Abstract
Temporal and tissue-specific alterations in gene expression have profound effects on aging of multicellular organisms. However, much remains unknown about the patterns of molecular changes in different tissues and how different tissues interact with each other during aging. Previous genomic studies on invertebrate aging mostly utilized the whole body or body parts and limited age-points, and failed to address tissue-specific aging. Here we measured genome-wide expression profiles of aging in Drosophila melanogaster for seven tissues representing nervous, muscular, digestive, renal, reproductive, and storage systems at six adult ages. In each tissue, we identified hundreds of age-related genes exhibiting significant changes of transcript levels with age. The age-related genes showed clear tissue-specific patterns: <10% of them in each tissue were in common with any other tissue; <20% of the biological processes enriched with the age-related genes were in common between any two tissues. A significant portion of the age-related genes were those involved in physiological functions regulated by the corresponding tissue. Nevertheless, we identified some overlaps of the age-related functional groups among tissues, suggesting certain common molecular mechanisms that regulate aging in different tissues. This study is one of the first that defined global, temporal, and spatial changes associated with aging from multiple tissues at multiple ages, showing that different tissues age in different patterns in an organism. The spatial and temporal transcriptome data presented in this study provide a basis and a valuable resource for further genetic and genomic investigation of tissue-specific regulation of aging.
For multicellular organisms, different tissues or organs perform distinct but coordinated physiological functions to ensure proper function of the whole organism. Virtually all physiological functions of tissues or organs in a multicellular organism decline with increasing age (Timiras 2003). Despite the apparent complexity of aging, a number of genetic and environmental factors have been identified to play an important role in aging in organisms ranging from the yeast, Saccharomyces cerevisiae, the nematode, Caenorhabitis elegans, the fly, Drosophila melanogaster, to the mice and rats (Kenyon 2005). The lifespan-related genetic factors fall into a number of biological pathways, including the evolutionarily conserved insulin/insulin-like growth factor signaling (IIS) pathway, the Jun kinase (JNK) pathway, and the sirtuin pathway. Lifespan is modulated not only at the molecular level but also at the tissue level in a temporal manner. For instance, reducing expression of genes in the IIS pathway, such as the insulin-like receptor daf-2, in the adult stage alone extends lifespan in C. elegans (Dillin et al. 2002). Modifications of the IIS pathway by overexpression of the forkhead transcription factor foxo in the adult adipose tissue alone prolong lifespan in D. melanogaster (Giannakou et al. 2004); Hwangbo et al. 2004). These findings suggest that tissue-specific alterations of gene expression in a temporal fashion can dramatically influence aging of the whole organism.
The multifactorial and temporal features of aging can be analyzed efficiently by genome-wide transcriptional profiling, which has been conducted in various model organisms and humans (Melov and Hubbard 2004). Aging is associated with alterations in transcript levels of many genes, including those involved in evolutionarily conserved mitochondrial and proteasomal functions (McCarroll et al. 2004), some of which have been shown to be directly involved in regulating lifespan in C. elegans (Dillin et al. 2002). However, little is known about expression patterns of aging across different tissues or organs and how they interact with each other during aging.
In all the previous genome-wide transcriptional studies on aging in invertebrates, including C. elegans and D. melanogaster, gene expression was monitored with the whole body or body parts, such as fly head and thorax (Zou et al. 2000; Lund et al. 2002; Pletcher et al. 2002; Landis et al. 2004; Wang et al. 2004; Kim et al. 2005; Girardot et al. 2006). Many of these studies sampled only two age-points, e.g., young and old. Although a number of age-related genes with diverse functions were identified, these previous studies missed a significant part of spatial and/or temporal expression profiles of aging. Since individual tissues were not examined separately, these studies offered little opportunities for comparative analyses among different tissues and with mammalian data, which largely focused on individual tissues. Even in mammals, only a few studies compared expression profiles of aging among different tissues (Fraser et al. 2005; Zahn et al. 2006). Therefore, it is important to build up a resource with temporal and spatial expression profiles of aging from multiple tissues at multiple ages from one organism, especially a genetically tractable model system to facilitate further genetic studies. Here we made a first step toward systematic investigation of tissue-specific aspects of aging by measuring genome-wide changes of gene expression in seven individual tissues or organs at six adult ages in D. melanogaster. This study provides a valuable resource for aging research and particularly for cross-species comparison with mammals to identify shared or public aging signatures.
