Abstract
We have reconstructed the gene content and order of the last common ancestor of the human pathogens Mycobacterium leprae and Mycobacterium tuberculosis. During the reductive evolution of M. leprae, 1537 of 2977 ancestral genes were lost, among which we found 177 previously unnoticed pseudogenes. We find evidence that a massive gene inactivation took place very recently in the M. leprae lineage, leading to the loss of hundreds of ancestral genes. A large proportion of their nucleotide content (∼89%) still remains in the genome, which allowed us to characterize and date them. The age of the pseudogenes was computed using a new methodology based on the rates and patterns of substitution in the pseudogenes and functional orthologous genes of closely related genomes. The position of the genes that were lost in the ancestor’s genome revealed that the process of function loss and degradation mainly took place through a gene-to-gene inactivation process, followed by the gradual loss of their DNA. This suggests a scenario of massive genome reduction through many nearly simultaneous pseudogenization events, leading to a highly specialized pathogen.
Reductive genome evolution has taken place in most bacterial lineages with lifestyles strictly requiring association to a host, either as parasites, commensals, or mutualistic symbionts (Andersson and Kurland 1998; Cole et al. 2001; Silva et al. 2001; Wernegreen 2002; Parkhill et al. 2003). The reduction involves the loss of many genes and their associated functions and results in the shrinkage of the genome due to the DNA loss of these inactive genes and genomic regions (Gómez-Valero et al. 2004). Several causes have been suggested to start reductive genome evolution, including diverse types of change in lifestyle: (1) from a free-living to a strictly intracellular or host-associated life, (2) the restriction from multiple to a specific host, or even (3) from multiple to specific host tissues. These changes in lifestyle produce a relaxation of the natural selection pressure, resulting in individuals accumulating detrimental or loss-of-function mutations. Eventually these mutations become fixed in the populations, which is favored by the small population size of bacterial mutualists (Mira and Moran 2002). As time goes by, some DNA repair functions are lost and this leads to a further increase in the mutation rate and a concomitant increase in the production of deleterious mutations. All of these phenomena lead to the loss of many genes and, in bacteria, to the reduction of the genome size.
The loss of genes is not necessarily associated with the loss of DNA. In fact, the half-life of a pseudogene in some eukaryotic species may be hundreds of millions of years (Graur et al. 1989), but it has been observed in bacteria that the tendency of these nonfunctional regions is to disappear from the genome in short periods of time (Gómez-Valero et al. 2004). Several reasons have been proposed, such as a systematic mutational bias toward deletion events (Mira et al. 2001) or the effect of natural selection favoring small genome sizes due to their faster replication and small metabolic cost (Cavalier-Smith 2005).
The most striking case of reductive genome evolution among published pathogens has probably occurred in the causative agent of leprosy, Mycobacterium leprae (Cole et al. 2001). Not only is its genome small (3.2 Mb) when compared with other mycobacterial species, but it also has a small number of active genes (∼1600) (Cole et al. 2001) as compared with closely related species (>4000) (Cole et al. 1998; Fleischmann et al. 2002; Garnier et al. 2003; Li et al. 2005). Strikingly, M. leprae contains the highest number of pseudogenes (>1000) among published genomes. The temporal dynamics of the massive gene decay of this species has been analyzed in several studies. Based on the frequency of in-frame stop codons, it has been proposed that there are two populations of pseudogenes associated with two large independent gene decay events caused by the loss of two sets of sigma factors (Madan Babu 2003). These events were tentatively explained by the “domino theory” (Dagan et al. 2006). This process starts with a gradual gene-by-gene death (Silva et al. 2001), but eventually a crucial gene within a complex pathway is lost, simultaneously producing multiple gene losses. However, these results must be considered with care because the number of in-frame stop codons is a very inaccurate measure of evolutionary distance, and pseudogenization usually involves small deletions that automatically introduce a large number of stop codons.
In this study, we have analyzed the dynamics of the reductive process by reconstructing the order and gene content of the genome of the last common ancestor of M. leprae (Mle) and Mycobacterium tuberculosis (Mtu) and comparing it with the present M. leprae genome. We have identified all of the ancestral genes that were lost during the lineage evolution whose present status is either pseudogene or absent gene. We have also determined the age of each individual pseudogene with a new methodology, the distribution of gene losses on the ancestor’s genome, the rearrangement of the genome during the reductive evolution, and the proportion of lost DNA in either pseudogenes or absent genes.
Results
Reconstruction of the gene content and order of the M. leprae ancestor’s genome
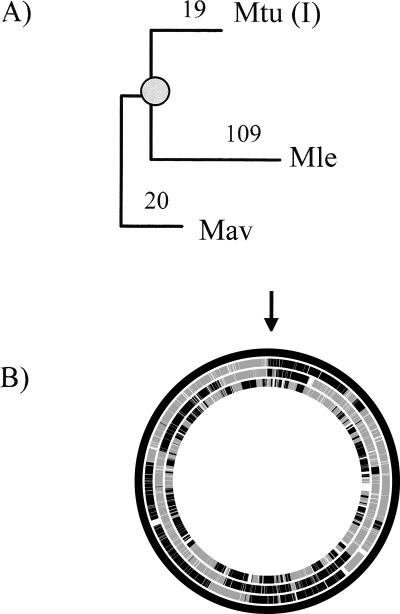
We started by positioning Mle and other completely sequenced mycobacterial species available at the start of the present study in the reconstructed phylogenetic tree (see Methods). This showed that Mle is closer to Mtu, while Mycobacterium avium (Mav) is an outgroup (Fig. 1). Since the ancestral gene order could not be completely reconstructed using only these genomes, we added to this study the available information on the ongoing genome of Mycobacterium marinum (Mma), which is an outgroup relative to Mle and Mtu.
