Abstract
Exonization of retroposed mobile elements, a process whereby new exons are generated following changes in non-protein-coding regions of a gene, is thought to have great potential for generating proteins with novel domains. Our previous analysis of primate-specific Alu-short interspersed elements (SINEs) showed, however, that during their 60 million years of evolution, SINE exonizations occurred in some primates, only to be lost again in some of the descendent lineages. This dynamic gain and loss makes it difficult to ascertain the contribution of exonization to genomic novelty. It was speculated that Alu-SINEs are too young to reveal persistent protein exaptation. In the present study we examined older mobile elements, mammalian-wide interspersed repeats (MIRs) that underwent active retroposition prior to the placental mammalian radiation ∼130 million years ago, to determine their contribution to protein-coding sequences. Of 107 potential cases of MIR exonizations in human, an analysis of splice sites substantiates a mechanism that benefits from 3′ splice site selection in MIR sequences. We retraced in detail the evolution of five MIR elements that exonized at different times during mammalian evolution. Four of these are expressed as alternatively spliced transcripts; three in species throughout the mammalian phylogenetic tree and one solely in primates. The fifth is the first experimentally verified, constitutively expressed retroposed SINE element in mammals. This pattern of highly conserved, alternatively and constitutively spliced MIR sequences evinces the potential of exonized transposed elements to evolve beyond the transient state found in Alu-SINEs and persist as important parts of functional proteins.
Genomic plasticity has contributed significantly to the dynamic generation of novel features in evolution. In this context, retroposed genetic elements, which are sequences of DNA that amplify via RNA to different positions within the genome, play a decisive role as inducers or substrates of novel evolutionary building blocks (Brosius and Gould 1992). Discernible retroposed elements compose up to 42% of the human genome (Lander et al. 2001) and, in an appropriate genomic context, have the potential to provide alternative splice sites and/or polyadenylation signals or may modify gene expression as parts of promoters or enhancers (Brosius 2005). Retroposed sequences can even be exapted (i.e., assume a new role) as protein-coding modules in a process that requires exonization, and when expressed via alternative splicing, they increase the complexity of the proteome (Xing and Lee 2006). In rare cases, when their sequences or parts thereof are under strong negative selection, retroposed elements can be identified as very ancient exaptations dating as far back as half a billion years (Bejerano et al. 2006). In primates, Alu-short interspersed elements (SINEs) are the prevailing retroposed elements found in open reading frames (ORFs) of mRNAs and expressed sequence tags (ESTs) (Sorek et al. 2002).
Following the path of Alu exonizations along a phylogenetic tree of primates indicates a dynamic gain and loss of exonizations, processes that may embrace >60 million years (Myr) of evolution (Krull et al. 2005). Thus, it appears that the time frame of primate evolution might not yet be sufficiently long to ascertain whether exonized elements have been exapted as persistent modules of the proteome (Gotea and Makalowski 2006). Although transcribed, exonized sequences are destined to negotiate a grueling course along the way to becoming part of functional proteins that only a very few ever survive. Various mRNA surveillance mechanisms might degrade alternatively exonized transcripts before they become essential parts of functioning proteins (Wagner and Lykke-Andersen 2002; Hillman et al. 2004). Survivors of the degradation mechanism are further exposed to natural selection in maintaining their status as alternative splice products or in replacing the ancestral splice variant. A number of Alu exonizations in primates might not have reached that status yet. A retrospective view of much older retropositions, ones active during the Mesozoic era of mammalian evolution, promises a more definitive picture of the exonization process, especially regarding their contribution to protein plasticity.
Good candidates for illuminating these older exonization processes are the retroposed mammalian-wide interspersed repeat (MIR) elements. MIR elements amplified ∼130 million years ago (Mya), and they number, for example, in human, ∼368,000 discernible copies (Lander et al. 2001). Their age and their abundant distribution in mammals make MIRs an excellent source to elucidate the history of their exonizations. MIRs are usually truncated at either or both ends (Smit and Riggs 1995). The 5′ tRNA part of the MIR is fused to a tRNA-unrelated sequence, and the 3′ end features a 50-nt fragment that is similar to the 3′ end of long interspersed elements (LINEs), a likely binding site of the LINE reverse transcriptase needed for retroposition (Kapitonov et al. 2004). The conserved central domain includes a 15-nt core sequence (Fig. 1). However, it is not known why the core region is highly conserved in so-called CORE-SINEs like MIRs. Possibly, CORE-SINEs have some as yet not readily understood genomic function (Gilbert and Labuda 1999; Nishihara et al. 2006). MIR elements underwent active retroposition before monotremes and placental mammals diverged, but this activity was silenced after the required LINE-specific reverse transcriptase was lost.