Results
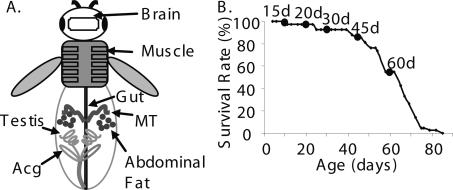
In this study, we analyzed gene expression profiles of aging for seven fly tissues or organs, collectively referred as tissues. These tissues represented various physiological systems in flies: brain representing the nervous system, thoracic muscle representing the muscular system, gut representing the digestive system, malpighian tubule (MT) equivalent to the mammalian kidney representing the renal system, accessory gland (Acg) equivalent to the mammalian prostate and testis representing the reproductive system, and, finally, abdominal adipose tissue (i.e., fat) representing the storage and secretory system (Fig. 1A) (Demerec 1994). In our design of microarray experiments, the RNA sample for each array came from four individual flies, which allowed efficient cover of genetic variations and minimized the masking of intra-individual variability associated with the stochastic nature of aging (Golden and Melov 2004). As shown in Figure 1B, each type of tissue was examined by measuring temporal changes of gene expression for w1118 adult males at five ages, 15-, 20-, 30-, 45-, and 60-d-old flies, compared with 3-d-old flies. The survival rates of males at these age-points ranged from 100% to ∼50% based on the observation that the mean lifespan of w1118 males was ∼60 d under the culture conditions used in this study (Fig. 1B).
Figure 1.
Fly tissues for microarray experiments. (A) RNA samples for microarray experiments were prepared from seven tissues shown in the fly image. (B) Lifespan curve of w1118. The mean lifespan of w1118 at 25°C was 59.2 ± 2.3 d. The age-points labeled on the curve represent ages of males when tissues were collected for microarray experiments.
Age-related transcriptional and functional signatures
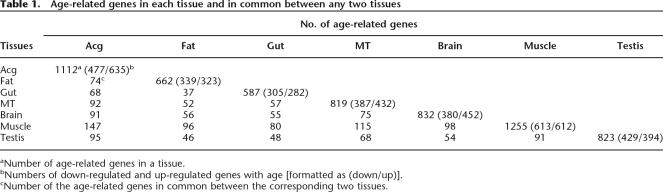
We first identified age-related genes as those showing significant changes with age (P < 0.05). The identification was made using the Extraction of Differential Gene Expression (EDGE) algorithm, which is suitable for analysis of time-series array data (Storey et al. 2005). The muscle had the largest number of age-related genes (1255 genes), followed by Acg (1112), brain (832), testis (823), MT (819), and fat (662), while the gut had the smallest number (587) (Table 1; Supplemental Tables 1–7). The age-related genes represented 4%–9% of all the genes (∼14,000) recognized from the Drosophila genome (Adams et al. 2000), suggesting that a significant portion of genes in the genome change their expression with age in different tissues, especially when considering that not all the genes are expressed in every tissue.
Table 1.
Age-related genes in each tissue and in common between any two tissues
aNumber of age-related genes in a tissue.
bNumbers of down-regulated and up-regulated genes with age [formatted as (down/up)].
cNumber of the age-related genes in common between the corresponding two tissues.
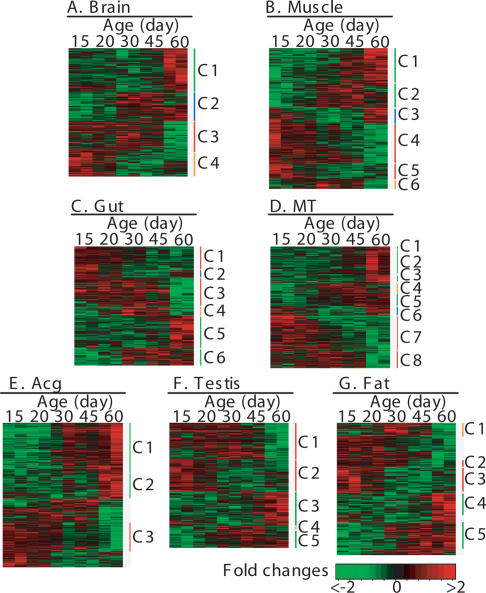
Next, we examined the temporal patterns of tissue-specific gene expression by conducting unsupervised hierarchical clustering analysis on the age-related genes. In each tissue, the age-related genes formed three to eight clusters that showed a gradual or transient increase or decrease of transcript levels from young to old ages (Fig. 2; Supplemental Tables 8–14). In general, approximately half of these genes were up-regulated and the other half were down-regulated with age in each tissue (Table 1). The critical changes of the transcriptional levels generally occurred after 30 d of age, some as late as 60 d, suggesting that alteration of gene expression in aging is a gradual process.
Figure 2.
Heat map presentation of expression patterns of the age-related genes in each tissue. (A) Brain; (B) muscle; (C) gut; (D) MT; (E) Acg; (F) testis; (G) adipose tissue. Each column represents an age-point. Each row represents the expression pattern of one gene across all the age-points. The ratios of transcript levels between experiment and reference samples are color-coded in red and green. Red represents an increase of the transcript level of a gene in the experiment samples aged 15–60 d compared with the 3-d-old reference sample, and green represents a decrease. The age-related genes in each tissue are grouped to three to eight clusters (labeled as C1 to C8) by hierarchical clustering analysis based on their similarities in expressional profiles.
We then conducted functional analysis for each cluster of the age-related genes based on Gene Ontology (GO) (Ashburner et al. 2000). We found that >500 biological processes were altered in aging in at least one tissue (P < 0.05) (Supplemental Table 17). A significant part of these biological processes correlated with functional or physiological roles specific to the corresponding tissue where the alternations were identified. The age-related genes and GO terms provided a basis to identify molecular signatures of aging in each tissue as follows.