Figure 1.
Phylogenetic relationships among mycobacterial species. See Methods for abbreviations. The numbers to the left of each node are bootstrap values. A line of length 0.1 amino acid substitutions per site is shown.
We identified the orthologous genes and pseudogenes in the genomes of Mle, Mav, and three genomes of the Mtu complex: M. tuberculosis strain H37Rv [Mtu(I)], M. tuberculosis strain CDC1551 [Mtu(II)], and M. bovis (Mbo). For this, we performed an extensive search for the presence of putative pseudogenes (even those that were extremely degraded) in every genome. This allowed the detection of 177 new pseudogenes (see Supplemental Table S2) not included in previously published annotations (Cole et al. 2001; Leproma, http://genolist.pasteur.fr/Leproma). Most of these new pseudogenes were detected by their sequence similarity to the genes of the recently sequenced Mav genome (Li et al. 2005). The ancestral genome content was predicted based on the parsimony criterion that if a gene was present in at least two of the three mycobacterial lineages (Mle, Mav, and Mtu complex), it was probably ancestral. Using this strategy we inferred that the ancestor of M. leprae had 2977 genes at the moment when its lineage diverged from that of Mtu. We further inferred that 1537 of these genes were lost in Mle. Lost genes were classified into the 952 equivalents to previously annotated pseudogenes, 177 newly annotated pseudogenes and 408 absent genes. The absent genes showed no significant sequence similarity in the Mle genome. To establish the order of these genes in the ancestor’s chromosome, we applied the same parsimony criterion. Thus, the ancestral order of two genes was inferred when their orthologs were included in the ancestor’s genome and were present in the same order in two of the analyzed genomes. When the two ancestral genes were contiguous in one genome, noncontiguous in the other, and at least one of them was absent in the third genome, we applied the parsimony criterion after including the comparison with Mma. We were able to establish the ancestral order for 2975 of the 2977 genes. The comparison of the order of the ancestor’s genome and those of the other mycobacterial species allowed us to estimate the number of breakpoints in the three lineages, which is directly related with the rearrangement rates (Fig. 2). For the same period of time, the number of rearrangements was more than fivefold higher in the Mle lineage than in the Mtu.
Figure 2.
Chromosomal rearrangements between Mav, Mtu(I), Mle, and the ancestor’s genome. (A) Number of breakpoints from the ancestor’s genome to each species used. The position of the ancestor’s genome is marked with a shaded circle. (B) Graphic representation of the genome rearrangements. From the outside: ancestor, Mav, Mtu(I), and Mle. White spaces represent absent genes in the corresponding genome. The black line in the ancestral genome and in the genomes of Mav and Mtu(I) (top, marked by the arrow) corresponds to two genes of unknown order in the ancestor. Each change of color from dark to light gray indicates a breakpoint.
Age of M. leprae pseudogenes
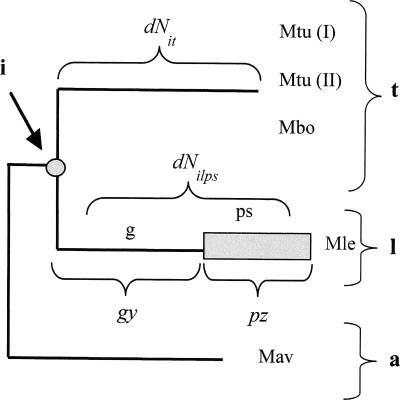
To estimate the age of the pseudogenes and to determine whether they arose from one single massive pseudogenization event or from two, as previously proposed (Madan Babu 2003), we developed a method to estimate the age of each individual pseudogene based on the number of nonsynonymous nucleotide substitutions per site. The method is based on the idea that the number of substitutions accumulated from the ancestor’s gene to the present pseudogene is the sum of those produced during two periods of time: while evolving as a gene and while evolving as a pseudogene. The rates of change at nonsynonymous sites are very different in the two cases, because in the former most changes will be purged by natural selection, whereas in the latter they are neutral. It is an abuse of language to talk about nonsynonymous changes in pseudogenes, but here we shall use this term to refer to changes at positions that, while the gene was functional, would lead to nonsynonymous changes. Our method accounts for the diversity of nonsynonymous substitutions among different genes, but can only be applied to the pseudogenes having orthologs in both Mtu and Mav. The older the pseudogenization event, the larger the number of nucleotide substitutions that pseudogenes have accumulated. The age of pseudogenes is represented by a parameter (p), which indicates the relative period of time when the element evolved as a pseudogene in the Mle evolutionary branch (Fig. 3).
Figure 3.
Phylogenetic tree of Mav (a), Mle (l) and the M. tuberculosis group (t). The common ancestor to t and l is designed as i. Two periods are considered in the branch of Mle: a first period as an active gene (g) and a second period as a pseudogene (ps). Parameters y and z are the numbers of nonsynonymous substitutions per nucleotide site in the Mle branch, in the event that the active gene had reached the present time or it had evolved as a pseudogene from i until the present, respectively. dNit is the number of nonsynonymous substitutions from the ancestor to Mtu.