Figure 1.
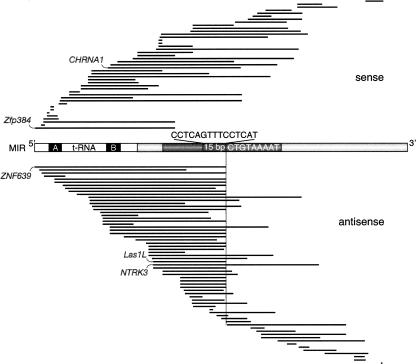
Distribution and orientation of exonized MIR sequences located in protein-coding sequences (CDS). The 107 human exonized MIR sequence regions are aligned against a schematic of a MIR consensus element. The tRNA-related part, including the internal promoter A and B boxes (black), is shown without shading (white). The region shown in dark gray comprises the 70 nt conserved central domain including a highly conserved 15-bp core sequence. The location of the MIR internal, antisense cryptic splice site AG is indicated on the sense strands (CT) by a vertical line. The 9-nt natural 3′ MIR splice site (5′-ATTTTACAG-3′) is shown as the inverse consensus sequence (Supplemental Fig. S3). The exonized MIR regions are represented as black lines; intronic or untranslated region (UTR) portions of the MIRs are not shown. The five experimentally analyzed examples are indicated (CHRNA1, Zfp384, ZNF639, LAS1L, and NTRK3).
In this paper, we focus on MIR exonizations in a phylogenetic context by characterizing novel gene modules in representatives of all mammalian clades (Kriegs et al. 2006). We examine the exonization patterns of MIR elements in species of all major branches of mammals, which evolved over a period of >100 Myr, and compare these to the relatively young processes involved in Alu exonizations in primates.
Results
Selection of the data set and examples
To identify MIR elements in protein-coding sequences of human, mouse, and rat, we screened a compilation of mammalian mRNAs presumably harboring transposable element-cassettes assembled by Makalowski and co-workers (Genomic ScrapYard; http://warta.bio.psu.edu/SYDB/database.html), performing separate searches for the listed species. From 372 ScrapYard records with potential MIR element-cassettes identified in this initial search, 126 MIR sequences were present in protein-coding sequences (CDS), the remaining were either redundant entries or otherwise artifactual (Supplemental Table S1; Supplemental Fig. S1). Of these 126 loci, 107 were found in human (one of which was initially identified in rat) and were used to investigate the distribution and orientation of exonized MIR sequences (Fig. 1).
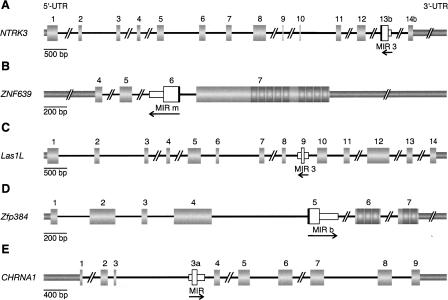
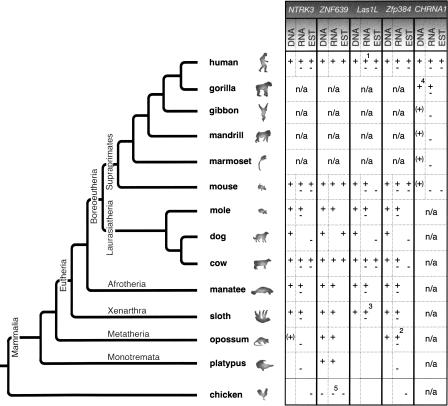
Five of the above 126 cases were (1) supported by ESTs or other indications of expression, (2) flanked by conserved sequence regions facilitating mammalian wide PCR amplification of fragments not exceeding 2 kb, (3) expressed in available tissue, and (4) embedded in introns, and thereby suitable for intensive phylogenetic reconstruction (Fig. 2; Supplemental Data Set S1). The five MIR cassettes were found in the following genes: (1) neurotrophic tyrosine kinase receptor type 3 gene (NTRK3; GenBank accession no. BT007291), (2) zinc finger protein 639 gene (ZNF639; NM_016331), (3) LAS1-like gene (LAS1L; NM_031206), (4) zinc finger protein 384 gene (Zfp384; AF216807), and (5) cholinergic receptor nicotinic alpha 1 gene (CHRNA1; NM_001039523). The mammalian-wide evolutionary distribution and state of expression of these exonized sequences is summarized in Figure 3.
