Abstract
The roles of bacterial RecA in the evolution and transmission of antibiotic resistance genes make it an attractive target for inhibition by small molecules. We report two complementary fluorescence-based ATPase assays that were used to screen for inhibitors of RecA. We elected to employ the ADP-linked variation of the assay, with a Z′ factor of 0.83 in 96-well microplates, to assess whether 18 select compounds could inhibit ATP hydrolyis by RecA. The compounds represented five sets of related inhibitor scaffolds, each of which had the potential to cross-inhibit RecA. Although nucleotide analogs, known inhibitors of GHL ATPases, and known protein kinase inhibitors were not active against RecA, we found that three suramin-like agents substantially inhibited RecA's ATPase activity.
Drug resistance is an ever-increasing problem in the chemotherapy of bacterial infectious diseases. The de novo development and clonal spread of drug-resistant bacteria, and the horizontal transfer of resistance factors among bacteria have resulted in a dramatic increase in the incidence of drug-resistant infections. One strategy to improve the efficacy of existing antibacterial drugs involves countering bacterial mechanisms of drug resistance. In this context, RecA has emerged as a potential target because its activities allow bacteria to overcome the metabolic stress induced by a range of antibacterial agents, and promote the de novo development and transmission of antibiotic resistance genes.1-5 Although potent and selective inhibitors of RecA could be used to modulate its activities in the development of antibiotic resistance, no small-molecule natural product inhibitor of RecA's activities has been reported. Herein, we report two rapid, microvolume molecular screening assays and their implementation in the directed screening of prospective inhibitors of RecA's ATPase activity.
We have previously demonstrated that select NDP and NTP analogs inhibit RecA ATP hydrolysis.6,7 Because nucleotide analogs are largely unsuited for use in cell-based assays, we screened a small, focused set of commercially available compounds to discover non-nucleotide inhibitors of RecA. The compounds we elected to study can be ordered in five groups (Figure 1). The first group comprises vanillin,8,9 cinnamaldehyde,9 curcumin,10 and the soy-derived compounds genistin and genistein,11 all of which may inhibit RecA based on their activities in microbiological assays. The second group includes adenosine nucleotide-like compounds12,13 that may extend upon our previous success with ADP analogs. The third group is composed of inhibitors of the gyrase-Hsp90-like (GHL) family of ATPases.14,15 The fourth group includes adenine-like inhibitors of protein kinases.16 The fifth group comprises compounds related to the non-nucleotide inhibitors of purine nucleotide receptors, suramin and PPADS.17,18
Figure 1.
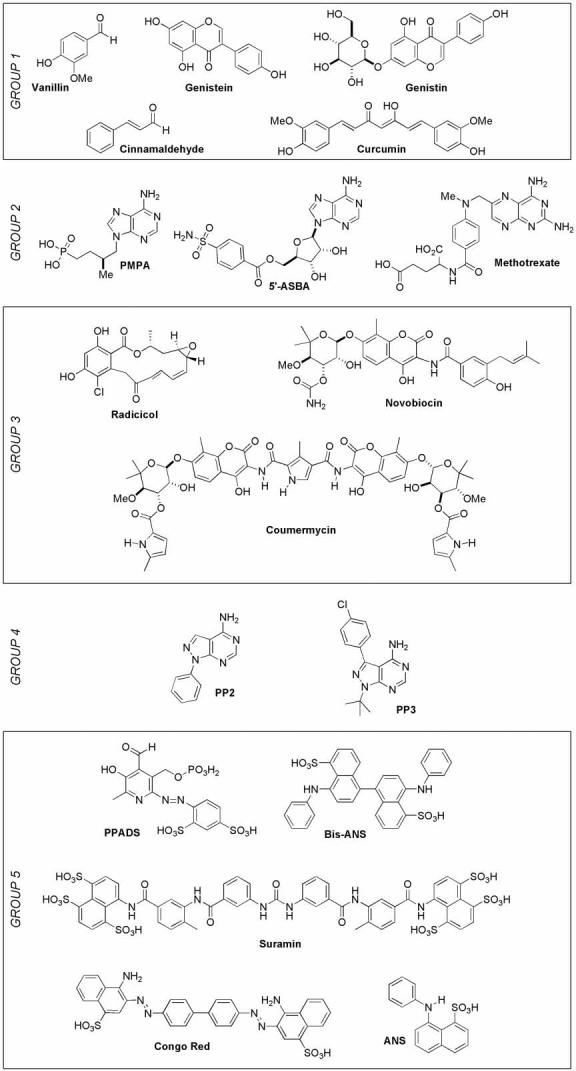
Five classes of compounds screened for RecA inhibition.
High-throughput screening is a useful method for the identification of novel inhibitory scaffolds. Recently, we reported a coupled enzyme assay that was optimized for determination of E. coli RecA's ssDNA-dependent ATPase activity, which is a useful indicator of active RecA-DNA filament assembly.7 It was undesirable to use this assay to screen a larger, more diverse library because many of the compounds may be UV active at 360 nm. This interfering absorbance would lead to false negatives in a high-throughput screening project.