In order to estimate the parameter p for each pseudogene, we first need to estimate dNilps (the actual number of nonsynonymous substitutions from the ancestral gene (i) to the present Mle pseudogene [lps]), which is obtained by means of the following formula:
where g is the relative period of evolution as a gene, y is the number of expected nonsynonymous substitutions per site if the sequence had been evolving as a gene through the complete period, and z is the number of substitutions per nonsynonymous site if the sequence had been evolving as a pseudogene through the complete period (we refer to those sites of the pseudogene that were nonsynonymous in the gene). Since g is equal to 1 − p, we have:
and then we obtain:
For each gene, the first value (dNilps) is easily estimated from the pairwise distances obtained from the nucleotide alignment involving the Mtu (t) and Mav (a) genes and the Mle pseudogene:
The second value (y) would be identical to dNit (the number of nonsynonymous substitutions per site in the Mtu lineage) in the case that the rate of gene evolution was identical in the Mle and Mtu lineages. To test this assumption, we performed an analysis of dN on 1281 orthologous genes present in the three genomes. This showed that the average dNit (0.045) was smaller than dNil (0.065), indicating that even Mle genes were evolving faster than Mtu genes (Wilcoxon P < 0.001). This different average evolutionary rate in the Mle lineage was probably not homogeneous over the complete period of time, and may be associated with a recent acceleration due to the same type of changes that produced the massive gene inactivation. For that reason, we searched for a function f(dNit) that produced the best correlation between dNit and dNil values. After removing a few outliers, the best adjustment (R2 = 0.48) was obtained using the formula (see Supplemental text, section 1):
The third value (z) should be the same value for all pseudogenes, considering that the rate of substitution for any pseudogene is the same as the rate of evolution of the nonfunctional DNA. The easiest way to estimate the value of z is to obtain the average number of synonymous substitutions in the Mle lineage, because it is easier to accurately align pseudogenes than putatively nonfunctional intergenic regions. This method is expected to slightly underestimate the z value, because selection for codon usage may purge some synonymous substitutions. However, we have observed that in mycobacterial genes there is no significant bias indicating selection for codon usage (data not shown). From the alignments of Mle pseudogenes and Mtu and Mav genes, we estimated the average number of synonymous substitutions per site (dSilps = 0.94), which we will use as the value of the z parameter. Thus the final formula was:
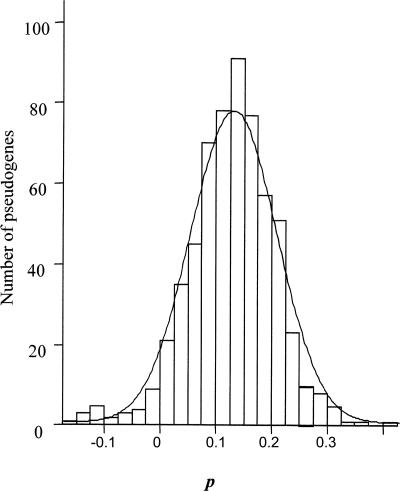
The formula was applied to the 611 pseudogenes aligned with Genewise (see Methods). The distribution of p values showed a normal distribution (Kolmogorov-Smirnov, P = 0.373) of mean 0.13 and standard deviation 0.08 (Fig. 4). It is apparently pointing to the fact that a single and recent event led to the formation of all of the analyzed pseudogenes. We have compared the pseudogene P values with the decrease in nucleotide identity and the decrease in the GC content against the Mtu and Mav genes, finding low, but significant (p = 0.01) correlation coefficients (R = 0.545 and R = 0.329 for identity and GC content, respectively) (Supplemental text, section 2). As many studies with intracellular endosymbionts have shown, the pseudogene DNA sequences show a tendency with time toward the loss of identity and decrease in GC content (Silva et al. 2007). For that reason, the observation of a low correlation between these factors and p may suggest that a part of parameter p variation is not random but due to differences in the moment of pseudogenization. Accordingly, those genes that inactivated early would have larger reductions in similarity and GC content. Thus, we find evidence that a massive gene inactivation took place very recently (P = 0.13), with gene inactivation events possibly taking place over a short period of time.
Figure 4.
Frequency distribution for the age of Mle pseudogenes (n = 611).
We estimated p for the two proposed sets of pseudogenes derived from two independent inactivation events (Madan Babu 2003), and the values we obtained were 0.12 ± 0.07 and 0.14 ± 0.08 (no significant differences, Mann-Whitney U P = 0.05), respectively. We also failed to find significant differences for the dN/dS ratio, computed among the orthologous genes in Mtu and Mav (average values of 0.0606 and 0.0619, P = 0.14, Mann-Whitney U test). Finally, we found a small, but significant difference in the GC content of the two sets of Mle pseudogenes (0.5734 and 0.5606, P < 0.001, Mann-Whitney U test). Hence, according to our estimation, there is no statistical evidence for two large pseudogenization events when the evolutionary history of the pseudogenes is analyzed precisely. To understand why we obtained a different result from that published (Madan Babu 2003), we estimated the numbers of stop codons of the two sets of pseudogenes in our alignments and observed that, after correcting the alignments for frameshifts, the number of stop codons decreased drastically, indicating that most of the stop codons were not of the original gene open reading frame (data not shown). This means that counting stop codons is a very inaccurate way to estimate evolutionary time, but also that the quality of the alignments must be very high to make this type of inference.
We tried to translate the value of p in years to have a rough idea of the time when the massive pseudogenization took place. There is no information about the time of divergence between Mle and Mtu, but as an approach we used the previously estimated time of divergence between Escherichia coli and Salmonella typhimurium and assumed that the rate of nucleotide substitution in the 16S rRNA gene is similar in these two enteric bacteria and in the Mtu lineage. Naturally, since both assumptions are inaccurate, the resulting value is only meant to provide an order of magnitude, not a precise measure of evolutionary time. Based on the number of nucleotide substitutions in the 16S rRNA gene, the divergence between E. coli and S. typhimurium was estimated at 140 million years (Ochman and Wilson 1987). We estimated the average number of nucleotide substitutions per site between the seven E. coli and seven S. typhimurium 16S rRNA genes (0.0295). Then we estimated the number of substitutions between the single 16S rRNA genes of Mle, Mtu, and Mav and used these pairwise distances to estimate the number of substitutions between the ancestor and Mtu (dit = 0.00691). Considering that 0.0295 nucleotide substitutions per site took place over 280 million years (140 in each lineage), we estimated at ∼66 million years the age of the last common ancestor of Mle and Mtu. This means that p is equivalent to around nine million years. Taking into account the different errors associated with this estimation, including those previously mentioned and the standard errors indicated by the nucleotide distance method, we may assume that a massive gene inactivation event took place in the last 20 million years.