Signatures of aging in brain
In brain or the nervous system, four distinctive clusters of age-related genes were identified (Fig. 2A; Supplemental Table 8). Aging in brain was associated with up-regulation of genes (Clusters 1 and 2) involved in neurotransmitter secretion and ubiquitin-dependent protein catabolism as represented by Plenty of SH3s (POSH). POSH was previously shown to extend lifespan of flies by 14% when it was overexpressed specifically in neurons (Seong et al. 2001). Aging in brain was also associated with down-regulation of genes (Clusters 3 and 4) participating in microtubule-based processes, immune functions, oxidative stress response, and tricarboxylic acid cycle (TCA). The results indicate that neuronal functions related to neurotransmitter release, protein degradation, and energy production are preferentially affected during aging of brain in D. melanogaster.
Signatures of aging in muscle
For the muscular system, six clusters of age-related genes with significant enrichment of functional annotation were identified (Fig. 2B; Supplemental Table 9). Aging in muscle was associated with an increase of transcript levels of genes (Clusters 1, 2, and 3) involved in a number of biological processes, including antimicrobial humoral response, ubiquitin-dependent protein catabolism, autophagic cell death, prosthetic group metabolism, protein membrane targeting, secretion pathway, transmembrane receptor protein tyrosine kinase signaling pathway, cell motility, and response to toxin as represented by glutathione S transferase. On the other hand, aging in muscle was found to be associated with decreased transcript levels of genes (Clusters 4–6) involved in generation of energy derived by oxidation of organic compounds as represented by succinate dehydrogenase B (SdhB), in oxidative phosphorylation as represented by ATPase coupling factor 6, in protein kinase cascade as represented by Jun-related antigen, and in metal ion transport as represented by ferritin 1 heavy chain homolog and I’m not dead yet (Indy). It has been shown that SdhB, ATP synthase, ferritin, and aconitase in C. elegans (Hamilton et al. 2005; Hansen et al. 2005) and Indy and SdhB in D. melanogaster (Rogina et al. 2000; Walker et al. 2006) modulate lifespan in these organisms, respectively. Overall, these findings suggest that a prominent feature of aging in muscle is the alteration of expression of genes involved in proteasomal and mitochondrial functions.
Signatures of aging in gut
For gut or the digestive system, six clusters of age-associated genes had significant enrichment of functional annotations (Fig. 2C; Supplemental Table 10). Aging in gut was found to be associated with down-regulation of genes (Clusters 1, 2, 3, and 4) participating in oxidative phosphorylation, aromatic compound metabolism, muscle contraction, amino sugar metabolism, regulation of apoptosis, and vesicle transport. Aging was also associated with up-regulation of genes (Clusters 5 and 6) involved in regulating various physiological processes, amino acid metabolism, and regulation of transport. These results suggest that metabolic pathways, especially nutrient intake and energy production, are primarily affected during aging of gut, which are the fundamental function of the digestive system.
Signatures of aging in malpighian tubule
For MT or the renal system, eight clusters of age-related genes had significantly enriched functional annotations (Fig. 2D; Supplemental Table 11). Aging in MT was associated with up-regulation of genes (Clusters 1–6) involved in anion transport, metal ion homeostasis, tRNA metabolism, chromosome organization and biogenesis, and amino acid metabolism. In contrast, aging was associated with down-regulation of genes (Cluster 7) involved in aromatic compound metabolism, vesicle exocytosis, and polysaccharide metabolism. This suggests that nutrient transport and excretion are prominently altered during aging of MT, which are related to kidney-like functions of MT.
Signatures of aging in Acg and testis
The reproductive system was represented by Acg and testis in this study. In Acg, three clusters of age-related genes showed significant enrichment of functional categories (Fig. 2E; Supplemental Table 12). The Acg-related genes (Clusters 1 and 2) up-regulated in aged flies were those involved in cytoskeleton organization and biogenesis, cell homeostasis, protein targeting, stress-activated protein kinase signaling pathway, oxygen and reactive oxygen species metabolism, cytoskeleton-dependent intracellular transport, and anti-microbial humoral. On the other hand, the Acg-specific genes (Cluster 3) down-regulated in older flies included those involved in protein targeting and glycoprotein biogenesis. These patterns suggest that cytoskeleton structure, nutrient transport, and stress resistance are significantly changed during aging of Acg.
In testis, five clusters of age-related genes had significantly enriched functional terms (Fig. 2F; Supplemental Table 13). Aging in testis was associated with down-regulation of genes (Clusters 1 and 2) participating in regulation of catabolism and aromatic compound metabolism. Aging in testis was also associated with up-regulation of genes (Clusters 3, 4, and 5) involved in cellular morphogenesis, mRNA metabolism, protein folding, and tRNA aminoacylation. These findings suggest that metabolism and cell cycle–related processes are prominently altered during aging of testis, which is correlated well with a decrease of reproduction during aging.
