Abstract
Alternative pre-mRNA splicing increases proteomic diversity and provides a potential mechanism underlying both phenotypic diversity and susceptibility to genetic disorders in human populations. To investigate the variation in splicing among humans on a genome-wide scale, we use a comprehensive exon-targeted microarray to examine alternative splicing in lymphoblastoid cell lines (LCLs) derived from the CEPH HapMap population. We show the identification of transcripts containing sequence verified exon skipping, intron retention, and cryptic splice site usage that are specific between individuals. A number of novel alternative splicing events with no previous annotations in either the RefSeq and EST databases were identified, indicating that we are able to discover de novo splicing events. Using family-based linkage analysis, we demonstrate Mendelian inheritance and segregation of specific splice isoforms with regulatory haplotypes for three genes: OAS1, CAST, and CRTAP. Allelic association was further used to identify individual SNPs or regulatory haplotype blocks linked to the alternative splicing event, taking advantage of the high-resolution genotype information from the CEPH HapMap population. In one candidate, we identified a regulatory polymorphism that disrupts a 5′ splice site of an exon in the CAST gene, resulting in its exclusion in the mutant allele. This report illustrates that our approach can detect both annotated and novel alternatively spliced variants, and that such variation among individuals is heritable and genetically controlled.
The human genome is estimated to contain ∼20,000–25,000 genes, and recent studies suggest that ∼50%–75% of multi-exon genes undergo alternative splicing (AS), generating multiple mRNA isoforms and greatly increasing human proteomic diversity (Lander et al. 2001; Modrek et al. 2001). The splicing of mRNA is a highly regulated process involving the interactions of trans-acting splicing factors and cis-acting regulatory motifs. Disruptions of this process through mutations within these factors and regulatory signals may play an important role in phenotypic diversity and genetic disorders (Faustino and Cooper 2003; Nissim-Rafinia and Kerem 2005; Black and Graveley 2006).
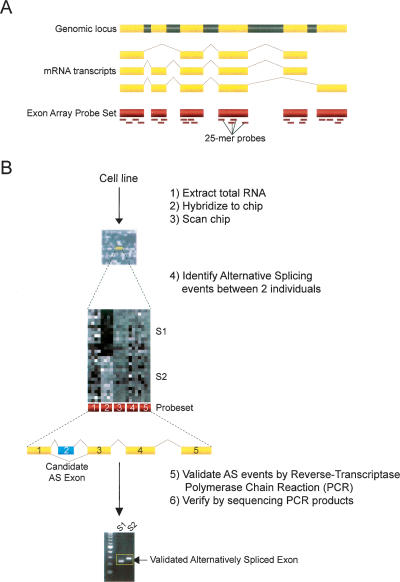
Recent advances in microarray technology hold great promise for the genome-wide detection of AS events (Lee and Roy 2004). Small to large-scale microarrays have been designed using probes spanning predicted exon junctions (Modrek et al. 2001; Johnson et al. 2003; Ule et al. 2005; Sugnet et al. 2006; Zhang et al. 2006), probes targeted toward individual exons (Frey et al. 2005), or a combination thereof (Srinivasan et al. 2005) and applied to identification of AS events that are tissue-specific, for the most part. However, one caveat of these studies utilizing customized arrays is a bias toward genes with solid EST and cDNA evidence for known AS events and that are therefore limited in their usefulness as a discovery tool for de novo splicing events. Here, we have chosen to use an alternative array design, the Affymetrix GeneChip Human Exon 1.0 ST Array, which is less biased toward known AS events by targeting multiple probes to individual exons and allowing simultaneous, exon-level detection of expression levels for 1.4 million probe sets covering over one million known and predicted human exons (Fig. 1). Exon-tiling arrays have several advantages over exon-junction arrays: flexibility of probe placement, exact transcript structures do not need to be known a priori, and most AS events can be monitored without designing probes specific to all possible junctions. However, it should be noted that exon arrays do not provide immediate information on transcript structures containing candidate alternative events.
Figure 1.
(A) Schematic for coverage of probe sets across the entire length of the transcript. Yellow regions are exons, whereas gray regions represent introns. The short dashes underneath the exon regions indicate individual probes of 25 nucleotides in length representing the probe set. The Affymetrix GeneChip Human Exon 1.0 ST Array allows for exon-level expression profiling in a single chip, and can interrogate over one million predicted exons within the human genome. (B) Flowchart for processing and analysis of chips to validation of alternative splicing events. Total RNA is extracted from the two cell lines (n = 15 replicates per individual) and is transcribed to cDNA and labeled with biotin. The total cDNA is then hybridized to the exon chip, followed by washing and staining with an anti-streptavidin antibody. Chips are then scanned, and hybridization data are processed and analyzed by the Affymetrix Power Tools (version 1.6) software package. A splicing index is calculated for ∼1.4 million probe sets covering one million exons. A subset of 20 alternative splicing events predicted between the two individuals using an unpaired t-test (P < 8.915 × 10−4) on the splicing index and other criteria (see Methods), are then validated by (1) RT-PCR using exon body primers flanking the probe set of interest and (2) sequencing of the RT-PCR products.
We show that (1) the Exon Array is able to detect AS at a level that is comparable in sensitivity as other microarray methods, and (2) we can identify quantitative and qualitative variations in splicing among individuals. Preliminary analysis estimates that up to 5% of all RefSeq exons are differentially spliced between individuals. Our approach for establishing a genetic basis for the variation in splicing uses lymphoblasts derived from individuals of the CEPH population (Cohen et al. 1993), where we take advantage of the high resolution HapMap genotype information from these samples (Altshuler et al. 2005) to perform allelic association studies.