Analysis of dN/dS ratio in the three ancestral gene categories
We estimated the functional importance of the three ancestral gene categories in the Mle genome (retained, pseudogenized, and absent) by estimating the strength of purifying selection through the dN/dS ratios. These values were estimated for Mtu–Mav orthologous gene pairs. Retained genes showed the lowest average value (0.0528 ± 0.0396), followed by pseudogenized (0.0643 ± 0.0728), and by absent (0.0823 ± 0.0717). All three pairwise comparisons were significantly different (Mann-Whitney U P < 0.001). These results show that, although differences were small, a significant gradation was observed in the strength of the selective pressure for the three categories.
Gene-loss distribution along the ancestor’s genome
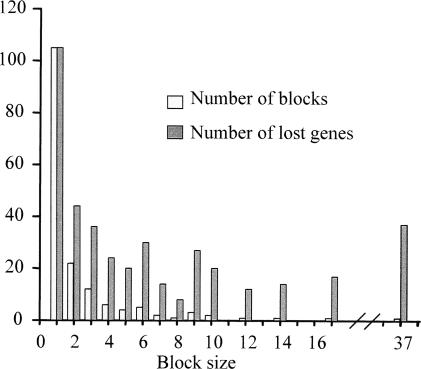
The positions of the genes in the inferred ancestor’s and Mle genomes were compared to estimate whether ancestral genes were mainly lost individually (gene by gene) or in blocks. The complete or partial sequence of 1129 pseudogenes present as genes in the ancestral genome was detected in Mle. This strongly suggests that in this lineage, inactivation gene by gene, and not in large chunks, was the most frequent form of genome degradation. Moreover, the analysis of 408 ancestral genes that were not detected by sequence similarity in the present Mle genome has revealed that an important fraction was also lost individually. In fact, the sizes of lost adjacent gene blocks were in general small (mean ± SD = 2.5 ± 3.7) with only one block of 37 contiguous lost genes in Mle (Fig. 5). Since the individual loss of several contiguous genes may be mistaken for a large deletion removing a block of contiguous genes, the previous values are in fact an overestimation of the size of the deletions that we can infer. Hence, genome degradation in Mle is mainly taking place through small, mostly individual gene inactivation events, followed by gradual nucleotide loss.
Figure 5.
Distribution of genes with the status of absent genes in Mle. The chromosomal position of these genes in the ancestor’s genome has been analyzed. Absent genes may be isolated (block size = 1, although they are not blocks, we called them blocks of a single gene with the aim of simplifying the figure) or in blocks of several contiguous absent genes (size range 2–37).
Nucleotide loss in M. leprae
Two types of genes have been lost in the Mle lineage after separation from Mtu. Some maintain sufficient levels of nucleotide similarity and for that reason are classified as pseudogenes, whereas others have either been totally deleted or have diverged beyond recognition and are classified as absent genes. We estimated the fraction of DNA lost after becoming nonfunctional in a similar way to that previously described for the bacterial endosymbiont Buchnera aphidicola (Gómez-Valero et al. 2004). However, because mycobacterial genomes are much less stable than B. aphidicola (horizontal gene transfer, transposable elements, genome rearrangements), we focus our study only on lost genes flanked by the same functional genes in Mle, Mtu, or Mav. For this we measured the variation in sequence size of the region between these two flanking conserved genes. The average fraction of DNA lost in the regions of pseudogenes was 11% (Fig. 6A). This value showed a large variation, including several genes with negative values, suggestive of sequence insertion in some regions. The percentage of lost DNA in the absent gene regions was much higher, with close to 50% of the regions having a loss higher than 80% (Fig. 6B).
Figure 6.
Percentage of lost DNA in Mle pseudogenes or absent genes. (A) Proportion of lost DNA in pseudogenes. (B) Proportion of lost DNA in absent genes.
Discussion
The sequencing of the Mle genome revealed an extreme case of reductive evolution that was based on the comparison with the available Mtu(I) genome (Cole et al. 1998, 2001). The recent reporting of the genomes of other mycobacteria, especially the most distantly related Mav (Li et al. 2005), opened the possibility of using comparative genomic analysis to gain in-depth understanding on this subject. For example, recently published papers on the metabolic pathways retained in these Mycobacterium spp. (Marri et al. 2006) showed that Mle evolved by retaining a minimal gene set for most of the gene families. In our study, we aimed to reconstruct the gene content and order of the ancestor of Mle. Because the closest sequenced genome corresponds to those of the Mtu complex, we were only able to reconstruct the structure of the genome of the ancestor of both species that lived around 66 million years ago. We estimate that this ancestor had a minimum of around 3000 genes. This value is a minimal estimate, because convergent gene losses in the Mle and Mtu lineages cannot be detected and included in the ancestral gene inventory. In addition, genes acquired through horizontal transfer events during the Mle lineage evolution were not included in the ancestor’s genome. This is the reason why the ancestor’s genome contains as genes only 952 of the more than 1100 pseudogenes previously annotated in Mle.