Figure 2.
Structures of five selected genes harboring internal exonized MIR elements. Numbered, thick gray boxes represent protein-coding sequences (CDS). The 5′ and 3′ untranslated regions (UTRs) are shown as medium thick gray bars. Introns are indicated as black lines; double slashes denote gaps in larger introns. Orientations and recognizable lengths of the MIR elements are indicated by arrows. The short white boxes represent the intronic parts of the MIR elements, and the adjacent tall white boxes the exonized regions. In three cases the exonization exceeds the MIR boundaries and includes anonymous intronic sequences (black bars adjacent to the exonized regions). (A) Human neurotrophic tyrosine kinase receptor type 3 gene (NTRK3; BT007291). (B) Human zinc finger protein 639 gene (ZNF639; NM_016331). (C) Human LAS1-like gene (LAS1L; NM_031206). (D) Rat zinc finger protein 384 gene (Zfp384; AF216807). (E) Human cholinergic receptor nicotinic alpha1 gene (CHRNA1; NM_001039523). The substructures in exon 7 of ZNF639 and in exons 6 and 7 of Zfp384 represent sequences encoding zinc finger domains.
A natural splice site in MIR elements
Because the presence or acquisition of alternative splice sites recognized by the splicing machinery is crucial for intronic elements to be exonized, we examined the nature of the splice sites flanking the 107 MIR exonizations identified in human by compiling and comparing their potential protein-coding sequences (Fig. 1). Significantly more of the potential exonized MIR elements (64 of 107; χ2 test, P < 0.05) were inserted in the antisense orientation, presumably favored by an internal oligopyrimidine tract in the appropriate distance, an additional component of splice sites. Surprisingly, although one might argue that there is a general insertion preference for antisense MIRs, just the opposite is true. We analyzed MIR elements in all human introns and found significantly more MIR elements inserted in the sense orientation (36,120) relative to the transcription of the host gene than in the antisense orientation (34,414; χ2 test, P < 0.001). By comparison, from 492,344 intronic human Alu sequences, 222,597 were located in the sense and 269,747 in the antisense orientation. Thus, contrary to MIR intronic insertions, significantly more Alu insertions were located in the antisense orientation (χ2 test, P ≤ 0.001).
Twenty of the 64 antisense exonized MIR elements feature a MIR-contributed AG splice site that is preceded by a MIR-contributed oligopyrimidine tract (Fig. 1). This configuration is similar to the prevalent 3′ splice site in the right arm of antisense-oriented Alu elements (Makalowski et al. 1994; Lev-Maor et al. 2003). Interestingly, the MIR-contributed splice sites are located just 3′ adjacent to their highly conserved core regions (Fig. 1). The prevalent singular consensus splice site for the 20 antisense exonized MIR elements is 5′-ATTTTACAG-3′ (Fig. 1). The splice site is similar to the major dual proximal and distal Alu consensus splice sites (5′-TTTTTTGAG-3′ and 5′-TTGAGACAG-3′, respectively) (Lev-Maor et al. 2003). However, we detected no other prevalent splice sites in MIR sequences. Two of the phylogenetically analyzed loci (ZNF639 and LAS1L) display the MIR-contributed splice site plus the oligopyrimidine tract.
Alternative expression of MIR sequences
Three of the five experimentally analyzed MIR elements (NTRK3, LAS1L, Zfp384) exhibit stable alternative splice forms in representative species of the major mammalian branches evidenced both in DNA (conservation of splice sites and maintenance of ORFs) and mRNA, demonstrating that the theoretical splice variants actually exist (Fig. 3). In the case of CHRNA1, stable alternative splice forms in mRNA were found in human and gorilla. However, available DNA sequence data also suggest the exonization of the intronic MIR element in chimpanzee and orangutan but not in gibbon, mandrill, or marmoset, suggesting a great ape specificity of this exonization. Demonstrable expression in human and gorilla and conservation in other great apes indicate selection pressure and probably functionality for this exonization >100 Myr after insertion (Supplemental Fig. S2). Because of the low sequence divergence between exonized MIRs of the compared species, calculated Ka/Ks values, frequently used to describe functional selection pressure for alternative splicing events (Xing and Lee 2005), were not meaningful for these examples (Supplemental Data Set S2).
Figure 3.