Results
Examination of splicing differences between two CEPH HapMap individuals
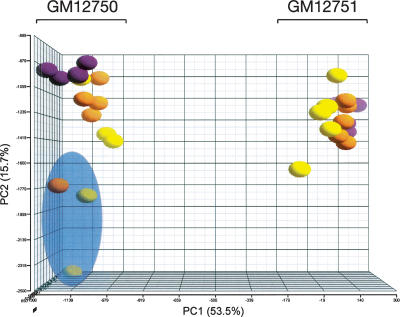
We investigated differences in exon-level expression in lymphoblastoid cell lines (LCLs; three biological and five technical replicates, for a total of 15 replicates per individual) from two unrelated individuals from the CEPH HapMap population (GM12750 and GM12751). We defined the splicing index (SI) as the expression level of a given probe set (representing one exon) divided by the expression of the corresponding meta-probe set (representing the gene), to control for differences in gene expression levels between samples (Clark et al. 2002; Srinivasan et al. 2005). Principal component analysis (PCA) indicates that the majority of the variance in SI is due to individual differences, while the remainder is due to biological and technical factors, suggesting that splicing variation between the two cell lines is frequent (Fig. 2). Three of the replicates from individual GM12750 appear to be outliers and were removed from all subsequent analyses.
Figure 2.
Principal component analysis. A three-dimensional plot of the splicing index data showing the three passages of five technical replicates each of individuals GM12750 and GM12751, on the left and right sides, respectively. The three biological replicates are shown as purple, orange, and yellow spheres, respectively. The three outliers that were removed from all subsequent analyses are shaded in a blue sphere. The percentage of variance attributed to principal components one and two is shown on the X- and Y-axes, respectively. Plots were created using the Partek Genomics Suite software package (Partek).
The array contains sequences from two main sources: high confidence mRNAs from RefSeq and GenBank databases and ESTs from dbEST, and a lower confidence set of speculative gene structures predicted using software such as GENSCAN (Burge and Karlin 1997), TWINSCAN (Korf et al. 2001), and Exoniphy (Siepel and Haussler 2004). For this study, we restricted our analyses to the high confidence set of mRNAs and probe sets. Inclusion of the low confidence theoretical probe sets may contribute expression values that go toward the overall summary and calculations of the meta-probe set score and may adversely affect the SI and all subsequent analyses. In doing so, the number of probe sets has been reduced approximately fivefold, from 1.4 million to 277,000 probe sets belonging to core RefSeq transcripts.
One of the potential issues regarding the use of microarrays, particularly with respect to our study of looking at differences in splicing between individuals, is the effect of polymorphisms within the probes that potentially affect binding affinities. Single nucleotide polymorphisms (SNPs) are very common genetic variations and occur at a frequency of one in 1000 bp in the human genome (Sachidanandam et al. 2001). Considering such a high frequency of SNPs, we would expect a large number of the probes to contain SNPs and, in some of the cases, to be polymorphic between the individuals that we are examining. In the comparison of two individuals, if a SNP exists within the target sequence in only one of the individuals, probe binding and intensity will most likely be negatively affected in this sample. This would result in an apparent lower SI relative to the individual with the wild-type allele, potentially leading to a false-positive identification of differential probe set expression. We circumvent this issue by conservatively masking out all probes containing SNPs from the dbSNP database (release 126) and all HapMap SNPs polymorphic between our two samples, from the calculation of probe set and meta-probe set summaries. However, there are most likely unknown SNPs that are not yet annotated that may be present within the probes on the array, and all candidate probe sets will be dealt with on a case by case basis, examining the probe set for any discordant probes within them. Probes showing below background intensities in all samples were also masked out before calculation of probe set summaries in order to avoid potential influences of these low intensity probes on the estimated exon and transcript expression levels. After masking out all of these SNP-containing and background intensity probes, 234K probe sets remain for analysis.
After summarizing probe set scores, ∼76K probe sets did not pass the statistical DABG (detected above background) criteria (see Methods) and therefore were not included in subsequent analyses. In order to identify candidates from the remaining 158K probe sets suggestive of differential splicing between the two individuals, we performed a t-test comparing the log-transformed SI scores on replicates of the two groups. Since there is no clear method for optimal determination of statistical cutoffs (Thomas et al. 2005), we applied three different methods for multiple testing correction. The Bonferroni correction provided the most conservative estimate (P = 3.159 × 10−7, significance threshold P = 0.05), yielding 1892 potential probe sets (1.2% of expressed “core” probe sets) showing differential splicing. The false discovery rate (FDR) (Benjamini and Hochberg 1995; Storey and Tibshirani 2003) at a 0.01 significance level provided the least conservative estimate (P = 8.915 × 10−4), with 8771 (5.7%) potential splicing events. We also ascertained the significance values using an empirical null distribution of P-values from the observed data, by shuffling the SI scores for all samples of each probe set (Churchill and Doerge 1994). For each probe set, we calculated an empirical P-value by comparing our observed, nonpermuted P-value to the distribution of permuted P-values, followed by Bonferroni correction on the permuted P-values. This method estimates 4020 (2.6%) differentially spliced probe sets between the two individuals. The average fold change in SI of all significant probe sets at the Bonferroni, permuted, and FDR corrected cutoffs are 1.85-fold, 1.48-fold, and 1.45-fold, respectively, showing a positive correlation between significance and fold-change expression.