To address this concern, we developed two robust and reproducible microplate assays for RecA's ATPase activity that are suitable for screening collections of small molecules as prospective RecA inhibitors without the potential for signal interference generated by UV-active compounds (Figure 2). Each variation of the assay utilizes one product of ATP hydrolysis, either ADP or Pi, as a substrate for commercially available enzymes and, for every molecule of ATP hydrolyzed by RecA, one molecule of amplex red is ultimately oxidized to resorufin, which has a unique fluorescence emission at 595 nm.19 In one variant of the assay, Pi and inosine serve as substrates for PNP in the production of hypoxanthine and ribose-1-phosphate. In turn, the O2-dependent oxidation of hypoxanthine by xanthine oxidase produces uric acid and H2O2, the latter of which is used by horseradish peroxidase to oxidize amplex red to resorufin. In the other assay variant, ADP and phosphoenolpyruvate serve as substrates for the commercially available enzyme pyruvate kinase to produce ATP and pyruvate, the latter of which is a substrate for O2-dependent oxidization by pyruvate oxidase in the production of acetylphosphate and H2O2.20 Identical to the first assay, horseradish peroxidase uses H2O2 to catalyze the oxidation of amplex red to resorufin.
Figure 2.
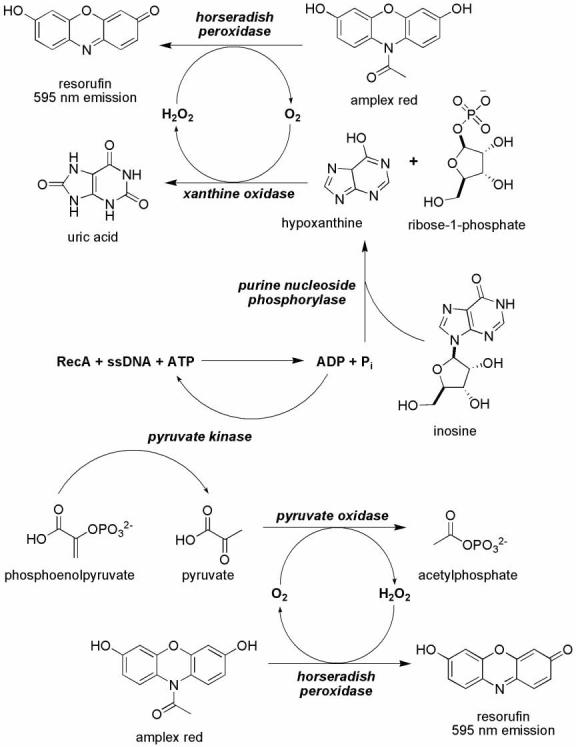
Two fluorescent ATPase assay schemes used to monitor ATP hydrolysis by RecA .
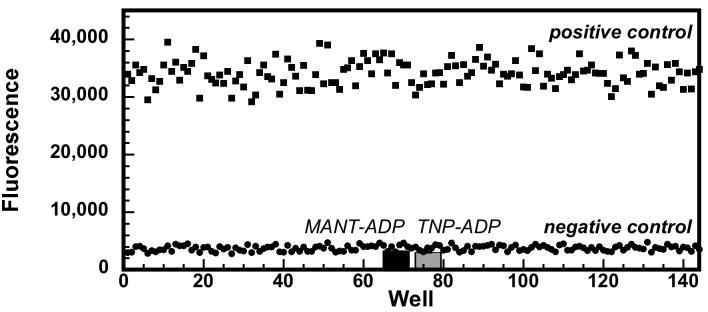
To determine if these assays were suitable for high-throughput screening, we assessed their robustness and reproducibility using a statistical analysis.21 In our hands, the ADP-linked ATPase assay was more useful as a screening assay because the Pi-linked assay was sensitive to variations in the residual phosphate contaminating enzyme and DNA preparations. For the ADP-linked ATPase assay optimized for 96-well microplates,22 positive and negative control experiments were performed on three different days with 48 wells per condition to simulate the day-to-day and well-to-well variability between assays (Figure 3). Statistical evaluation of the results yielded a reproducible Z′ factor of 0.83, demonstrating the excellent utility of the assay for reproducibly differentiating normal activity from inhibition. Furthermore, the inclusion of the two most potent NTP-analog inhibitors discovered in our previous work7 verified that known RecA ATPase inhibitors would prevent resorufin production using this assay system (Figure 3).
Figure 3.
Interday and intercolumn precision for the ADP-linked fluorescent ATPase assay.