Phylogenetic tree of investigated mammals showing the species distributions of the five internal MIR exonizations. In DNA, “+” indicates the presence of the intronic MIR element leaving the splice sites and reading frame intact, “(+)” its presence, without leaving the reading frame intact, and “−” its absence. In RNA, “+” indicates the presence of the exonized MIR sequence in cDNA derived by RT-PCR, and “−” the presence of a cDNA that does not include the exonized MIR sequence. Both symbols indicate that both splice forms are expressed. EST, expressed sequence tag, “+” including or “−” without the exonized MIR region, available at GenBank (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Empty boxes indicate that the corresponding sequence information is not available or not PCR amplifiable; n/a, not analyzed. 1A third splice variant exists, derived from the human MegaMan Human Transcriptome Library (Stratagene). 2Five splice variants exist, three of which include the exonized MIR element. 3RT-PCR performed in the nine-banded armadillo instead of sloth. 4Not shown are “+” symbols under the DNA for chimpanzee and orangutan. 5Unrelated exonized intronic sequence in chicken and a corresponding exonization in the ostrich.
Constitutive expression of a MIR sequence
Zhang and Chasin (2006) proposed that advantageous new exons are eventually expressed constitutively. For SINEs, we provide the first evidence of this prediction. Of the five genes containing internal MIR exonizations, ZNF639 is the only one in which the MIR sequence is constitutively exonized in protein-coding sequences. All analyzed mammalian representatives of Eutheria (Supraprimates: human and mouse; Laurasiatheria: mole, dog, and cow; Afrotheria: manatee; and Xenarthra: sloth), Metatheria (Didelphimorphia: opossum), and Monotremata (Ornithorhynchidae: platypus) express ZNF639 mRNAs only with, and none without, the MIR contribution. The Ka/Ks value of 0.19 indicates purifying selection. The exonized MIR component encodes 45 amino acids located within the 205 amino acid N-terminal part of the ZNF639 protein. This region (amino acids 58–102; Imoto et al. 2003) contains no recognizable protein motifs, while the 280 C-terminal amino acids contain nine zinc finger motifs (Fig. 2). The protein is exclusively expressed in cell nuclei and probably has transcription repressor activity (Bogaerts et al. 2005). ZNF639 is overexpressed in squamous cell carcinomas and over-expression correlates with a shorter survival of these patients. However, the stable MIR inclusion in the 56-kDa protein in all mammalian lineages indicates a significant function of the corresponding 45 additional amino acids.
For ZNF639, all necessary events from insertion of the MIR element to recruitment of parts of the MIR as a novel protein-coding exon occurred on the phylogenetic branch leading from the common ancestor of amniotes (mammals, reptiles, dinosaurs, and birds) to the mammalian ancestor. However, there are no living animals that diverged in this time period that would enable us to reconstruct, step by step, the successive evolution of molecular changes that were necessary to facilitate the 5′ functional splice site and an intact ORF, or to show possible intermediate alternative splice variants.
Discussion
Mammalian-wide detection of MIR sequence exonizations
From a total of 372 potential exonized MIR elements, we investigated 126 elements with strong indications of exonization in human, mouse, or rat (Supplemental Fig. S1). On five of these, we performed an extensive retrospective analysis reconstructing >100 Myr of mammalian history. To establish a complete mammalian exonization pattern, we limited our analyses to those loci that could be amplified in the greatest number of species, sampling for both DNA and RNA analyses (introns ≤ 2 kb). From our extensive experience analyzing 160,000 genomic loci in the evolutionary history of mammalian species (Kriegs et al. 2006), we concluded that larger introns in some compared species render PCR amplification and comparative analyses in the rest of the species difficult. However, in addition to the five experimentally accessible loci presented in this study, we attempted to amplify additional exonizations with larger intronic regions (up to 5.8 kb; Supplemental Table S1, e.g., NM_145065, NM_015424, NM_010058). None of these cases was amplifiable in all essential key mammalian species. To gain full species sampling for those examples and to add some additional loci with lower conservation of flanking regions, we must await the upcoming data from genome sequencing projects.
Expression of MIR sequences
Relative to gene orientation, significantly more MIR exonized sequences are located in the antisense orientation (60%); this preference is even more pronounced for Alu exonizations (85%; Sorek et al. 2002). The difference might be explained by the significant predominance of antisense intronic Alu insertions compared to MIR insertions that are found preferentially in the sense orientation. Furthermore, there are two prevalent 3′ splice sites in the right arm of exonized Alu sequences (proximal and distal; Lev-Maor et al. 2003), whereas we could detect only one such prevalent 3′ splice site for the potential MIR exonizations (Fig. 1; Supplemental Figs. S2, S3).