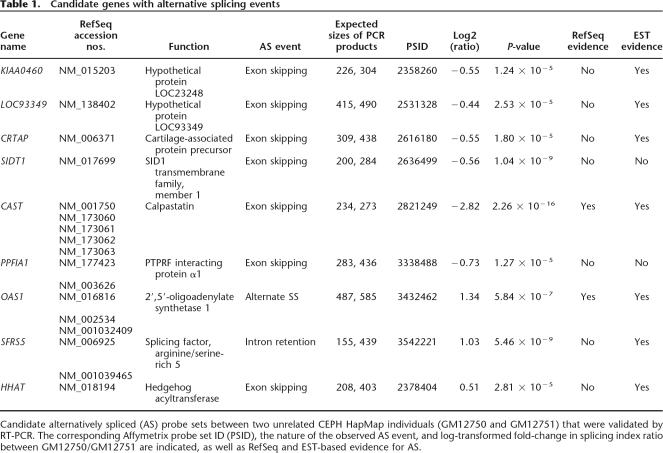
We applied some additional biological and statistical criteria to the data set (see Methods), reducing the number of candidate probe sets to 1028. From this list, we proceeded to test a random selection of probe sets ranging from the highest significance level to those near the FDR cutoff. A small subset of 20 candidates were subjected to validation by reverse transcriptase–polymerase chain reaction (RT-PCR) using a pair of primers in two distinct exons flanking a third exon containing the predicted probe set. The presence of alternative isoforms for nine transcripts was confirmed by RT-PCR (Table 1; Supplemental Fig. 1), which translates into a 45% validation rate. However, our study evaluates the ability of this microarray technology to identify alternative AS events de novo in genetically diverse populations. Restricting our candidates to those showing EST and cDNA evidence of AS in sequence databases reduces the number of cases from 20 to 12, thereby increasing our success rate to 60% (seven out of 12). This is similar to the observed rates in a genome wide junction array study (73/153 = 48%) (Johnson et al. 2003) and a smaller custom array of both exon and junction primers (11/20 = 55%) based on a priori knowledge of AS events (Le et al. 2004).
Table 1.
Candidate genes with alternative splicing events
Candidate alternatively spliced (AS) probe sets between two unrelated CEPH HapMap individuals (GM12750 and GM12751) that were validated by RT-PCR. The corresponding Affymetrix probe set ID (PSID), the nature of the observed AS event, and log-transformed fold-change in splicing index ratio between GM12750/GM12751 are indicated, as well as RefSeq and EST-based evidence for AS.
Analysis of validated AS events
Based on EST and RefSeq evidence, seven of the nine probe sets with confirmed AS are predicted to confer exon-skipping events, with the exception of the OAS1 and SFRS5 genes. Two OAS1 splice variants (RefSeq accession nos. NM_016816 and NM_002534) are predicted to encode isoforms with alternative 3′ splice site (ss) usage of the last downstream coding exon. The probe set identified in the SFRS5 gene is located within an intron between exons 4 and 5 and represents an intron-retention event. In total, seven of the nine probe sets that were identified in this study show annotated evidence in EST and RefSeq databases of AS. Probe sets corresponding to exons from the PPFIA1 and SIDT1 genes show no previous evidence of AS, demonstrating that the array can detect novel splicing events.
In three (CAST, PPFIA1, OAS1) of the top four validated splicing events with the highest degree of fold-change in SI between individuals, we observe a clear predominance of one isoform in one individual versus the alternate variant in the second individual. The majority of candidates with lesser fold changes show the presence of both splice variants in each of the individuals. From a biological perspective, the presence or absence of one of the two splice variants between individuals is more likely to have a functional consequence than are cases where two splice variants are expressed in all individuals with subtle differences in relative ratios. Loss of function from one variant without compensatory effects from expression of the alternative splice isoform may have drastic differences in downstream effects. However, until a complete validation of all candidate probe sets is performed, we cannot estimate how many of these “all-or-none” splicing events are present compared with the observation of both isoforms in each individual.
In one of our candidate genes, sequence analysis of the RT-PCR products identified a variant using a cryptic splice site within the predicted exon. Two OAS1 transcripts show alternative 3′ ss usage in the predicted last exon of the gene, resulting in differential stop codon usage and a longer 3′ UTR in one transcript. In the future, sequence analysis of all validated probe sets will be necessary to accurately determine cryptic splice site usage, especially those in close proximity to the annotated splice site, which may be beyond the resolution of standard gel electrophoresis.
The available EST and mRNA-based evidence of AS in most of our candidate genes provides support and validation for our array-based discovery of known alternatively spliced transcripts. More importantly, the identification of new PPFIA1 and SIDT1 splice variants provide confidence that we may be able to discover novel AS events and increase the catalog of the human transcriptome.
Association of splicing to cis-regulatory haplotypes
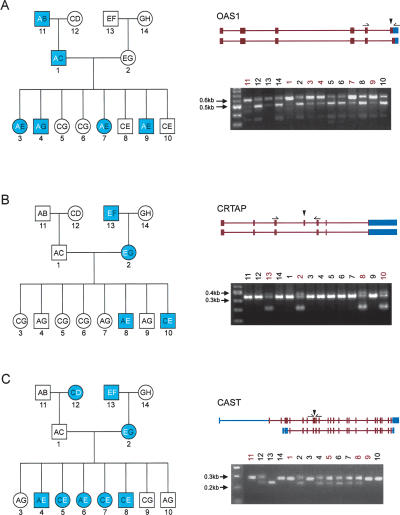
An important goal of this study was to demonstrate the genetic component of AS, specifically the inheritance of a splicing pattern and its association to a cis-regulatory haplotype. Using the SI of an exon as a quantitative trait, we performed regression-based linkage analysis (implemented in Merlin) (Abecasis et al. 2002) within a three-generation family (CEPH 1444) for the nine verified AS events detected in this study. At a nominal level of LOD > 0.59, corresponding to P < 0.05, we observed evidence of linkage between SI scores and the corresponding chromosomal region in the OAS1 (LOD = 0.76), CRTAP (LOD = 1.29), and CAST (LOD = 1.98) genes. RT-PCR based analysis confirmed segregation of the splicing pattern with the associated haplotype through all three generations of this pedigree (Fig. 3).
