| Forward | GTCAGTGGCACAAAGCCAAG | 62 | 64 | |
| Reverse | AGCTCATAAGTGCCAAGTCCA | |||
| Forward | CCTGCTGTGAACCGGATA | 60 | 60 | |
| Reverse | GGCCAACTGTCTGTCTGAGG | |||
| Forward | GCCAGCTATTGCCTGACAT | 60 | 60 | |
| Reverse | CGCATACTCTCAGTTCAAAGACA | |||
| Forward | GCCAAATGGTATTCCCACAA | 59 | 57 | |
| Reverse | ATTGGACTAGCCTGGGTGCT |
Table 3B.
Primers designed to screen the 5'flanking sequence of ENaC α, β, and γ genes. Optimum temperatures (°C) for PCR and DHPLC are included in the table.
| Sequence | PCR | DHPLC | ||
| ENaC α | ||||
| Forward | CAGATTCAGCTGCCATGC | 60 | 62 | |
| Reverse | AGGCCCTGCAGAACTTGCT | |||
| Forward | GGTAGGAAGAGGCAGTGTGC | 61 | 62 | |
| Reverse | ACAGGGTTGCAGGAATGTG | |||
| Forward | GCTGGTGGTCTCTCCCAGTA | 64 | 62 | |
| Reverse | GGTCCCTTCTGATGGAGGTC | |||
| Forward | GGGGGACCCATTCTCCTT | 62 | 59 | |
| Reverse | GGACTGATGGCGAACTGACT | |||
| Forward | CCTTCCTGGCTTTTGTGTGT | 61 | 62 | |
| Reverse | GTGTGGCTGGGCTGTGAC | |||
| Forward | CAGGCACTGCACCTGTCA | 62 | 62 | |
| Reverse | GGCTTAGCGTCTCTCCCTCT | |||
| Forward | CACAGAAAGGGAGACCCAAA | 60 | 63 | |
| Reverse | GGTCTTGCTCCTTGAATTGG | |||
| Forward | GAGGGCAGCCTGGGATGCGG | 61 | 62 | |
| Reverse | TTATAATAGCAATAGCCCCA | |||
| Forward | TCCAAGGAGAAGGCGCCCCCA | 61 | 62 | |
| Reverse | AGGGCTGGGTGAGAGGAT | |||
| Forward | GGTTTAAGGATTTGCTTGATTC | 61 | 61 | |
| Reverse | TGTTCTGCAAGGACAGCATC | |||
| Forward | CAAAGTACCCAATATCTATT | 61 | 61 | |
| Reverse | AGGGCTGGGTGAGAGGAT | |||
| Forward | CCTGGTTTTGGGGTGTGT | 61 | 61 | |
| Reverse | TGTTCTGCAAGGACAGCATC | |||
| Forward | GCTCTCTTTGGGCTGTGGGGAC | 61 | 61 | |
| Reverse | TGTTCTGCAAGGACAGCATC | |||
| ENaC β | ||||
| Forward | GGTCTTCTGGGAAGAGCAAA | 61 | 62 | |
| Reverse | CACGGTTCCATTTCCTGTGT | |||
| Forward | CAGCCATGCACATGAAGACT | 61 | 57 | |
| Reverse | TTGCAGCCAACTTGACTAGAT | |||
| Forward | TGGTGATACCCTTCTTGTTGG | 61 | 60 | |
| Reverse | CCTCCGTGTATCCTCACTCC | |||
| Forward | AAAATAGAACGGGGCTGTGA | 61 | 62 | |
| Reverse | GGAGCTGGGTCACCTGTAGA | |||
| Forward | TGACCTGGAGCCTGTCAACT | 62 | 64 | |
| Reverse | GACGGAACTGCGGTCATT | |||
| ENaC γ | ||||
| Forward | CTTGACATGTTTCTACCCACCA | 61 | 58 | |
| Reverse | TTAGAACGCTGAAACCGTGA | |||
| Forward | CATCCTCAACTCAAGAATCTGC | 60 | 60 | |
| Reverse | CCACTCTGCAAGCTGCATTA | |||
| Forward | TGCAGAAGCAGCAGTAAGAGA | 61 | 62 | |
| Reverse | CTGGCGTGTGTACAGTCTGG | |||
| Forward | CTTCTGGCCGTGCCACTT | 61 | 62 | |
| Reverse | TCGGGCTCCTCTTTCAACTA | |||
| Forward | CGCGGGAAAGCTTTTCTTTA | 61 | 63 | |
| Reverse | CTTGCTCCCTTCCCTCTCAC | |||
| Forward | ATGAACAGAGCCTCGCAAAC | 61 | 62 | |
| Reverse | GATAGTGGTCGCAGGTGACA | |||
| Forward | GGTGGAAACGTTGGAATAGG | 62 | 58 | |
| Reverse | AAAGCAAGGCTGTCGCTCTA |