An intriguing observation is that the natural, MIR-specific splice site is just 3′ adjacent to the MIR conserved core region. However, there are hundreds of thousands of MIR elements that are not associated with splice sites but still bear the conserved core region. Given their origins dating back at least 130 Mya, it is still unclear why, in contrast to the rest of the MIR sequence, the core sequences remain so highly conserved. In coding sequences the 5′ domain of MIR elements appears to be preferentially exonized (Fig. 1). On the one hand, this bias may be due to the naturally occurring splice site in the consensus sequence, but it also might reflect a more efficient recognition of exonized conserved 5′ MIR parts compared with more diverged, but possibly as well exonized, 3′ parts by the RepeatMasker. The documented persistence of MIR element exonizations appears to be different from that of Alu-SINEs. In our detailed analysis of selected Alu exonizations, we found examples in which apparently functional splice sites in ancestral species were not conserved in all related primate lineages, indicating a loss of functional splice forms (Krull et al. 2005). This implies that many Alu-derived exons still have a selection status close to neutrality; in other words, they are not yet indispensable. Due to the relatively short time that has transpired since their exonization, the alternative exons might not yet be capable of making a significant evolutionary contribution. Gotea and Makalowski (2006) propose that such “young” exonized sequences might have functions other than protein coding, such as regulatory functions, including nonsense-mediated decay (NMD) to control gene expression. Another explanation may be the loss of temporarily exapted (historically constrained) alternative splice forms in response to divergent functional needs in different taxa, comparable to the loss of historically constrained duplicated genes (Greenberg et al. 2006).
Time point of MIR exonization
MIR elements were actively mobile prior to the mammalian radiation ∼130 Mya (Smit and Riggs 1995). In four of the five phylogenetically analyzed cases, exonization of MIR elements also took place very early in the evolution of mammals (Fig. 3). However, in the case of CHRNA1 we found a more recent, great ape-specific exonization pattern. Although Saini et al. (2005) suggested, based on RT-PCR fragment size not verified by sequencing, that the CHRNA1 isoform (containing the MIR element) is also present in mouse, we can clearly exclude its expression in mouse. By sequencing both DNA and RNA, we clearly observed that the human orthologous exonized MIR region is missing in mouse (Supplemental Fig. S2; Supplemental Data Set S3). This underscores once more that exaptation in general and exonization in particular of retroposed sequences, can occur at any age and consequently at any stage of decay (Brosius 2005). In the other four cases, five of eight splice sites were part of the MIR consensus sequence, a fact that may have facilitated their exonization immediately after integration. Depending on the evolutionary time point of exonization or the degree of initial positive selection, the exonized part of the retroposed SINE more or less reflects the ancestral sequence of the MIR element.
Nearly 30 years ago, Walter Gilbert recognized in alternative splicing a process that allows evolution to try out new solutions without destroying the old (Gilbert 1978), a variation of a “strategy” that had been recognized in gene and genome duplication (Bridges 1936; Ohno 1970). Particularly in mammalian genomes, exonized transposed elements contribute significantly to alternative splicing.
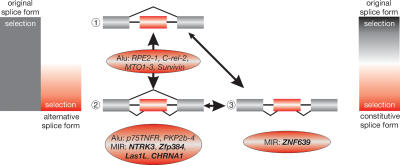
Evolutionary time seems to be a critical factor in establishing essential key mutations required for exonization. Three examples endorse this assumption (Fig. 4): (1) Alu exonizations that are present in specific primate lineages but not in others (RPE2–1, C-rel-2, MTO1–3, survivin) (Krull et al. 2005; Mola et al. 2007). A relatively short evolutionary period (<60 Myr) led to exonizations, the gain and loss of which fluctuated greatly in various species of given taxonomic groups. During early stages of exonization, the newly acquired exons (perhaps present only in a small percentage of the alternative transcripts) probably ranged from slightly detrimental to slightly advantageous. In any event, selection pressure was perhaps either not present or too modest for persistence of the exaptation. (2) MIR exonizations (NTRK3, LAS1L, and Zfp384) are present in all mammals. Selection over longer time periods favored exonizations that are now stably established and expressed in all representative taxa. In many cases, both the ancestral and the new alternative splice forms were advantageous and therefore expression of both forms persisted. (3) The MIR exonization in ZNF639 is constitutively expressed in all analyzed mammalian species. Expression shifted completely, possibly via co-existence of two alternative splice forms for a time, from 100% of the ancient form (prior to MIR exonization) to 100% of the form harboring the MIR exon, such that the latter is now constitutive. We cannot discern whether the constitutive inclusion of the exonized sequence in ZNF639 was achieved via a transient alternative splicing or directly after acquisition of the necessary splice sites (Fig. 4).