Figure 3.
Heritability of alternative splicing. Inheritance of alternative splicing for genes (A) OAS1, (B) CRTAP, and (C) CAST. Left panel shows pedigree structure of CEPH/UTAH family 1444 with the autosomal dominant inherited splice pattern as blue symbols. Haplotypes for each of the eight founder chromosomes are labeled A, B, C, D, E, F, G, and H, and the two inherited haplotypes of each family member are indicated within the symbol. The regulatory haplotype is shown as bold white text. Squares represent males, and circles represent females. CEPH/UTAH 1444 pedigree is labeled as follows: 1 (GM12739), 2 (GM12740), 3 (GM12741), 4 (GM12742), 5 (GM12743), 6 (GM12744), 7 (GM12745), 8 (GM12746), 9 (GM12847), 10 (GM12747), 11 (GM12748), 12 (GM12749), 13 (GM12750), and 14 (GM12751). The right panel shows the two transcript isoforms of the genes. Exon-body primers are shown above the flanking exons of the predicted alternatively spliced exons. Shown below the transcript isoforms are the RT-PCR results. Lanes are numbered from 1–14 according to the pedigree on the left.
The association between alternatively spliced isoforms and genetic variation was examined further by testing our nine candidates on a larger panel of 60 unrelated HapMap CEU individuals. In many cases, both splice variants are expressed in different ratios in various individuals, but the RT-PCR approach that was used here was not sensitive enough to quantify the relative isoform levels and establish a statistical association with a regulatory haplotype. Other methods based on the use of fluorescent dyes such as TaqMan PCR (Gibson et al. 1996) may be more sensitive in detecting relative amounts of each isoform, although the cost associated with this technology is prohibitive for large-scale validation of predicted AS events. In clear cases where only one of the isoforms or the other is expressed, classical RT-PCR is a more suitable method. We were able to confirm the previously described association of OAS1 variants to a candidate regulatory polymorphism (Field et al. 2005), and establish that the CRTAP splicing variant is rare and does not occur outside of members of CEPH family 1444 (data not shown).
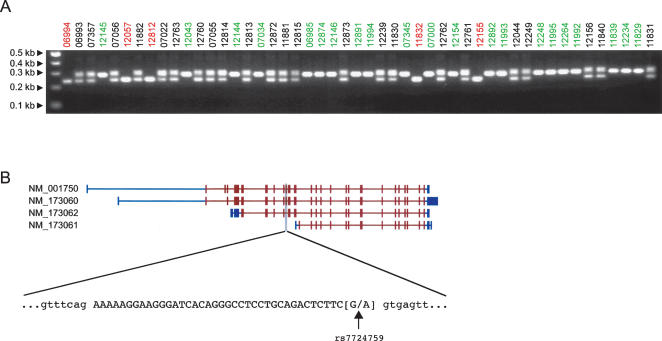
The most interesting example of allelic association was identified in the CAST gene, which encodes for calpastatin, a calpain protease inhibitor. There are at least 11 known isoforms of calpastatin, all differing in their N-terminal regions (Fig. 4B) (Lee et al. 1992). The predicted alternatively spliced exon of the CAST gene is supported by RefSeq and EST evidence of AS and encodes a portion of the first of four repetitive protease-inhibition domains. Consequently, removal or disruption of these calpain-inhibition domains may affect functionality and/or tissue specificity of the protein (Takano et al. 1993). The splicing pattern in the entire panel was correlated to a single SNP (rs7724759) that is most likely the causative polymorphism resulting in our differentially spliced isoforms. The SNP is located at the 3′ end of the exon and involves a G to A substitution that abates the weak consensus 5′ ss sequence. All individuals genotyped as homozygous GG for rs7724759 have an intact 5′ ss sequence and properly splice the exon, resulting in the larger PCR product. Individuals homozygous for AA at this position have a nonfunctional 5′ ss on both alleles that is improperly recognized by the splicing machinery; as such, the exon is excluded and accounts for the shorter, lower molecular weight band. When both isoforms are observed, the individual is heterozygous for this SNP and has both wild-type and polymorphic alleles. This exon also demonstrated linkage in the CEPH 1444 family, as previously mentioned, and examination of the pedigree clearly shows the inheritance of the two haplotypes through the three generations (Fig. 3C).
Figure 4.
Association of alternative splicing and genotypes for the CAST gene. (A) RT-PCR of CAST exon against a panel of unrelated parents from each of the 30 HapMap CEU trios. Sample names are colored according to their genotype for SNP rs7724759: homozygous GG (green), homozygous AA (red), and heterozygous AG (black). (B) Four known isoforms of the CAST gene are shown with their RefSeq accession numbers on the left and the candidate probe set shaded in gray. Shown below is the sequence of the exon in capital letters and flanked by the intronic sequence in lower case. The SNP rs7724759 is located at the last position of the exon and is a G to A substitution that disrupts the consensus splice site sequence.