Signatures of aging in adipose tissue
In fat or adipose tissue, five clusters showed significant enrichment of functional annotation (Fig. 2G; Supplemental Table 14). Aging in fat was associated with down-regulation of genes (Clusters 1, 2, and 3) involved in JNK cascade, glucose catabolism, ubiquitin-dependent protein catabolism, and regulation of signal transduction. Aging in fat was also associated with up-regulation of genes (Clusters 4 and 5) involved in humoral immune response, second messenger mediated signaling, and coenzyme metabolism. Overall, these patterns suggest that protein degradation, immune response, and energy metabolism pathways are prominently altered in aging of adipose tissue.
Comparison of transcriptomes of aging among different tissues
Differences and similarities of transcriptional profiles among tissues
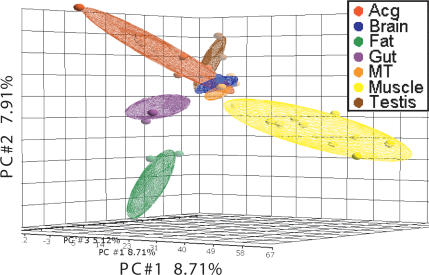
To investigate the relatedness of transcriptional profiles of aging among different tissues, we first conducted principle component analysis (PCA) on age-related genes along the time course (age-points) in all the tissues. PCA, as an unsupervised dimension reduction method, is robust in capturing and presenting the major variations of expression profiles. The distance between data-points shown on the PCA maps visually reflects the relatedness of the tissue samples. As shown in the PCA map (Fig. 3) and from the pairwise correlation coefficients of expression profiles between any two tissues (Supplemental Table 15), Acg, testis, brain, and MT were relatively close to each other, while gut, fat, and muscle were distant from others and formed three distinct clusters. The PCA based on all the expressed genes showed similar results to that based on the age-related genes (Supplemental Fig. 3). These results suggest that different tissues in general have distinct age-related transcriptomes.
Figure 3.
PCA maps were generated with the age-related genes showing relationships of transcriptional profiles of aging among different tissues. The amount of variation covered by the top three principle components are as follows: (#1) 8.71%; (#2) 7.91%; (#3) 5.12%. The samples of seven tissues are color-coded. Each small sphere represents projection of one age sample from one tissue in the three-dimensional space formed by the top three principle components. Each large oval represents the area covered by all the samples of one tissue formed by the small spheres.
Age-related genes in common among tissues
To further examine differences and similarities of age-related changes among tissues, we sought to identify the age-related genes that were shared by two or more tissues (Table 1). We found that muscle and Acg had the largest number of age-related genes in common (147 genes), while gut and fat tissue had the smallest common age-related genes (37 genes). Overall, only 3%–10% of age-related genes in any given tissue overlapped with those in any other tissue. Only 16 genes were found in common in four or more tissues (Supplemental Table 16). We confirmed the expression patterns for three of these 16 genes with quantitative RT-PCR (Supplemental Fig. 4). Overall, this suggests that aging is associated with a high degree of tissue-specific regulation of gene expression.
Age-related biological processes in common among tissues
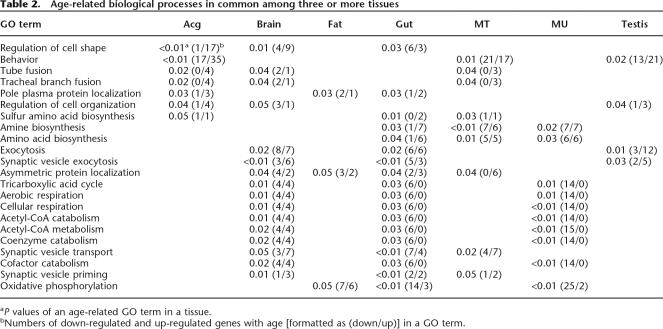
Next we compared age-related GO terms identified in each tissue. Among 531 age-related biological processes in all tissues (P < 0.05), none of them were shared among four or more tissues, only 22 were in common among three tissues (Table 2), and 103 were in common among at least two tissues (Supplemental Table 17).
Table 2.
Age-related biological processes in common among three or more tissues
aP values of an age-related GO term in a tissue.
bNumbers of down-regulated and up-regulated genes with age [formatted as (down/up)] in a GO term.
Brain shared its age-related GO terms mostly with Acg and gut but none with adipose tissue. Most GO terms shared between brain and Acg belonged to categories of morphogenesis, iron and protein transport, and immune response. Most GO terms shared between brain and gut contained genes participating in aerobic respiration, coenzyme metabolism, and exocytosis.
Muscle shared its significant GO terms relatively evenly with other tissues except testis. Muscle shared the aerobic respiration and coenzyme catabolism categories with brain; the ion homeostasis and immune response categories with Acg; stress response and protein degradation categories with fat; amine biogenesis and amino acid biogenesis categories with MT; and amine biogenesis, amino acid biogenesis, coenzyme catabolism, ATP synthesis, and muscle contraction categories with gut. These findings indicate that changes in expression of genes involved in metabolism are the prominent feature shared between muscle and other tissue.