The comparison of the order of the orthologous genes in the ancestor, Mle, Mtu, and Mav, allowed the quantification of the level of rearrangement in those genomes. Thus, for the same period of time, the lineage of Mle presents 109 breakpoints compared with the ancestor, while Mtu presents only 19. This lineage-specific heterogeneity in the level of genome rearrangement has been previously described in gamma-Proteobacteria, with fast-evolving species such as Pasteurellaceae having many rearrangements and others with an almost complete absence of rearrangements, such as the endosymbionts B. aphidicola or Blochmannia spp. (Tamas et al. 2002; Gil et al. 2003; Silva et al. 2003; Belda et al. 2005; Degnan et al. 2005). The increase in the level of genome rearrangement detected in this study probably did not happen homogeneously during the evolution of the Mle lineage, but was probably associated with the same cause that produced the massive gene inactivation. This is probably the same situation that occurred in the lineage of B. aphidicola after its divergence from enterics, where most of the genome rearrangements took place in an early period, probably associated with the change from a free to an endosymbiont way of life (Belda et al. 2005).
The analysis of 1537 gene losses from the ancestor to Mle showed that 1129 ancestral genes were present in the Mle genome as pseudogenes, while 408 were not. To clarify the dynamics of the losses, we carried out several analyses, including the estimation of the age of the pseudogenes, based on a new methodology and the estimation of the proportion of lost DNA compared with the ancestral genome. Several methods for the estimation of the age of pseudogenes have been previously reported in the literature: (1) the calculation of the proportion of stop codons (Madan Babu 2003), (2) the assumption that a relation exists between the fraction of disablements in a pseudogene (frameshifts and stop codons) and the number of matching residues with the orthologous gene (Liu et al. 2004), and (3) the estimation of genetic distances (the number of nucleotide substitutions per site) between the gene and the pseudogene, but without correcting for the variable evolutionary rate of each gene (Dagan et al. 2006). Our method presents an important improvement. First, more precise alignments are used. Second, the system requires the presence of two orthologous genes and a pseudogene. The estimation of the number of nucleotide substitutions between the two genes allows us to determine the rate of evolution of each gene and the increase produced as a consequence of the higher evolutionary rate of the pseudogene. Third, the number of synonymous and nonsynonymous substitutions, which are very different and gene specific, are estimated and used separately.
With our method we have estimated the age of the pseudogenes through the estimation of the parameter p. In its formula, we consider that the rate of nucleotide substitutions for nonfunctional DNA regions is identical for any pseudogene independently of its position in the genome. We have estimated this value as the average of synonymous nucleotide substitutions per site in the Mle branch (dSilps). This may not be completely true, because a significant but small difference in the dS of genes placed at different distances from the origin of replication was detected for several pairs of bacterial species (Mira and Ochman 2002). Additional analyses using the specific dSilps values of each pseudogene produced a very similar average p value (see Supplemental text, section 3). In addition, we analyzed whether the among-gene variation of the purifying selection pressure would be able to affect our estimations of the age of the pseudogenes. None of the terms of the formula (Equation 6) can affect our estimations, although we cannot discard the possibility that dSilps was slightly underestimating the number of substitutions for nonfunctional DNA and producing a small underestimation of p as a consequence (see Supplemental text, section 4).
Our results show a recent massive gene inactivation event that we estimate took place in the last 20 million years. The normal distribution of p and the analysis of the two populations of pseudogenes previously described (Madan Babu 2003) corroborates the hypothesis that a single cause produced the simultaneous inactivation of many genes through multiple independent events. However, the detection of low-correlation coefficients with the decrease in identity and in GC content suggests small differences at the time of gene inactivation.
The loss of DNA of the ancestral genes classified as absent could occur through two mechanisms: (1) deletion mutations spanning the complete gene or (2) point mutations and gradual erosion of the DNA. Our analysis of Mle absent gene syntenic regions has shown that, in some cases, there is still some remnant DNA that does not show sequence similarity with the corresponding orthologous gene. These gene losses would indicate that the pseudogenization event took place soon after divergence from Mtu, and because of the high number of nucleotide substitutions (around one per site) no sequence similarity would be detected at present. For that reason, some absent genes would correspond to old pseudogenization events, while others could be associated with large deletion mutations. Recently, however, a massive gene inactivation took place and many genes were lost simultaneously. Their sequences have recently started to be enriched in A and T, to become more divergent from the orthologs in other genomes and to accumulate deletions.
The presence of Mle genes and pseudogenes without orthologs in the other mycobacterial genomes suggests that several horizontal transfer events may have occurred in the Mle lineage after divergence from Mtu. A large proportion of these insertion events failed to provide an advantage and quickly led to further pseudogenes (Liu et al. 2004). A minimum of about 200 Mle pseudogenes were inserted after divergence from Mtu and, for that reason, they were not included in the ancestor’s genome.
The cause that started the massive process of gene inactivation, as suggested by several authors, might be a niche change with the adaptation to life in highly specialized cells, such as Schwann’s cells, free from the competitive pressure of other microbes (Young and Robertson 2001; Marri et al. 2006).
The rhythm and types of gene losses that occurred in the early steps of a change of lifestyle is a matter of controversy, as was exemplified by the early studies of comparative genomics of B. aphidicola and other enterobacteria, which proposed two nonmutually exclusive scenarios: multiple events of gene disintegration dispersed through the genome (Silva et al. 2001) or deletions of large sets of contiguous genes (Moran and Mira 2001). Evidence of both types of events has been reported recently. In an interesting genome reduction experimental study with Salmonella enterica, several deletions of up to 202 kb were detected, indicating that extensive genome reduction can occur over a short evolutionary timescale (Nilsson et al. 2005). However, an observation of the position of the nine deletions detected in the experiment showed that none of these large deletions had taken place in a large region of 3 Mb around the origin of replication (1.4 Mb upstream and 1.6 Mb downstream), indicating that this part of the genome, which tends to contain a higher fraction of highly expressed and housekeeping genes, is less tolerant to large deletions (Couturier and Rocha 2006). Other recent comparative genome analyses of insect endosymbiotic bacteria have shown that gene losses mostly occurred through the loss of rather small blocks of genes (Delmotte et al. 2006; Pérez-Brocal et al. 2006).