We also examined the remaining eight AS events for both functional domains encoded within the respective exons and also for putative cis-acting SNPs that may control the splicing patterns. We did not identify any domains for any of the exons except a putative transmembrane domain within the HHAT exon. In most of the cases, the closest polymorphic SNPs between individuals GM12750 and GM12751 were all located either in the 5′ or 3′ flanking introns but at significant distances (>100 bp) from the splice site. We were able to identify SNPs either within or in close proximity (<100 bp) to the putative AS exon for the SIDT1 and OAS1 genes and within the retained SFRS5 intron. SNP rs2271494 is located 25 bp upstream of the SIDT1 exon and is found within the polypyrimidine tract. Mutations within this region may alter binding between the large subunit of the U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor, U2AF, to this motif (Singh et al. 1995). The SNP rs151042 is located within exon 7 of the OAS1 gene and is part of a haplotype block where another SNP marker, hCV2567433, at the exon 7 splice-acceptor site, has been shown to result in the usage of an internal splice site in the mutant allele (Bonnevie-Nielsen et al. 2005). In the one example of intron retention for the SFRS5 gene, we identified a SNP (rs3104) centrally located within the intron; however, it does not appear to disrupt any known intronic splice enhancer or silencer. These results demonstrate that association studies of alternatively spliced exons with well-genotyped individuals are valuable in identifying the potential polymorphisms linked to the splicing event.
Discussion
Identifying AS events is important to understanding the diversity and complexity of the human genome, and we report on the use of a comprehensive exon-tiling array in our experimental design to discover such events between individuals. The same microarray design has also been recently used for a complete analysis of tissue-specific differences in splicing (Gardina et al. 2006; Clark et al. 2007) and is potentially useful for many pairwise comparisons of splicing. Since the design of this array is not biased toward a priori knowledge of AS events, there is more potential for detecting novel splicing events. We demonstrated that novel isoforms can be discovered using this microarray, and others have recently shown the same (Gardina et al. 2006; Clark et al. 2007). A number of different types of splicing events were identified, including exon exclusion, intron retention, and the use of cryptic splice sites. Exon-tiling arrays provide an advantage over exon junction arrays in their ability to identify the use of cryptic splice sites due to the design of probes within an exon. Exon-junction probes can detect the joining of two exons at specific, known splice sites and are not as effective at the detection of novel, unannotated cryptic splice site usage. However, one disadvantage of tiling probes only within exons is its inability to provide information on how all the individual exons are linked within the different splice isoforms of a particular gene, a feature more suited to an exon-junction probe array. Proper design of an exon junction array for the entire human genome to interrogate all possible gene structures requires too many probes for every possible joining event. Such a design is more suitable for the examination of a smaller number of events, as demonstrated recently (Ben-Ari et al. 2006; Valverde et al. 2006; Zhang et al. 2006). Each of these array designs possesses advantages and disadvantages, and given comparable false-positive rates obtained in this study and other splicing microarray studies, both are useful and informative in the identification of AS events. A follow-up study using a custom microarray consisting of a combination of exon and exon-junction probes may prove useful for confirming AS events and examining all possible transcript structures for a smaller subset of genes. This study focused on differentially expressed probe sets located within in-frame coding exons. Validation of probe sets corresponding to out-of-frame exons were not looked at, but these may introduce an upstream stop codon through cryptic splice site usage. This may confer differences in post-transcriptional regulation through nonsense-mediated decay. Probe sets located within 5′/3′ UTRs can also have widely varying biological functional consequences, such as changes in promoter regions or polyadenylation and transcript termination differences.
Exactly how much differential splicing is occurring between any two individuals is still unknown. We estimated that up to 2.5% of all RefSeq exons expressed in lymphoblasts may show differential expression between the two samples tested, after factoring in our current validation rate, although a more accurate determination on the amount of differential splicing events will require a proper ROC-type analysis. However, this study examines splicing in lymphoblasts, and this estimate may change depending on the tissue tested. Alternative splice variants of the same gene can be expressed in multiple cell types to exert different functional and regulatory effects, which may also be individual specific. Neuronal tissues are known to have high levels of splicing (Yeo et al. 2004), and it is not unreasonable to assume that the amount of splicing between individuals may be higher in brain tissues than in lymphoblasts. A more complete picture may be ascertained by pairwise comparison of splicing in many tissues between individuals.
The large amount of genotyping information within identified populations from the HapMap project provides a tremendous resource for associating known SNPs or regions of linkage disequilibrium with genetic differences such as copy number variation, allelic imbalance, and AS, or phenotypic traits that may convey an increased risk of disease. Here, we have shown that this approach can be used to identify one or more SNPs associated with some of the splicing events identified. Further examination of the nature of the polymorphisms and their location relative to the spliced exon can give insight as to whether it is part of a larger cis-regulatory haplotype or in fact the causative SNP disrupting a splice site consensus sequence, an exonic splicing enhancer (ESE) or silencer (ESS), an intronic splicing enhancer (ISE) or silencer (ISS), or other splice regulatory motifs such as the branch point or the polypyrimidine tract. Assigning a definitive causative effect of the SNP will require further experimental validation in vitro, such as monitoring splicing activity in cells using splice reporter constructs (Mayeda and Krainer 1999). However, it is quite possible that there are unannotated SNPs proximal to the exon that are responsible for the differential splicing, and resequencing of the genomic regions neighboring the exons will be necessary to identify these polymorphisms.
Although we identify a candidate exon from the CAST gene showing genetic association with expression level changes, we do not know how often this occurs in a human population on a genome-wide scale. One method of properly assessing how common inherited splicing occurs would be to perform a whole-genome association study with more individuals from the HapMap population, using the SI scores as a quantitative trait. This is very similar to recent whole-genome association studies that suggest that common genetic variation explains much of the gene expression differences among individuals (Stranger et al. 2005, 2007). Carrying out a similar analysis at the exon level will yield better estimates of how common this heritability and genetic association is in humans.