Figure 4.
Evolutionary scenario after exonization of sequences, from the ancestral constitutive to alternative and, occasionally, to novel constitutive splicing. Intensities of selection are indicated at the left and right bars. For evolution of alternative splice sites, the intensities of selection vary over time. Initially, inclusion of the novel exon might be slightly deleterious, neutral, or slightly advantageous. Subsequently, selection changes, perhaps during a period of positive selection to increasing levels of negative selection (bar at the left with a gradient from white to red). The numeration refers to intronic insertion of a transposed element (red box) (1), followed by acquisition of necessary components for alternative splicing (2). Under relaxed selective pressure, the exonized condition might revert or become fixed in different lineages (vertical double arrow). In an extreme case, (3), as shown for ZNF639, the novel alternative splice form might, over time, completely replace the ancestral constitutive splice form, thus representing a different constitutive form (bar at the right side, with gradients from black via white to red). However, the constitutive inclusion of the exonized sequence in ZNF639 could also have been acquired directly after acquisition of the necessary splice sites. Examples corresponding to different stages of exonization taken from previous works (Singer et al. 2004; Krull et al. 2005; Mola et al. 2007) or from the present work (bold letters) are presented in ovals.
The low Ka/Ks value emphasizes the moderate selection pressure acting on the exonized MIR sequence of ZNF639. Interestingly, although we did not detect a MIR element in the corresponding locus in chicken, we found an exonized intronic sequence of the same size in this locus. The same sequence region (one additional triplet with respect to chicken) is also exonized in the ostrich, which represents another major branch of the bird phylogenetic tree. A DNA sequence alignment shows no apparent relationship between the two independently exonized sequences in mammals and birds and only ∼50% random similarity (Supplemental Fig. S4A). However, although the additional sequences of the proteins do display some similarities in charges and hydrophobicity (Supplemental Fig. S4B,C), protein structural information is necessary to understand if the exonized sequences might play any beneficial role at all in separating neighboring protein domains. The orthologous gene in Xenopus lacks this additional exon, and consequently the protein lacks the extra 45-amino acid segment.
Although Ka/Ks values indicate that the exonized part of the MIR element itself is under moderate selection pressure, the 291-nt adjacent protein coding flanks (parts of exons 5 and 7) show even lower Ka/Ks values (0.17 vs. 0.19 for the exonized MIR). This, at most, suggests a possibly lower selection pressure on the MIR exonized sequence than on the flanks. This difference is even greater when compared to the nine functional zinc finger domains of ZNF639 in exon 7 (582 nt; Ka/Ks = 0.03; data not shown). However, more information about functional domains of the N-terminal region of the protein is necessary to present more conclusive information about a potential spacer function of the exonized MIR sequence. There is also another report of two independent exonizations, although of different lengths and origin, in the same intron of the ADARB1 gene in different taxonomic groups (human and mouse; Slavov and Gardiner 2002).
In theory, the first transition from random insertion to alternative splicing is reversible and is either not at all under purifying selection or under relaxed negative selection (Xing and Lee 2006). Novel exons might be generated and shaped for “testing” under such relaxed selection (this period might include a phase of positive selection as well). Through a gradient of purifying selection, the second transition leads to stable alternative splice forms and in rare cases to constitutive splicing that includes the exonized form. ZNF639 is the first gene experimentally verified to contain such a constitutively expressed MIR exonization and exaptation. Of course, at this time we cannot exclude the possibility that the ZNF639 exonization was constitutive from the start and did not pass through this transitional state.
Conclusion
The contribution of transposed elements to gene structures is more or less coincidental. Their persistence is usually transient. If not deleted, they fade beyond recognition over longer evolutionary periods. However, a notable fraction of transposed elements escapes transience, for example, by integrating into protein-coding parts of genes, facilitated by internal components providing splice sites and oligopyrimidine tracts and “reprogramming” the splicing system of a targeted gene. Once proven worthy in the struggle of survival, they endure recognizably over hundreds of millions of years and contribute to significant tasks. We have identified and analyzed some of these candidates, thus shedding light on their >100 million-year-old evolutionary histories that show they have clearly stood the test of evolutionary time and persisted in mammalian lineages. We showed that 3′ splice site selection in exonized transposed elements is not restricted to Alu elements but seems to be an older and significant mechanism for MIR exonization as well. Alternative splicing of exonized MIRs is exemplarily shown to be a stable process retained >100 Myr in all major groups of mammals. Functional persistence shown by constitutive splicing of an exonized MIR sequence was evidenced for the first time and demonstrates that this evolutionary pathway is not necessary correlated with genetic disorder, as has been suggested for Alu exonizations by Lev-Maor et al. (2003). The present data are ample evidence of the value of exonization, given enough evolutionary time, in generating genomic novelties.