The nucleotides representing the entire coding sequences and the 3'UTR and flanking regions that were screened in ENaC genes are as follows (numbering starts at the A nucleotide of the primary initiation codon): for ENaC α, nt. 1 to 590, 9199 to 9652, 10553 to 10887, 15934 to 16281, 16442 to 16777, 17027 to 17271, 17246 to 17536, 20442 to 20730, 20663 to 20874, 20927 to 21134, 21117 to 21372, and 21582 to 23041; for ENaC β, nt. 1 to 413, 3287 to 3686, 5622 to 6012, 23724 to 23969, 25539 to 25839, 25974 to 26213, 27914 to 28154, 28987 to 29221, 29116 to 29351, 29910 to 30157, 30857 to 31100, and 31078 to 32049; for ENaC-γ nt, 1 to 378, 2108 to 2552, 3781 to 4141, 5153 to 5403, 8093 to 8385, 22437 to 22674, 24560 to 24797, 25439 to 25676, 25616 to 25849, 25842 to 26091, 28671 to 28918, 29065 to 30547. As for the 5' flanking regions of ENaC genes, -2078 to +1 bp, -34812 to -33582 bp, -4359 to +1 bp of ENaC α, β, and γ genes respectively were amplified.
Sequence Analysis
a) DHPLC (Helix, Varian, Palo Alto, CA) analysis. DHPLC runs were performed as recommended by Varian using buffer A and buffer B (Varian) and a flow rate of 0.45 ml/min. Freshly prepared PCR products were denatured at 95°C for 3 min and re-annealed by decreasing the temperature from 95°C to 64°C at a rate of 1°C/min. Optimal melting temperatures for the PCR products were determined using the Stanford University website [27]. The chosen temperatures correspond to the point at which the retention time was 75% of (trmax -trmin). b) Automatic sequencing (ABI 310, PE Applied Biosystems, Foster City, CA). Sequencing was performed using the DYEnamic ET Terminator kit according to the instructions provided by the manufacturer (PE Applied Biosystems, Foster City, CA). Sequencing products were purified (DyeEx 2.0 spin kit columns; Qiagen Canada, Mississauga, ON, Canada) and analysed on 310 DNA analyser (Applied Biosystems, Foster City, CA). Resulting sequences were compared to the Brown Norway sequence [15]. c) ENaC 5' flanking regions analysis. A comprehensive analysis of putative kidney or brain transcription factor binding sites was performed using both literature reports [6,7,13] and two well-known and large-scale databases, TRANSFAC [28]® and TFSEARCH® [29]. Using the cell selectivity track, the database searches were refined to transcription factors active in the rat kidney and brain where ENaC contributes to BP control.
Authors' contributions
MS performed the all experiments and sequences analysis, participated in experimental design, drafted the manuscript.
FL conceived the project, finalized the manuscript.
FT designed the experimental strategy, supervised the study, and finalized the manuscript.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are grateful for the generous advice and assistance of Drs. M. Ahmad, N. Sylvius and K. Adamo and Mrs. P Bolongo.