The chromosomal distribution of the gene losses over the ancestral genome supports the scenario that most of the pseudogenization events took place through a gene-by-gene process. Thus, at least in the Mle lineage, large deletions have not been necessary to reduce the coding capacity of the species drastically. Many lineages of pathogenic bacteria, such as Chlamydiaceae, Spirochetes, and Mollicutes, show very small genomes that presumably resulted from the reduction of larger ones. Our work suggests that this transition can be extremely fast in the evolutionary timescale without requiring singular dramatic events of genome change.
Methods
Genomes
The following Mycobacterium genomes were used in this study: M. avium subsp. paratuberculosis strain K-10 (Mav), M. tuberculosis strain H37Rv [Mtu(I)], M. tuberculosis strain CDC1551 [Mtu(II)], M. bovis strain AF2122/97 (Mbo), and M. leprae TN (Mle) (Cole et al. 1998, 2001; Fleischmann et al. 2002; Garnier et al. 2003; Li et al. 2005). Pseudogenes were retrieved from August-2005 GenBank annotations. Several reannotated M. leprae pseudogenes were obtained from the database Leproma (http://genolist.pasteur.fr/Leproma). Other genomes used to resolve the phylogenetic reconstruction, the orthology, or the ancestral order of some genes were: Mycobacterium marinum (Mma), Nocardia farcinica strain IFM10152 (Nfa), Corynebacterium glutamicum strain ATCC13032 (Cgl), Corynebacterium efficiens YS-314 (Cef), and Corynebacterium diptheriae NCTC13129 (Cdi). The sequence data for Mma were produced by a collaborative project of the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/mm/.
Phylogenetic analysis
A concatenated amino acid sequence alignment of 12 proteins involved in informative processes was used to obtain the phylogenetic tree (AlaS, DnaE, GyrA, IleS, InfB, LeuS, PheT, PolA, TopA, UvrD, ValS, and RpoC). The phylogenetic reconstruction was carried out by maximum likelihood using the PHYML program (Guindon and Gascuel 2003). The model applied was JTT optimizing gamma, and we estimated the proportion of invariant sites. The support of the nodes was estimated with 300 bootstrap pseudosamples.
Reconstruction of the ancestor’s genome
With the aim of reconstructing the last common ancestral genome of Mle and the Mtu complex, we produced a table containing not only the orthologous genes, but also the orthologous pseudogenes in the genomes of Mav, Mle, and Mtu complex. We considered a pseudogene as any remnant of the gene having a detectable sequence similarity. An initial orthologous table for the genomes of Mycobacterium spp. and N. farcinica was extracted from MBGD (Microbial Genome Database, http://mbgd.genome.ad.jp/doc/intro.html). Insertion sequences were removed from the table due to the difficulty of assigning the correct orthology for these elements. The table was ordered following the position of orthologs in the genome of M. tuberculosis. To incorporate orthologous pseudogenes (i.e., elements that are genes in one genome and pseudogenes in another) into the table, we made BLASTX searches for the pseudogenes against the proteins of the remaining Mycobacterium species (BLASTX). Additionally, we used TBLASTN to search for similarities of proteins from each Mycobacterium genome in intergenic DNA sequences of the remaining genomes. We only analyzed TBLASTN hits with an E-value < 0.05 further if the intergenic region had >30 nt.
To avoid the incorporation of false orthologous sequences derived from the presence of large protein-coding gene families, we also used a criterion of conservation of gene order according to which the putative orthologous pseudogenes were only selected when the gene and the pseudogene BLAST hit were flanked by the same orthologous genes in the compared genomes. This criterion incorporates the observation that the probability of finding an ortholog in a nonsyntenic position is low when the genomes are closely related (Rocha 2006). However, this probability increases as the number of contiguous nondetected orthologs in a nonexpected position is larger, as this may reflect large rearrangements. To establish a limit, we calculated the probability of obtaining consecutive genes after the shuffling of a genome of 3000 genes (this number was taken as an approximation of the final number in the ancestral genome). This probability is smaller than 0.05 for three or more genes. Thus, only when three or more contiguous genes showed a positive BLAST hit were they incorporated as orthologs, even though they were not in an expected position. Finally, phylogenetic validation was performed to solve the most complex cases.
We considered that genes or pseudogenes belonging to the orthologous table and present in at least two of the three lineages studied were present in the ancestral genome. To establish the ancestral states for the rare cases of tandem duplications, we used a parsimony criterion taking as the ancestral state the situation corresponding to the majority found in the Mycobacterium species. In the case of genes only present in two of the three Mycobacterium lineages, we used the genome of N. farcinica as an additional reference. In the case of fissions and fusions, we took the same criterion to establish the ancestral state (See Supplemental Table S1 for a list of ancestral genes and Table S2 for the new pseudogenes detected in the analyzed mycobacterial genomes).
Reconstruction of the ancestral genome order
When we found genes in the same order in two of the three lineages, we considered that such a situation corresponded to the ancestral state. This is the most parsimonious scenario. In the regions where each genome had a different order or where there were only genes for two of the three Mycobacterium lineages, we used an additional reference genome (Mma). In these cases, the ancestral order was that shared by Mma and one of these species. To determine the level of rearrangement from the ancestor’s to a present Mycobacterium genome, we ordered the table by ancestor with only the genes and pseudogenes shared with the present genome, and calculated the number of breakpoints. We considered a breakpoint each time that, in the table, two consecutive genes in the ancestor were separated in the present genome. The transcriptional direction of the genes was not considered. Thus, inversions affecting single genes were not taken into account.