Gut and MT shared seven GO terms. Most of these GO terms were related to amine biosynthesis, amino acid biosynthesis, and vesicle transport, which are part of nutrient absorption and process functions in the digestive and renal systems. Acg and testis, representing the reproductive system, shared 11 GO terms, nine of which were not found in any other tissues. Genes in these terms were mostly involved in germ line biogenesis, intercellular bridge organization and biogenesis, and regulation of actin filament formation. This suggests that expression of genes involved in cell cycle and cytoskeleton structure is coordinately altered in different tissues of the reproductive system.
Comparison of temporal expression profiles
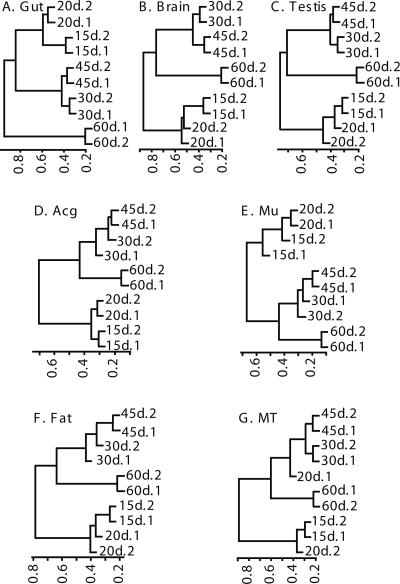
As indicated above, most of the age-related genes displayed gradual alterations of expression from young to old ages. To further examine the temporal patterns, we performed hierarchical cluster analysis and PCA for the time course data in each tissue. The results are shown in Figure 4. Three distinct aging clusters were visible in each tissue in most cases: The first was formed by day 15 and 20 data, the second by day 30 and 45 data, and the third by day 60 data. However, the relatedness among the three clusters was different in different tissues as indicated by the distances between clusters. The farther the distance is between clusters, the lesser correlation or the more difference the clusters have. In gut, the second cluster with day 30 and 45 data was farthest away from the third cluster with day 60 data (distance >0.8, Fig. 4A). This was followed by brain and testis, in which the second cluster, to a lesser degree, was also far from the third cluster (∼0.8, Fig. 4B,C). On the other hand, in Acg and muscle, the second cluster was close to the third cluster (∼0.4, Fig. 4D,E). In fat or MT, the distances between the second and third clusters were modest in between (∼0.6, Fig. 4F,G). Similar temporal patterns of age-related transcriptomes in individual tissues were observed by the PCA (Supplemental Fig. 2). This finding indicates that the expression profiles at the middle ages (30 and 45 d) were more similar to those at the old age (60 d) in Acg and muscle and less similar in brain and gut. The results suggest that different tissues have different temporal patterns of transcriptomes during aging, physiological ages are not the same among different tissues of the same chronological ages, and diverse molecular mechanisms regulate aging in different tissues. Zheng et al. (2005) found that muscle cells showed a gradual increase of apoptosis with age, but brain-containing heads showed no or little signs of apoptosis. The temporal and spatial expression profiles of aging revealed in our study are consistent with the age-related patterns of apoptosis and provide possible molecular mechanisms for tissue-specific regulation of aging processes.
Figure 4.
Hierarchical clustering analysis of temporal transcriptional profiles of individual tissues using the age-related genes. In each tissue, the difference of transcriptomic data at different ages is reflected by the distance scale bar shown at the bottom of each cluster dendrogram. Each branch represents one age-point labeled in the order of age, tissue type, and data set identification. (A) Gut; (B) brain; (C) testis; (D) Acg; (E) muscle; (F) fat; (G) MT.
Discussion
In this study, we measured genome-wide expression profiles of aging in seven tissues representing the nervous, muscular, digestive, renal, reproductive, and storage systems at six adult ages in D. melanogaster. Hundreds of the age-related genes were identified in each tissue, most of which displayed a gradual increase or decrease of transcript levels with age, which is consistent with the gradual decline of physiological functions in aging. In each tissue, a significant portion of the age-related genes was composed of those involved in physiological functions specific to the corresponding tissue. We found that a number of biological processes were prominently affected during aging in a tissue-specific fashion, which included neuronal function, protein degradation, and energy production in brain; proteasome and mitochondrial function in muscle; nutrient intake and energy production in gut; nutrient transport and excretion in MT; cytoskeleton structure, nutrient transport, and stress resistance in Acg; metabolism and cell cycle–related processes in testis; and protein degradation, immune response, and energy metabolism in adipose tissue. The multi-tissue data allowed us to examine differences and similarities of age-related changes among tissues. We found that <10% of the age-related genes were common between different tissues, and <20% of the age-related biological processes were in common among no more than three tissues. Our results suggest that the age-related genes are generally regulated in a tissue-specific manner, and different tissues have different temporal patterns of transcriptomes in aging.