The identification of SNPs within specific individuals in a population that affect splicing is an important issue to address when considering its relevance to possible resistance or susceptibility to disease states. An estimated 20%–30% of disease-causing mutations is believed to affect pre-mRNA splicing (Faustino and Cooper 2003), through the disruption of splice sites, exonic and intronic splicing enhancers and silencers, or RNA secondary structure. In this study, the two OAS1 splice variants identified have been previously associated with a SNP at an exon splice-acceptor site. This polymorphism results in the usage of an internal splice site in the mutant allele, which is thought to confer differences in host susceptibility to viral infection in type I diabetes patients (Field et al. 2005). A genome-wide analysis with well-genotyped CEPH HapMap individuals will be an important starting point in identifying many more AS events and the causative polymorphisms involved in human diseases.
Methods
Cell line preparation
RNA samples were obtained from 74 Epstein-Barr virus-transformed LCLs belonging to the CEPH (Center d’étude du polymorphisme humain) reference individuals from the state of Utah in the United States (CEU). For this study, we used DNA samples from 60 unrelated individuals that have been genotyped for approximately four million SNPs by the International HapMap Project (Altshuler et al. 2005). Additionally, LCLs from CEPH pedigree 1444 (14 samples) were included to examine genetic influences on AS in a three-generation family. Cells were grown at 37°C and 5% CO2 in RPMI 1640 medium (Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum (Sigma-Aldrich), 2 mM L-glutamine (Invitrogen), and penicillin/streptomycin (Invitrogen). Cell growth was monitored with a hemocytometer, and cells were harvested at a density of 0.8 × 106 to 1.1 × 106 cells/mL. Cells were then resuspended and lysed in TRIzol reagent (Invitrogen). For all LCLs, three successive growths were performed (corresponding to the second, fourth, and sixth passages) after thawing frozen cell aliquots.
Affymetrix exon arrays
RNA was isolated using TRIzol reagent following the manufacturer’s instructions (Invitrogen). The RNA quality was assessed using RNA 6000 NanoChips with the Agilent 2100 Bioanalyzer (Agilent). Biotin-labeled target for the microarray experiment were prepared using 1 μg of total RNA. The RNA was subjected to a rRNA removal procedure with the RiboMinus Human/Mouse Transcriptome Isolation Kit (Invitrogen), and cDNA was synthesized using the GeneChip WT (Whole Transcript) Sense Target Labeling and Control Reagents kit as described by the manufacturer (Affymetrix). The sense cDNA was then fragmented by UDG (uracil DNA glycosylase) and APE 1 (apurinic/apyrimidic endonuclease 1) and biotin-labeled with TdT (terminal deoxynucleotidyl transferase) using the GeneChip WT Terminal labeling kit (Affymetrix). Hybridization was performed using 5 μg of biotinylated target, which was incubated with the GeneChip Human Exon 1.0 ST array (Affymetrix) at 45°C for 16–20 h. Following hybridization, nonspecifically bound material was removed by washing and detection of specifically bound target was performed using the GeneChip Hybridization, Wash and Stain kit, and the GeneChip Fluidics Station 450 (Affymetrix). The arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix), and raw data was extracted from the scanned images and analyzed with the Affymetrix Power Tools software package (Affymetrix).
For the initial study, three separate passages of two unrelated individuals, GM12750 and GM12751, from the CEPH 1444 pedigree were used, with five technical replicates of each growth, for a total of 15 arrays hybridized for each sample. Multiple replicates were used to assess the relative contributions of biological and technical noise to the observed exon and transcript levels. In particular, since this array uses probe cells with a feature size that is only one-quarter of previous expression array designs, we aimed to determine whether they showed greater technical variability or higher background noise and also to identify a minimum number of biological and technical replicates required for an acceptable signal-to-noise ratio. For the linkage studies of the CEPH 1444 pedigree, three passages for each of GM12739, GM12740, GM12750, and GM12751 were used along with single replicates for the remaining 10 individuals.
Analysis of array hybridization data
The Affymetrix Power Tools software package (Affymetrix) was used to quantile normalize the probe fluorescence intensities and to summarize the probe set (representing exon expression) and meta-probe set (representing gene expression) intensities using a probe logarithmic intensity error model (see http://www.affymetrix.com/support/technical/technotes/plier_technote.pdf). Probe sequences that map to SNPs in a particular sample may give rise to altered binding affinities and influence intensity data and the resulting SI scores (data not shown); therefore, probe sets were cross-referenced to the dbSNP database (release 126) for the presence of polymorphisms within the probes, and SNP-containing probes were excluded from this analysis. Probes showing sub-background levels of expression in all samples were also removed to reduce the influence of these probes on total probe set and meta-probe set expression levels. We calculated mean probe intensities for a set of anti-genomic probes, which we designated as background expression. For each probe on the array, if the intensity for all samples was less than the background expression plus two standard deviations for the same GC content, then the probe was excluded from the summary calculations. The SI score was calculated by simply dividing the probe set intensity by the meta-probe set intensity (i.e., exon expression/gene expression) after the addition of a stabilization constant (13) to both the probe set and meta-probe set scores.
PCA was performed on the SI scores from all chips using the Partek Genomics Suite software package (Partek) in order to attribute the variance averaged over all exons to sources of variability, and to determine a confidence level in the consistency of expression profiles from biological and technical replicates. Comparison of expression data from individuals GM12750 and GM12751 identified outliers for three replicates of GM12750 (Fig. 2) that were excluded from all subsequent analyses.