Alignment of genes and pseudogenes
We aligned orthologous proteins with Clustal X (Thompson et al. 1997) and then back-translated the alignments to DNA. Pairwise alignments were performed with a total of 1281 Mle ancestral genes having an orthologous gene in Mav and Mtu(II). The Mle pseudogenes were aligned with the corresponding orthologous Mav and Mtu(II) genes [we took this representative of the M. tuberculosis complex composed of Mbo, Mtu(I), and Mtu(II)]. The alignment of pseudogenes is more delicate because insertions and deletions disrupt the reading frame. This was done in three steps: selection of orthologs, preliminary alignment, and exact alignment. Firstly, we retrieved the pseudogenes from the ortholog table that had an ortholog in both Mav and Mtu(II). A total of 714 pseudogenes were in this situation and 1428 pairwise alignments were thus carried out. The alignments were made using an adapted version of the Neddleman-Wusch algorithm (global alignment), where the nonaligned edges of the largest sequence are not penalized using the matrix BLOSUM62 and typical gap penalties (Erickson and Sellers 1983). This second step allowed the elimination of all the sequences that were aligned too poorly. We then made a second alignment with the program GeneWise (Birney et al. 2004) for the 611 pseudogenes with >65% identity in the previous DNA alignment. GeneWise uses a slow but optimal dynamic programming algorithm to align a protein query with a DNA sequence that may be frameshifted or partly deleted. Hence, GeneWise takes indels and frameshifts in the pseudogenes into account and produces a DNA alignment that is guided by the protein sequence. It is therefore much more robust and allows us to infer substitutions reliably.
Estimation of the number of substitutions per site
The numbers of synonymous (dS) and nonsynonymous substitutions per site (dN) for 1281 ancestral genes were estimated for each possible pair comparison: Mav-Mle, Mav-Mtu(II), and Mle-Mtu(II). Both numbers were computed using yn00 from PAML (Yang and Nielsen 2000). The numbers of substitutions in 16S rRNA genes between pairs of species were estimated with the method of Tamura-Nei implemented in MEGA (Kumar et al. 2004).
Estimation of the amount of DNA loss
To determine the proportion of nucleotides lost after gene inactivation, we computed the quotient between the length of the disintegrated DNA region after the reductive process (in Mle) and the number of nucleotides included between the upstream and downstream adjacent genes before the inactivation (taking Mtu complex and Mav as a reference) (see Supplemental text, section 5). When several adjacent genes were simultaneously lost in a lineage, we treated them as a block and estimated the quotient for the block. For a graphical representation (Fig. 6), we assigned the value obtained for a block to each of the genes that composed it. To avoid the problems associated with horizontal gene transfer and genome rearrangements, this analysis was performed with a sample of regions including pseudogenes or absent genes that were flanked by the same genes in Mle and the compared genome (Mtu or Mav).
Acknowledgments
Financial support was provided by projects BFU2006-06003/BMC from Ministerio de Educación y Ciencia and Grupos03/204 from the Generalitat Valenciana, Spain. L.G.V. was funded by a pre-doctoral fellowship from the Generalitat Valenciana (Spain). We thank José Bermúdez for his help with data analysis.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6360207
References
- Andersson S.G.E., Kurland C.G., Kurland C.G. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Belda E., Moya A., Silva F.J., Moya A., Silva F.J., Silva F.J. Genome rearrangement distances and gene order phylogeny in gamma-proteobacteria. Mol. Biol. Evol. 2005;22:1456–1467. doi: 10.1093/molbev/msi134. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: Evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. (Lond.) 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Eiglmeier K., Gas S., Barry C.E., Gas S., Barry C.E., Barry C.E., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Honore N., Garnier T., Churcher C., Harris D., Garnier T., Churcher C., Harris D., Churcher C., Harris D., Harris D., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Couturier E., Rocha E.P.C., Rocha E.P.C. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol. Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- Dagan T., Blekhman R., Graur D., Blekhman R., Graur D., Graur D. The “domino theory” of gene death: Gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol. Biol. Evol. 2006;23:310–316. doi: 10.1093/molbev/msj036. [DOI] [PubMed] [Google Scholar]
- Degnan P.H., Lazarus A.B., Wernegreen J.J., Lazarus A.B., Wernegreen J.J., Wernegreen J.J. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte F., Rispe C., Schaber J., Silva F.J., Moya A., Rispe C., Schaber J., Silva F.J., Moya A., Schaber J., Silva F.J., Moya A., Silva F.J., Moya A., Moya A. Tempo and mode of early gene loss in endosymbiotic bacteria from insects. BMC Evol. Biol. 2006;6:56. doi: 10.1186/1471-2148-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson B.W., Sellers P.H., Sellers P.H. Recognition of patterns in genetic sequences. In: Sankoff D., Kruskal J.B., Kruskal J.B., editors. Time warps, string edits, and macromolecules: The theory and practice of sequence comparison. Addison-Wesley; Boston, MA: 1983. pp. 55–91. [Google Scholar]