So far, we presented a comprehensive transcriptomic study on aging of invertebrates, which revealed spatial and temporal expression patterns of aging from multiple tissues at multiple ages. Previous genomic studies of aging used the whole body or body parts in invertebrates, including D. melanogaster (Golden and Melov 2004), and some of these studies only compared transcript levels of two ages, young and old, which missed valuable information regarding temporal and/or spatial expression patterns of aging. Girardot et al. (2006) attempted to investigate spatial expression profiles of aging by comparing age-related changes among the whole body and two body parts, head and thorax, in D. melanogaster between two age-points, 3 and 40 d. They identified more than 1000 age-related genes in each body part but found that only a small percentage (<7%) of the age-related genes were shared among the whole body and two body parts, and ∼18% of the age-related genes were common between head and thorax, despite the fact that these body parts share a number of similar tissue types, such as fat cells and muscles. The relative homogenous tissues used in our study offered an improved resolution for tissue comparison and showed much less overlaps of age-related genes (<10%) between any two tissues.
In mammals, including humans, although gene expression profiles of aging were normally measured with individual types of tissues, only a few studies compared age-related expression patterns among tissues. Fraser et al. (2005) compared age-related changes among different human and chimpanzee brain regions, including cerebellum, caudate nucleus, and several regions of cortex. Different regions of cortex displayed remarkable similarity in expression profiles of aging among each other. Cortex, however, showed dramatically different expression patterns from other brain regions, cerebellum, and caudate nucleus. This suggests that aging patterns are generally region specific in primate brains. This study, however, did not evaluate whether there are age-related genes or pathways in common among brain regions. In another study, Zahn et al. (2006) compared aging patterns of three different tissues, human muscle, kidney, and brain. Direct comparison of the age-related genes did not reveal any significant overlaps among these tissues. By analyzing 624 gene sets, Zahn and colleagues found that only six age-related gene sets were shared among the three human tissues. Our study in D. melanogaster supported and expanded these findings. We found very limited overlaps of the age-related genes and little overlaps of the age-related pathways among tissues. We also found that individual genes in the common age-related biological processes appeared to have different expression patterns during aging (Table 2). For example, all the age-related TCA genes were down-regulated in gut and muscle, while half of the age-related TCA genes were up-regulated and the other half were down-regulated in brain. These observations suggest that shared age-related regulation is mostly operated at the level of pathways and is not at the level of individual genes. The tissue-specific expression profiles of aging also stress the importance of using relatively homogenous tissues for gene expression studies.
A central question in aging research is what the public signatures of aging are among different species. Several studies addressed this question by comparing expression profiles between evolutionarily divergent species. One study compared C. elegans and D. melanogaster and found that alterations of mitochondrial energy production and proteasome pathways were conserved aging patterns (McCarroll et al. 2004). The other study compared human, mouse, and fly and found that the mitochondrial electron transfer chain pathway was shared among these three species (Zahn et al. 2006). The low commonalities observed in these studies may be due to the fact that the data from the whole body of flies and worms were used in the aging studies. The tissue-specific data in our study should offer a better resolution to identify public aging patterns between invertebrates and mammals by comparing tissues of similar types. The data generated by our study are valuable for future research on genetic and nongenetic interventions of aging and for a broad range of studies in systems biology in large.
Methods
Fly culture and RNA sample preparation
The fly strain w1118 was cultured on standard Caltech corn-meal fly food at 25°C (Ashburner 1989). Adult males were collected within 24 h after eclosion and maintained in vials with 20 flies per vial, transferred to fresh food once every 2–3 d. For the lifespan assay, the number of dead flies was recorded at the time of transfer. For microarray experiments, seven types of tissues—Acg, testis, brain, gut, MT, dorsal thoracic muscle, and abdominal fat body—were hand-dissected out of flies at ages of 3, 15, 20, 30, 45, and 60 d in the Ringer’s buffer under a dissection microscope (Ashburner 1989). Each tissue was carefully examined for morphology and rinsed twice with the Ringer’s buffer to minimize contamination of fat cells after hand-removing the fat cells attached to the nonfat tissues. To minimize the effect of circadian rhythm on gene expression and ensure consistence, the dissection was always conducted in the morning by one researcher (S. Zou). The oldest age (60 d) is comparable to the oldest age (61 d) for normal aging previously published in D. melanogaster (Landis et al. 2004). Individual tissues from four males of the same age were pooled together and used for each RNA sample preparation. Total RNA was extracted using miniRNA preparation kit from Zymo Research Inc. RNA was then amplified by a one-step linear amplification protocol to generate amplified RNA (aRNA) as described (Klebes et al. 2002). Quality and quantity of RNA was assessed using Agilent Bioanalyzer from Agilent Technologies Inc.