This study was supported by an operating grant MOP-74432 from the Canadian Institutes of Health Research, and by Program Grant PRG5275 from the Heart and Stroke Foundation of Ontario. F. H. H. Leenen holds the Pfizer Chair in Hypertension Research, an endowed chair supported by Pfizer Canada, the University of Ottawa Heart Institute Foundation, and the Canadian Institutes of Health Research. M Shehata was supported by an Ontario Graduate Scholarship and an Ontario Graduate Scholarship for Science and Technology.
Contributor Information
Marlene F Shehata, Email: mshehata@ottawaheart.ca.
Frans HH Leenen, Email: fleenen@ottawaheart.ca.
Frédérique Tesson, Email: ftesson@ottawaheart.ca.
References
- Benos D, Stanton B. Functional domains within the degenerin/epithelial sodium channel (Deg/ENaC) superfamily of ion channels. The journal of Physiology. 1999;520:631–644. doi: 10.1111/j.1469-7793.1999.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Grunder S, Hanukoglu A, Rösler A, Mathew P, Hanukoglu I, Schild L, Lu Y, Shimkets R, Nelson-Williams C. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type. Nature Genetics. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- Hansson J, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton R. Hypertension caused by a truncated epithelial sodium channel big gamma subunit: genetic heterogeneity of Liddle syndrome. Nature Genetics. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- Saxena A, Hanukoglu I, Strautnieks SS, Thompson RJ, Gardiner RM, Hanukoglu A. Gene structure of the human amiloride-sensitive epithelial sodium channel beta subunit. Biochem Biophys Res Commun. 1998;252:208–213. doi: 10.1006/bbrc.1998.9625. [DOI] [PubMed] [Google Scholar]
- Otulakowski G, Rafii B, Bremner H, O'Brodovich H. Structure and Hormone Responsiveness of the Gene Encoding the a-Subunit of the Rat Amiloride-Sensitive Epithelial Sodium Channel. American Journal of Respiratory Cell and Molecular Biology. 1999;20:1028–1040. doi: 10.1165/ajrcmb.20.5.3382. [DOI] [PubMed] [Google Scholar]
- Bremner H, Freywald T, O'Brodovich H, Otulakowski G. Promoter analysis of the gene encoding the β-subunit of the rat amiloride-sensitive epithelial sodium channel. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2002;282:124–134. doi: 10.1152/ajplung.2002.282.1.L124. [DOI] [PubMed] [Google Scholar]
- Thomas C, Auerbach S, Zhang C, Stokes J. The structure of the rat amiloride-sensitive epithelial sodium channel gamma subunit gene and functional analysis of its promoter. Gene. 1999;228:111–122. doi: 10.1016/S0378-1119(99)00016-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Leenen F. Brain sodium channels and central sodium-induced increases in brain ouabain-like compound and blood pressure. J Hypertens. 2003;21:1519–1524. doi: 10.1097/00004872-200308000-00016. [DOI] [PubMed] [Google Scholar]
- Huang H, Pravenec M, Wang J, Kren V, StLezin E, Szpirer C, Szpirer J, Kurtz T. Mapping and sequence analysis of the gene encoding the beta subunit of the epithelial sodium channel in experimental models of hypertension. J Hypertens. 1995;13:1247–1251. doi: 10.1097/00004872-199511000-00005. [DOI] [PubMed] [Google Scholar]
- Gründer S, Zagato L, Yagil C, Yagil Y, Sassard J, Rossier B. Polymorphisms in the carboxy-terminus of the epithelial sodium channel in rat models for hypertension. J Hypertens. 1997;15:173–179. doi: 10.1097/00004872-199715020-00008. [DOI] [PubMed] [Google Scholar]