To analyze splicing differences between the two samples for each probe set, an unpaired Student’s t-test was performed using the log-transformed SI values for all remaining replicates (12 of GM12750 and 15 of GM12751) of each individual (R statistical package, version 2.3.0). Probe sets showing significantly different SI scores were ranked by P-value. Linkage analysis tests of SI scores cosegregating with chromosomal regions for the CEPH 1444 family was carried out using MERLIN (version 1.0.1) with default settings (Abecasis et al. 2002). The scan was performed using a region spanning 20 SNP markers centered on the probe set.
Differentially spliced probe sets were filtered using a number of criteria including: (1) detectable level above background (DABG < 0.05) for both the probe set and the meta-probe set to which it belongs; (2) normalized meta-probe set scores with a minimum intensity score of 50; (3) the transcript defined by a minimum of three exons; and (4) size of the exon corresponding to the probe set is divisible by three. This last criterion was added to ensure that changes resulting from exon inclusion/exclusion would be in frame, which has been observed in a high percentage of conserved and species-specific alternative exons (Magen and Ast 2005). For two sample comparisons, we also required that transcript expression levels between samples was less than twofold.
RT-PCR and sequence analysis
Total RNA was treated with 4 U of DNase I (Ambion) for 30 min to remove any remaining genomic DNA. First-strand cDNA was synthesized using random hexamers (Invitrogen) and Superscript II reverse transcriptase (Invitrogen). For all candidate probe sets, locus-specific primers within the adjacent, flanking exons were designed using Primer3 (Rozen and Skaletsky 2000) software. Primers were designed within exons that had the following restrictions: (1) flanking exon expression level above background (DABG < 0.05) and (2) the flanking exon itself was not predicted to be alternatively spliced. Approximately 20 ng of total cDNA was then amplified by PCR using Hot Start Taq Polymerase (Qiagen) with an activation step of 15 min at 95°C followed by 35 cycles of 30 sec at 95°C, 30 sec at 58°C, and 40 sec at 72°C and a final extension step of 5 min at 72°C. Amplicons were visualized by electrophoresis on a 2.5% agarose gel. Sequencing of the two products whose sizes corresponded to the predicted larger exon/intron-inclusion and shorter exon-skipped forms confirmed the AS. We perfomed BLAST analysis of the two splice variants against the non-redundant and EST databases at the National Center for Biotechnology Information (NCBI) to verify if both sequences are known or whether a novel splice isoform has been identified.
Acknowledgments
We thank Eef Harmsen for helpful discussions. This work is supported by Genome Canada and Genome Québec. T.J.H. is the recipient of a Clinician-Scientist Award in Translational Research by the Burroughs Wellcome Fund and an Investigator Award from CIHR. J.M. is a Canada Research Chair holder.
Footnotes
[The microarray data from this study have been submitted to GEO under accession no. GES7952.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.6281007
References
- Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R., Cherny S.S., Cookson W.O., Cardon L.R., Cookson W.O., Cardon L.R., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Collins F.S., Daly M.J., Donnelly P., Consortium I.H., Daly M.J., Donnelly P., Consortium I.H., Donnelly P., Consortium I.H., Consortium I.H. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari S., Toiber D., Sas A.S., Soreq H., Ben-Shaul Y., Toiber D., Sas A.S., Soreq H., Ben-Shaul Y., Sas A.S., Soreq H., Ben-Shaul Y., Soreq H., Ben-Shaul Y., Ben-Shaul Y. Modulated splicing-associated gene expression in P19 cells expressing distinct acetylcholinesterase splice variants. J. Neurochem. 2006;97 (Suppl 1):24–34. doi: 10.1111/j.1471-4159.2006.03725.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Black D.L., Graveley B.R., Graveley B.R. Splicing bioinformatics to biology. Genome Biol. 2006;7:317. doi: 10.1186/gb-2006-7-5-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V., Field L.L., Lu S., Zheng D.J., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Field L.L., Lu S., Zheng D.J., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Lu S., Zheng D.J., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Zheng D.J., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F., Beck-Nielsen H., Lau Y.L., Pociot F., Lau Y.L., Pociot F., Pociot F. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am. J. Hum. Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Churchill G.A., Doerge R.W., Doerge R.W. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T.A., Sugnet C.W., Ares M., Sugnet C.W., Ares M., Ares M. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E., Staples M.K., Lu G., Wang H., Williams A., Blume J.E., Lu G., Wang H., Williams A., Blume J.E., Wang H., Williams A., Blume J.E., Williams A., Blume J.E., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Chumakov I., Weissenbach J., Chumakov I., Weissenbach J., Weissenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- Faustino N.A., Cooper T.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Field L.L., Bonnevie-Nielsen V., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Bonnevie-Nielsen V., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Lu S., Nielsen T.B., Beck-Nielsen H., Nielsen T.B., Beck-Nielsen H., Beck-Nielsen H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes. 2005;54:1588–1591. doi: 10.2337/diabetes.54.5.1588. [DOI] [PubMed] [Google Scholar]