Methods
Data selection
To identify MIR elements in protein-coding sequences, we screened a compilation of mammalian mRNAs with transposable element cassettes (Genomic ScrapYard; http://warta.bio.psu.edu/SYDB/database.html), performing independent searches for human, mouse, and rat. Out of 1091, 472, and 201 matching records we selected 314, 38, and 20 cases that included MIR elements in human, mouse, and rat, respectively. These 372 cases were then scrutinized to filter out duplications and other artifacts (246 cases; Supplemental Fig. S1). To search for cases that were suitable for experimental evaluation of their phylogeny, we screened the remaining 126 potential exonizations by applying the following criteria: (1) The MIR-derived sequence should not be located in the first or last protein-coding exon as they usually lack highly conserved flanking regions and hence are difficult to amplify by polymerase chain reaction (PCR). (2) To enable manageable PCRs of up to 2 kb in highly diverged mammalian species, the MIR-derived sequences should be flanked by conserved sequences. Conservation was determined by comparison of available genomic sequences (e.g., of human, mouse, and dog). (3) Indication for exonization should be supplied by such available information as EST data or other published information. (4) Relevant tissues should be available for specific alternative transcripts. Five of the 126 potential exonizations fulfilled all four criteria and were subjected to further phylogenetic examination using PCR, RT-PCR, and sequence analyses. Note that the ScrapYard database consists of GenBank entries from January 15, 2002. It is expected that an updated database would facilitate the recovery of additional cases of MIR exonizations.
Available sequence information was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat) and the NCBI trace archive (http://www.ncbi.nlm.nih.gov/blast/tracemb.shtml).
Experimental procedures concerning DNA and RNA extraction, PCR amplification, and reverse transcription are given in the Supplemental Protocol S1. PCR primers are listed in Supplemental Table S2 and are illustrated in Supplemental Fig. S5.
Sequence analyses
For detection and classification of the inserted and partially exonized MIR sequences, we used the RepeatMasker server (A.F.A. Smit, R. Hubley, and P. Green, unpubl.; RepeatMasker at http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). The available mammalian sequences of the five relevant loci were derived from the NCBI database, and all additionally amplified sequences were manually aligned. GenBank entries are compiled in Supplemental Table S3.
Ka/Ks values
We used the Yang–Nielsen maximum likelihood method (Yang and Nielsen 2000), implemented in the YN00 program of the PAML package (Yang 1997), to calculate the Ka/Ks values for aligned exonized parts of the MIR elements. Ka/Ks values were averaged over all investigated species pairs. It should be noted that in most cases the significance of the derived values was limited by the short length and low variation of the exonized regions.
Acknowledgments
We thank Frank Grützner, Rodney L. Honeycutt, Uwe Joite, Jan Ole Kriegs, Jörg Molten, Bernhard Neurohr, Christian Roos, Gertrud Scheele, Heike Weber, and Anja Zemann for providing us with tissue samples and Marsha Bundman for editorial assistance. We thank Valer Gotea for his help in selecting data from the Genomic ScrapYard database and Michael Haberl for his comments. We thank Django Sussman for introducing us to methods for analyzing structural features of the ZNF639 protein. J.S. thanks Matthias Schmitz for all his personal support. This work was supported by the Nationales Genomforschungsnetz (NGFN) (0313358A to J.B. and J.S.), the European Union (EU) (LSHG-CT-2003-503022 to J.B.), and the Deutsche Forschungsgemeinschaft (DFG) (SCHM1469 to J.S. and J.B.).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to NCBI under accession nos: DQ323592–DQ323661, DQ507223–DQ507235, DQ855908–DQ855913, EF418572, EF422277, EF520683, and EF520684.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6320607
References
- Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Rubin E.M., Kent W.J., Haussler D., Kent W.J., Haussler D., Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Bogaerts S., Vanlandschoot A., van Hengel J., van Roy F., Vanlandschoot A., van Hengel J., van Roy F., van Hengel J., van Roy F., van Roy F. Nuclear translocation of αN-catenin by the novel zinc finger transcriptional repressor ZASC1. Exp. Cell Res. 2005;311:1–13. doi: 10.1016/j.yexcr.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Bridges C.B. Genes and chromosomes. Teaching Biol. 1936;Nov:17–23. [Google Scholar]