Microarray experiments
A library of PCR amplicons, each with a 100- to 600-bp-long fragment representing 14,151 predicted or known genes in D. melanogaster, was kindly provided by Dr. Thomas Kornberg (University of California San Francisco) and used for constructing DNA microarrays as described (Klebes et al. 2002). PCR products were spotted on three-dimensional slides purchased from Full Moon Biosystems Inc. Experimental aRNA refers to amplified RNA from flies that were 15, 20, 30, 45, and 60 d old; reference aRNA, flies that were 3 d old using a modified one-round linear amplification protocol as described (Klebes et al. 2002). Two to four micrograms of experimental and reference aRNAs were used to generate cDNA for labeling with fluorescent dye Cy3 and Cy5, respectively, and purified using the PowerScript labeling kit from Clontech according to the protocol suggested by the manufacturer (Clontech). Concentrations of labeled cDNA and labeling efficiency were measured using Nanodrop ND-1000 (Nanodrop Technologies). Equal amount of labeled experimental and reference samples (1 or 2 μg) were then cohybridized to microarrays at 42°C using the cDNA Hyb Buffer from Full Moon Biosystems Inc. Hybridized slides were washed and then scanned with an Axon GenePix Scanner (Axon Instruments). For each tissue, RNA from the corresponding tissue of 3-d-old flies was used as the reference RNA, and expression profiles at each of the five age-points were measured twice by using two independently prepared RNA samples. Therefore, there are two biologically independent data-points for each age-point and a total of 10 time course samples for each tissue. In total, 70 sets of microarray data were collected for data analysis.
Microarray data analysis
Raw microarray data were first processed by conducting within-slide normalization of signal intensities using the LOWESS curve-fitting method followed by between-slide scaling using Median Absolute Deviation method using Bioconductor (Supplemental Fig. 1; Dudoit et al. 2003;Gentleman et al. 2004). Each microarray contained 17,664 probes. Only those with at most four missing values in the 10 time course samples for each tissue were included for further analysis. After filtering the low-quality data, there remained 15,263 probes for Acg, 14,292 for brain, 11,962 for adipose tissue, 10,950 for gut, 15,197 for MT, 14,205 for muscle, and 15,541 for testis. Remaining missing values were imputed using k-nearest neighbor algorithm (Troyanskaya et al. 2001). Age-related genes were identified by the EDGE algorithm with a P cutoff value of 0.05 (Storey et al. 2005). PCA was conducted using Partek (Partek Inc.), and hierarchical clustering was performed using the Euclidean distance and displayed using the Cluster and TreeView software (Eisen et al. 1998). Functional annotations and Fisher exact test for clustered genes were carried out using GoMiner, and age-related GO terms were defined as those with a P cutoff value of 0.05 (Zeeberg et al. 2003).
Acknowledgments
We thank Dr. Donald Ingram for his generous support of this project. We thank people in LEG at NIA, especially Tina Yu, Jingping Hu, Min Zhu, and Rafael de Cabo, for generous help and stimulating discussion. This work was supported by Intramural Research Program at National Institute on Aging, NIH.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6216607
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A laboratory handbook. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M. Biology of Drosophila. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1994. [Google Scholar]
- Dillin A., Crawford D.K., Kenyon C., Crawford D.K., Kenyon C., Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Dudoit S., Gentleman R.C., Quackenbush J., Gentleman R.C., Quackenbush J., Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;34:S45–S51. [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D., Spellman P.T., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B., Khaitovich P., Plotkin J.B., Paabo S., Eisen M.B., Khaitovich P., Plotkin J.B., Paabo S., Eisen M.B., Plotkin J.B., Paabo S., Eisen M.B., Paabo S., Eisen M.B., Eisen M.B. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Ellis B., Gautier L., Ge Y., Gentry J., Gautier L., Ge Y., Gentry J., Ge Y., Gentry J., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou M.E., Goss M., Junger M.A., Hafen E., Leevers S.J., Partridge L., Goss M., Junger M.A., Hafen E., Leevers S.J., Partridge L., Junger M.A., Hafen E., Leevers S.J., Partridge L., Hafen E., Leevers S.J., Partridge L., Leevers S.J., Partridge L., Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Girardot F., Lasbleiz C., Monnier V., Tricoire H., Lasbleiz C., Monnier V., Tricoire H., Monnier V., Tricoire H., Tricoire H. Specific age related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden T.R., Melov S., Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. 2004;3:111–124. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Liu W., Odell I., Ruvkun G., Lee S.S., Odell I., Ruvkun G., Lee S.S., Ruvkun G., Lee S.S., Lee S.S. A systematic RNAi screen for longevity genes in C. elegans. Genes & Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A.L., Dillin A., Kenyon C., Hsu A.L., Dillin A., Kenyon C., Dillin A., Kenyon C., Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:e17. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo D.S., Gershman B., Tu M.P., Palmer M., Tatar M., Gershman B., Tu M.P., Palmer M., Tatar M., Tu M.P., Palmer M., Tatar M., Palmer M., Tatar M., Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kim S.N., Rhee J.H., Song Y.H., Park D.Y., Hwang M., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Rhee J.H., Song Y.H., Park D.Y., Hwang M., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Song Y.H., Park D.Y., Hwang M., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Park D.Y., Hwang M., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Hwang M., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Lee S.L., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Kim J.E., Gim B.S., Yoon J.H., Kim Y.J., Gim B.S., Yoon J.H., Kim Y.J., Yoon J.H., Kim Y.J., Kim Y.J., et al. Age-dependent changes of gene expression in the Drosophila head. Neurobiol. Aging. 2005;26:1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Klebes A., Biehs B., Cifuentes F., Kornberg T.B., Biehs B., Cifuentes F., Kornberg T.B., Cifuentes F., Kornberg T.B., Kornberg T.B. Expression profiling of Drosophila imaginal discs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G.N., Abdueva D., Skvortsov D., Yang J., Rabin B.E., Carrick J., Tavare S., Tower J., Abdueva D., Skvortsov D., Yang J., Rabin B.E., Carrick J., Tavare S., Tower J., Skvortsov D., Yang J., Rabin B.E., Carrick J., Tavare S., Tower J., Yang J., Rabin B.E., Carrick J., Tavare S., Tower J., Rabin B.E., Carrick J., Tavare S., Tower J., Carrick J., Tavare S., Tower J., Tavare S., Tower J., Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J., Tedesco P., Duke K., Wang J., Kim S.K., Johnson T.E., Tedesco P., Duke K., Wang J., Kim S.K., Johnson T.E., Duke K., Wang J., Kim S.K., Johnson T.E., Wang J., Kim S.K., Johnson T.E., Kim S.K., Johnson T.E., Johnson T.E. Transcriptional profile of aging in C. elegans. Curr. Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- McCarroll S.A., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Kenyon C., Bargmann C.I., Li H., Bargmann C.I., Li H., Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Melov S., Hubbard A., Hubbard A. Microarrays as a tool to investigate the biology of aging: a retrospective and a look to the future. Sci. Aging Knowledge Environ. 2004;2004:re7. doi: 10.1126/sageke.2004.42.re7. [DOI] [PubMed] [Google Scholar]
- Pletcher S.D., Macdonald S.J., Marguerie R., Certa U., Stearns S.C., Goldstein D.B., Partridge L., Macdonald S.J., Marguerie R., Certa U., Stearns S.C., Goldstein D.B., Partridge L., Marguerie R., Certa U., Stearns S.C., Goldstein D.B., Partridge L., Certa U., Stearns S.C., Goldstein D.B., Partridge L., Stearns S.C., Goldstein D.B., Partridge L., Goldstein D.B., Partridge L., Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Rogina B., Reenan R.A., Nilsen S.P., Helfand S.L., Reenan R.A., Nilsen S.P., Helfand S.L., Nilsen S.P., Helfand S.L., Helfand S.L. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- Seong K.H., Matsuo T., Fuyama Y., Aigaki T., Matsuo T., Fuyama Y., Aigaki T., Fuyama Y., Aigaki T., Aigaki T. Neural-specific overexpression of Drosophila plenty of SH3s (DPOSH) extends the longevity of adult flies. Biogerontology. 2001;2:271–281. doi: 10.1023/a:1013249326285. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W., Xiao W., Leek J.T., Tompkins R.G., Davis R.W., Leek J.T., Tompkins R.G., Davis R.W., Tompkins R.G., Davis R.W., Davis R.W. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timiras P.S. Comparative and differential aging, geriatric functional assessment, aging and disease. In: Timiras P.S., editor. Physiological basis of aging and geriatrics. CRC Press; Boca Raton, FL: 2003. pp. 25–46. [Google Scholar]
- Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B., Hastie T., Tibshirani R., Botstein D., Altman R.B., Tibshirani R., Botstein D., Altman R.B., Botstein D., Altman R.B., Altman R.B. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Walker D.W., Hajek P., Muffat J., Knoepfle D., Cornelison S., Attardi G., Benzer S., Hajek P., Muffat J., Knoepfle D., Cornelison S., Attardi G., Benzer S., Muffat J., Knoepfle D., Cornelison S., Attardi G., Benzer S., Knoepfle D., Cornelison S., Attardi G., Benzer S., Cornelison S., Attardi G., Benzer S., Attardi G., Benzer S., Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc. Natl. Acad. Sci. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.D., Kazemi-Esfarjani P., Benzer S., Kazemi-Esfarjani P., Benzer S., Benzer S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc. Natl. Acad. Sci. 2004;101:12610–12615. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn J.M., Sonu R., Vogel H., Crane E., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Sonu R., Vogel H., Crane E., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Vogel H., Crane E., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Crane E., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Mazan-Mamczarz K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Rabkin R., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Davis R.W., Becker K.G., Owen A.B., Kim S.K., Becker K.G., Owen A.B., Kim S.K., Owen A.B., Kim S.K., Kim S.K. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115.eor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S., Kane D.W., Reinhold W.C., Lababidi S., Reinhold W.C., Lababidi S., Lababidi S., et al. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Edelman S.W., Tharmarajah G., Walker D.W., Pletcher S.D., Seroude L., Edelman S.W., Tharmarajah G., Walker D.W., Pletcher S.D., Seroude L., Tharmarajah G., Walker D.W., Pletcher S.D., Seroude L., Walker D.W., Pletcher S.D., Seroude L., Pletcher S.D., Seroude L., Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc. Natl. Acad. Sci. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Meadows S., Sharp L., Jan L.Y., Jan Y.N., Meadows S., Sharp L., Jan L.Y., Jan Y.N., Sharp L., Jan L.Y., Jan Y.N., Jan L.Y., Jan Y.N., Jan Y.N. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]