- Ellis L, Taylor C, Taylor G. A comparison of fluorescent SSCP and denaturing HPLC for high throughput mutation scanning. Human Mutation. 2000;15:556–564. doi: 10.1002/1098-1004(200006)15:6<556::AID-HUMU7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Eshleman S, Crutcher G, Petrauskene O, Kunstman K, Cunningham S, Trevino C, Davis C, Kennedy J, Fairmans J, Foley B. Sensitivity and Specificity of the ViroSeq Human Immunodeficiency Virus Type 1 (HIV-1) Genotyping System for Detection of HIV-1 Drug Resistance Mutations by Use of an ABI PRISM 3100 Genetic Analyzer. Journal of Clinical Microbiology. 2004;43:813–817. doi: 10.1128/JCM.43.2.813-817.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otulakowski G, Freywald T, Wen Y, O'Brodovich H. Translational activation and repression by distinct elements within the 5'-UTR of ENaC a-subunit mRNA. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2001;281:1219–1231. doi: 10.1152/ajplung.2001.281.5.L1219. [DOI] [PubMed] [Google Scholar]
- Berdiev B, Karlson K, Jovov B, Ripoll P, Morris R, Loffing-Cueni D, Halpin P, Stanton B, Kleyman T, Ismailov I. Subunit Stoichiometry of a Core Conduction Element in a Cloned Epithelial Amiloride-Sensitive Na+ Channel. Biophysical Journal. 1998;75:2292–2301. doi: 10.1016/S0006-3495(98)77673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat (Rattus norvegicus) Genome Browser Gateway http://www.genome.ucsc.edu/cgi-bin/hgGateway
- Rat Genome Database: VCMAP http://rgd.mcw.edu/VCMAP/
- Auerbach S, Loftus R, Itani O, Thomas C. Human amiloride-sensitive epithelial Na channel g subunit promoter: functional analysis and identification of a polypurine-polypyrimidine tract with the potential for triplex DNA formation. Biochem J. 2000;347:105–114. doi: 10.1042/0264-6021:3470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker J. Kruppel-like Factors: Three Fingers in Many Pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Kim M, Lesoon-Wood L, Weintraub B, Chung J. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Molecular and Cellular Biology. 1996;16:4366–4377. doi: 10.1128/mcb.16.8.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever-Chroneos Z, Bachurski C, Yan C, Whitsett J. Regulation of mouse SP-B gene promoter by AP-1 family members. American Journal of Physiology-Lung Cellular and Molecular Physiology. 1999;277:79–88. doi: 10.1152/ajplung.1999.277.1.L79. [DOI] [PubMed] [Google Scholar]
- Takimoto M, Quinn J, Farina A, Staudt L, Levens D. fos/jun and octamer-binding protein interact with a common site in a negative element of the human c-myc gene. Journal of Biological Chemistry. 1989;264:8992–8999. [PubMed] [Google Scholar]
- Volk K, Husted R, Sigmund R, Stokes J. Overexpression of the Epithelial Na Channel {gamma} Subunit in Collecting Duct Cells. Journal of Biological Chemistry. 2005;280:18348–18354. doi: 10.1074/jbc.M413689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannila-Handelberg T, Kontula K, Tikkanen I, Tikkanen T, Fyhrquist F, Helin K, Fodstad H, Piippo K, Miettinen H, Virtamo J. Common variants of the beta and gamma subunits of the epithelial sodium channel and their relation to plasma renin and aldosterone levels in essential hypertension. BMC Medical Genetics. 2005;25:4. doi: 10.1186/1471-2350-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjah M, Roxas B, Geenen D, Danziger R. Dietary Salt Regulates Renal SGK1 AbundanceRelevance to Salt Sensitivity in the Dahl Rat. Hypertension. 2003;41:874–878. doi: 10.1161/01.HYP.0000063885.48344.EA. [DOI] [PubMed] [Google Scholar]
- Aoi W, Niisato N, Sawabe Y, Miyazaki H, Marunaka Y. Aldosterone-induced abnormal regulation of ENaC and SGK1 in Dahl salt-sensitive rat. Biochem Biophys Res Commun. 2006 doi: 10.1016/j.bbrc.2005.12.194. [DOI] [PubMed] [Google Scholar]
- Automatic primer design http://primers.niob.knaw.nl
- DHPLC Melt Program http://insertion.stanford.edu/melt.html
- Matys V, Kel-Margoulis O, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K. TRANSFAC® and its module TRANSCompel®: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A, Kel O, Ignatieva E, Ananko E, Podkolodnaya O, Kolpakov F. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Research. 26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]