- Frey B.J., Mohammad N., Morris Q.D., Zhang W., Robinson M.D., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Mohammad N., Morris Q.D., Zhang W., Robinson M.D., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Morris Q.D., Zhang W., Robinson M.D., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Zhang W., Robinson M.D., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Robinson M.D., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Mnaimneh S., Chang R., Pan Q., Sat E., Rossant J., Chang R., Pan Q., Sat E., Rossant J., Pan Q., Sat E., Rossant J., Sat E., Rossant J., Rossant J., et al. Genome-wide analysis of mouse transcripts using exon microarrays and factor graphs. Nat. Genet. 2005;37:991–996. doi: 10.1038/ng1630. [DOI] [PubMed] [Google Scholar]
- Gardina P.J., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Schweitzer A., Awad T., Sugnet C., Dee S., Awad T., Sugnet C., Dee S., Sugnet C., Dee S., Dee S., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson U.E., Heid C.A., Williams P.M., Heid C.A., Williams P.M., Williams P.M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Schadt E.E., Stoughton R., Shoemaker D.D., Stoughton R., Shoemaker D.D., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M.R., Flicek P., Duan D., Brent M.R., Duan D., Brent M.R., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Le K., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C., Wang Q., Xu Q., Nelson S.F., Lee C., Xu Q., Nelson S.F., Lee C., Nelson S.F., Lee C., Lee C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Roy M., Roy M. Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 2004;5:231. doi: 10.1186/gb-2004-5-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.J., Ma H., Takano E., Yang H.Q., Hatanaka M., Maki M., Ma H., Takano E., Yang H.Q., Hatanaka M., Maki M., Takano E., Yang H.Q., Hatanaka M., Maki M., Yang H.Q., Hatanaka M., Maki M., Hatanaka M., Maki M., Maki M. Molecular diversity in amino-terminal domains of human calpastatin by exon skipping. J. Biol. Chem. 1992;267:8437–8442. [PubMed] [Google Scholar]
- Magen A., Ast G., Ast G. The importance of being divisible by three in alternative splicing. Nucleic Acids Res. 2005;33:5574–5582. doi: 10.1093/nar/gki858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- Modrek B., Resch A., Grasso C., Lee C., Resch A., Grasso C., Lee C., Grasso C., Lee C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim-Rafinia M., Kerem B., Kerem B. The splicing machinery is a genetic modifier of disease severity. Trends Genet. 2005;21:480–483. doi: 10.1016/j.tig.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Mullikin J.C., Mortimore B.J., Willey D.L., Mortimore B.J., Willey D.L., Willey D.L., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Proceedings of the eighth annual international conference on Resaerch in computational molecular biology. ACM Press; San Diego, CA: 2004. Computational identification of evolutionarily conserved exons. [Google Scholar]
- Singh R., Valcarcel J., Green M.R., Valcarcel J., Green M.R., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., Bryant J., Smith M., Rommelfanger C., Smith M., Rommelfanger C., Rommelfanger C., et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Kahl B., Antonarakis S.E., Tavare S., Antonarakis S.E., Tavare S., Tavare S., et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Redon R., Bird C.P., de Grassi A., Lee C., Bird C.P., de Grassi A., Lee C., de Grassi A., Lee C., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet C.W., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Srinivasan K., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Clark T.A., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., O’Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., Williams A., Kulp D., Blume J.E., Haussler D., Kulp D., Blume J.E., Haussler D., Blume J.E., Haussler D., Haussler D., et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal. pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E., Nosaka T., Lee W.J., Nakamura K., Takahashi T., Funaki M., Okada H., Hatanaka M., Maki M., Nosaka T., Lee W.J., Nakamura K., Takahashi T., Funaki M., Okada H., Hatanaka M., Maki M., Lee W.J., Nakamura K., Takahashi T., Funaki M., Okada H., Hatanaka M., Maki M., Nakamura K., Takahashi T., Funaki M., Okada H., Hatanaka M., Maki M., Takahashi T., Funaki M., Okada H., Hatanaka M., Maki M., Funaki M., Okada H., Hatanaka M., Maki M., Okada H., Hatanaka M., Maki M., Hatanaka M., Maki M., Maki M. Molecular diversity of calpastatin in human erythroid cells. Arch. Biochem. Biophys. 1993;303:349–354. doi: 10.1006/abbi.1993.1294. [DOI] [PubMed] [Google Scholar]
- Thomas D.C., Haile R.W., Duggan D., Haile R.W., Duggan D., Duggan D. Recent developments in genomewide association scans: A workshop summary and review. Am. J. Hum. Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Wang H., Clark T., Fraser C., Ruggiu M., Clark T., Fraser C., Ruggiu M., Fraser C., Ruggiu M., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Valverde D., Riveiro-Alvarez R., Bernal S., Jaakson K., Baiget M., Navarro R., Ayuso C., Riveiro-Alvarez R., Bernal S., Jaakson K., Baiget M., Navarro R., Ayuso C., Bernal S., Jaakson K., Baiget M., Navarro R., Ayuso C., Jaakson K., Baiget M., Navarro R., Ayuso C., Baiget M., Navarro R., Ayuso C., Navarro R., Ayuso C., Ayuso C. Microarray-based mutation analysis of the ABCA4 gene in Spanish patients with Stargardt disease: Evidence of a prevalent mutated allele. Mol. Vis. 2006;12:902–908. [PubMed] [Google Scholar]
- Yeo G., Holste D., Kreiman G., Burge C.B., Holste D., Kreiman G., Burge C.B., Kreiman G., Burge C.B., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li H.R., Fan J.B., Wang-Rodriguez J., Downs T., Fu X.D., Zhang M.Q., Li H.R., Fan J.B., Wang-Rodriguez J., Downs T., Fu X.D., Zhang M.Q., Fan J.B., Wang-Rodriguez J., Downs T., Fu X.D., Zhang M.Q., Wang-Rodriguez J., Downs T., Fu X.D., Zhang M.Q., Downs T., Fu X.D., Zhang M.Q., Fu X.D., Zhang M.Q., Zhang M.Q. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics. 2006;7:202. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]