- Fleischmann R.D., Alland D., Eisen J.A., Carpenter L., White O., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., Alland D., Eisen J.A., Carpenter L., White O., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., Eisen J.A., Carpenter L., White O., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., Carpenter L., White O., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., White O., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., Peterson J., Deboy R., Dodson R., Gwinn M., Haft D., Deboy R., Dodson R., Gwinn M., Haft D., Dodson R., Gwinn M., Haft D., Gwinn M., Haft D., Haft D., et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Eiglmeier K., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Eiglmeier K., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Duthoy S., Grondin S., Lacroix C., Monsempe C., Grondin S., Lacroix C., Monsempe C., Lacroix C., Monsempe C., Monsempe C., et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R., Silva F.J., Zientz E., Delmotte F., González-Candelas F., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Silva F.J., Zientz E., Delmotte F., González-Candelas F., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Zientz E., Delmotte F., González-Candelas F., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Delmotte F., González-Candelas F., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., González-Candelas F., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Latorre A., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Rausell C., Kamerbeek J., Gadau J., Holldobler B., Kamerbeek J., Gadau J., Holldobler B., Gadau J., Holldobler B., Holldobler B., et al. The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Valero L., Latorre A., Silva F.J., Latorre A., Silva F.J., Silva F.J. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 2004;21:2172–2181. doi: 10.1093/molbev/msh232. [DOI] [PubMed] [Google Scholar]
- Graur D., Shuali Y., Li W.H., Shuali Y., Li W.H., Li W.H. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J. Mol. Evol. 1989;28:279–285. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M., Tamura K., Nei M., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li L.L., Bannantine J.P., Zhang Q., Amonsin A., May B.J., Alt D., Banerji N., Kanjilal S., Kapur V., Bannantine J.P., Zhang Q., Amonsin A., May B.J., Alt D., Banerji N., Kanjilal S., Kapur V., Zhang Q., Amonsin A., May B.J., Alt D., Banerji N., Kanjilal S., Kapur V., Amonsin A., May B.J., Alt D., Banerji N., Kanjilal S., Kapur V., May B.J., Alt D., Banerji N., Kanjilal S., Kapur V., Alt D., Banerji N., Kanjilal S., Kapur V., Banerji N., Kanjilal S., Kapur V., Kanjilal S., Kapur V., Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Harrison P.M., Kunin V., Gerstein M., Harrison P.M., Kunin V., Gerstein M., Kunin V., Gerstein M., Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: Widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan Babu M. Did the loss of sigma factors initiate pseudogene accumulation in M. leprae? Trends Microbiol. 2003;11:59–61. doi: 10.1016/s0966-842x(02)00031-8. [DOI] [PubMed] [Google Scholar]
- Marri P.R., Bannantine J.P., Golding G.B., Bannantine J.P., Golding G.B., Golding G.B. Comparative genomics of metabolic pathways in Mycobacterium species: Gene duplication, gene decay and lateral gene transfer. FEMS Microbiol. Rev. 2006;30:906–925. doi: 10.1111/j.1574-6976.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- Mira A., Moran N.A., Moran N.A. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 2002;44:137–143. doi: 10.1007/s00248-002-0012-9. [DOI] [PubMed] [Google Scholar]
- Mira A., Ochman H., Ochman H. Gene location and bacterial sequence divergence. Mol. Biol. Evol. 2002;19:1350–1358. doi: 10.1093/oxfordjournals.molbev.a004196. [DOI] [PubMed] [Google Scholar]
- Mira A., Ochman H., Moran N.A., Ochman H., Moran N.A., Moran N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moran N.A., Mira A., Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:R54. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A.I., Koskiniemi S., Eriksson S., Kugelberg E., Hinton J.C.D., Andersson D.I., Koskiniemi S., Eriksson S., Kugelberg E., Hinton J.C.D., Andersson D.I., Eriksson S., Kugelberg E., Hinton J.C.D., Andersson D.I., Kugelberg E., Hinton J.C.D., Andersson D.I., Hinton J.C.D., Andersson D.I., Andersson D.I. Bacterial genome size reduction by experimental evolution. Proc. Natl. Acad. Sci. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Wilson A.C., Wilson A.C. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Murphy L.D., Thomson N., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Thomson N., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Harris D.E., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Holden M.T.G., Churcher C.M., Bentley S.D., Mungall K.L., Churcher C.M., Bentley S.D., Mungall K.L., Bentley S.D., Mungall K.L., Mungall K.L., et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V., Gil R., Ramos S., Lamelas A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A., Gil R., Ramos S., Lamelas A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A., Ramos S., Lamelas A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A., Lamelas A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A., Michelena J.M., Silva F.J., Moya A., Latorre A., Silva F.J., Moya A., Latorre A., Moya A., Latorre A., Latorre A. A small microbial genome: The end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Rocha E.P. Inference and analysis of the relative stability of bacterial chromosomes. Mol. Biol. Evol. 2006;23:513–522. doi: 10.1093/molbev/msj052. [DOI] [PubMed] [Google Scholar]
- Silva F.J., Latorre A., Moya A., Latorre A., Moya A., Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- Silva F.J., Latorre A., Moya A., Latorre A., Moya A., Moya A. Why are the genomes of endosymbiotic bacteria so stable? Trends Genet. 2003;19:176–180. doi: 10.1016/S0168-9525(03)00041-6. [DOI] [PubMed] [Google Scholar]
- Silva F.J., Latorre A., Gómez-Valero L., Moya A., Latorre A., Gómez-Valero L., Moya A., Gómez-Valero L., Moya A., Moya A. Genomic changes in bacteria: from free-living to endosymbiotic life. In: Bastolla U., et al., editors. Structural approaches to sequence evolution: Molecules, networks, populations. Springer; Berlin, Germany: 2007. pp. 151–167. [Google Scholar]
- Tamas I., Klasson L., Canback B., Naslund A.K., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Klasson L., Canback B., Naslund A.K., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Canback B., Naslund A.K., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Naslund A.K., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G.E., Sandstrom J.P., Moran N.A., Andersson S.G.E., Moran N.A., Andersson S.G.E., Andersson S.G.E. 50 Million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen J.J. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- Yang Z.H., Nielsen R., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Young D., Robertson B., Robertson B. Genomics: Leprosy—A degenerative disease of the genome. Curr. Biol. 2001;11:R381–R383. doi: 10.1016/s0960-9822(01)00213-5. [DOI] [PubMed] [Google Scholar]