- Brosius J. Echoes from the past—Are we still in an RNP world? Cytogenet. Genome Res. 2005;110:8–24. doi: 10.1159/000084934. [DOI] [PubMed] [Google Scholar]
- Brosius J., Gould S.J., Gould S.J. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Labuda D., Labuda D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc. Natl. Acad. Sci. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V., Makalowski W., Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Greenberg A.J., Moran J.R., Fang S., Wu C.-I., Moran J.R., Fang S., Wu C.-I., Fang S., Wu C.-I., Wu C.-I. Adaptive loss of an old duplicated gene during incipient speciation. Mol. Biol. Evol. 2006;23:401–410. doi: 10.1093/molbev/msj045. [DOI] [PubMed] [Google Scholar]
- Hillman R.T., Green R.E., Brenner S.E., Green R.E., Brenner S.E., Brenner S.E. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I., Yuki Y., Sonoda I., Ito T., Shimada Y., Imamura M., Inazawa J., Yuki Y., Sonoda I., Ito T., Shimada Y., Imamura M., Inazawa J., Sonoda I., Ito T., Shimada Y., Imamura M., Inazawa J., Ito T., Shimada Y., Imamura M., Inazawa J., Shimada Y., Imamura M., Inazawa J., Imamura M., Inazawa J., Inazawa J. Identification of ZASC1 encoding a Krüppel-like zinc finger protein as a novel target for 3q26 amplification in esophageal squamous cell carcinomas. Cancer Res. 2003;63:5691–5696. [PubMed] [Google Scholar]
- Kapitonov V.V., Pavlicek A., Jurka J., Pavlicek A., Jurka J., Jurka J. Anthology of human repetitive DNA. In: Meyers R.A., editor. Encyclopedia of molecular cell biology and molecular medicine. Wiley-VCH; Weinheim: 2004. pp. 251–305. [Google Scholar]
- Kriegs J.O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Kiefmann M., Jordan U., Brosius J., Schmitz J., Jordan U., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull M., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G., Sorek R., Shomron N., Ast G., Sorek R., Shomron N., Ast G., Shomron N., Ast G., Ast G. The birth of an alternatively spliced exon: 3′ Splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Makalowski W., Mitchell G.A., Labuda D., Mitchell G.A., Labuda D., Labuda D. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Mola G., Vela E., Fernández-Figueras M.T., Isamat M., Munoz-Mármol A.M., Vela E., Fernández-Figueras M.T., Isamat M., Munoz-Mármol A.M., Fernández-Figueras M.T., Isamat M., Munoz-Mármol A.M., Isamat M., Munoz-Mármol A.M., Munoz-Mármol A.M. Exonization of Alu-generated splice variants in the Survivin gene of human and non-human primates. J. Mol. Biol. 2007;366:1055–1063. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- Nishihara H., Smit A.F.A., Okada N., Smit A.F.A., Okada N., Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer; New York: 1970. [Google Scholar]
- Saini S.S., Tüzün E., Christadoss P., Tüzün E., Christadoss P., Christadoss P. The cDNA of mouse skeletal muscle transcribe for both isoforms 1 and 2 of acetylcholine receptor α subunit. J. Neuroimmunol. 2005;169:177–179. doi: 10.1016/j.jneuroim.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Singer S.S., Maennel D.N., Hehlgans T., Brosius J., Schmitz J., Maennel D.N., Hehlgans T., Brosius J., Schmitz J., Hehlgans T., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. From “junk” to gene: Curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- Slavov D., Gardiner K., Gardiner K. Phylogenetic comparison of the pre-mRNA adenosine deaminase ADAR2 genes and transcripts: Conservation and diversity in editing site sequence and alternative splicing patterns. Gene. 2002;299:83–94. doi: 10.1016/s0378-1119(02)01016-8. [DOI] [PubMed] [Google Scholar]
- Smit A.F., Riggs A.D., Riggs A.D. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995;23:98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G., Graur D., Ast G., Graur D., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E., Lykke-Andersen J., Lykke-Andersen J. mRNA surveillance: The perfect persist. J. Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- Xing Y., Lee C., Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl. Acad. Sci. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C., Lee C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang X.H.-F., Chasin L.A., Chasin L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl. Acad. Sci. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]