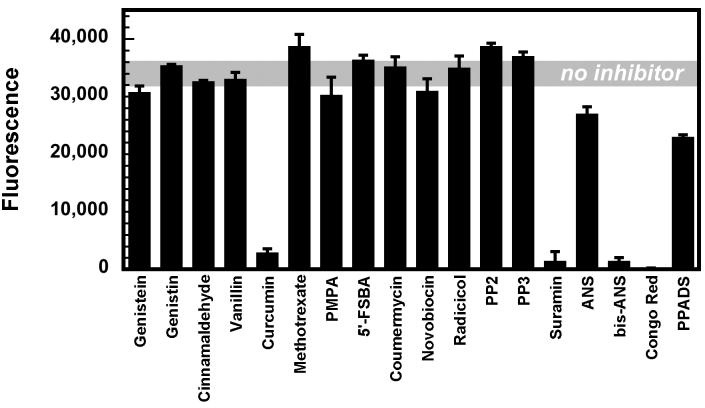
With a suitable ATPase assay in hand, we assessed the abilities of the 18 compounds in our directed mini-library (Figure 1) to inhibit RecA's ATPase activity at 100 μM. The fractional inhibition observed in the presence of each compound was obtained by comparing the total fluorescence in wells containing the reaction in the presence and absence of inhibitor (Figure 4). Only curcumin from Group 1 and the polysulfated naphthyl compounds suramin, Congo Red and bis-ANS, from Group 5, appeared to inhibit RecA's ATPase activity under these conditions.
Figure 4.
Results of the directed screen of 18 selected compounds against RecA ATP hydrolysis activity.
Upon the identification of molecules that attenuated resorufin production, it was necessary to determine whether these compounds were selectively inhibiting RecA or other components of the coupled assay system. Therefore, four compounds were evaluated in subsequent control reactions in the presence of 500 μM ADP, but in the absence of RecA. Because all four of these compounds also inhibited pyruvate kinase in the control assay, we evaluated their abilities to inhibit RecA's ATPase activity using the Pi-linked fluorescence ATPase assay described above. Suramin, Congo Red, and bis-ANS, but not curcumin, inhibited ATP hydrolysis by RecA.
It is important to note that none of the compounds expected to inhibit RecA based on prior biological activity studies (Group 1) inhibited RecA's ATPase activity in vitro. Possible explanations for the apparent inconsistency include the following: (1) these compounds may inhibit RecA-associated proteins rather than RecA itself; or (2) these compounds may interfere with RNA or protein synthesis. Of additional importance is the observation that no known inhibitor from Group 2, 3 or 4 substantially inhibited RecA's ATPase activity. The failure to discover a RecA inhibitor among these compounds suggests that the development of a potent RecA inhibitor will not be a trivial exercise. However, the same lack of cross-inhibition of RecA by known inhibitors of other ATP-dependent enzymes also suggests the likelihood of ultimately discovering specific inhibitors of RecA's activities.
Although we did not observe inhibition of RecA by any of the molecules in Groups 1, 2, 3, or 4, we did find that the polysulfated naphthyl compounds Congo Red, suramin, and bis-ANS strongly inhibited the ATPase activity of RecA. The nature of the inhibition by these compounds is reputed to be promiscuous. Indeed, it has been established that, in aqueous solution, Congo Red self-assembles into supramolecular aggregates23,24 that can result in apparent inhibition by the reversible sequestration of enzyme.25,26 In contrast, suramin, which is also active against many different enzymes27 and is structurally similar to Congo Red, does not form aggregates and does not inhibit model enzymes that are sensitive to supramolecular ligands.25,26 The possibility that suramin may be a structure- or mechanism-specific inhibitor of RecA is supported by the observation that PPADS does not inhibit RecA's ATPase activity, despite the fact that both compounds are potent antagonists of the P2X1 nucleotide receptors.18 Finally, it is noteworthy that select transition metal cations trap inactive RecA as insoluble aggregates,28 but no visible precipitates were formed in the presence of suramin, Congo Red, or bis-ANS.
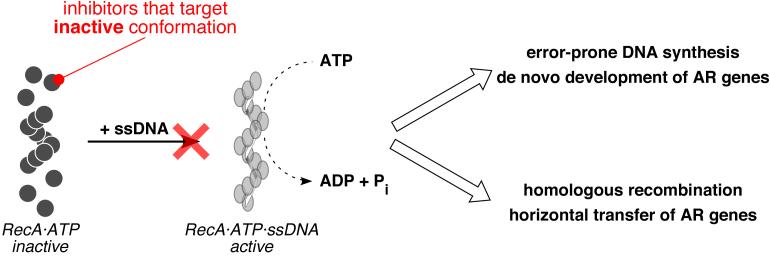
To probe this class of polysulfated naphthyl compounds further, we characterized the nature of the inhibition of RecA by suramin. While suramin inhibited RecA's ATPase activity with an IC50 of approximately 2 μM, the inhibition was not competitive with respect to ATP or ssDNA binding (data not shown). We speculate that suramin modulates RecA's activity by binding to an allosteric region of the protein and trapping the protein in its inactive conformation (Figure 5).
Figure 5.
Cartoons depicting the inactive and active conformation of RecA filaments and the roles of the latter in the de novo development and transmission of antibiotic resistance (AR) genes.
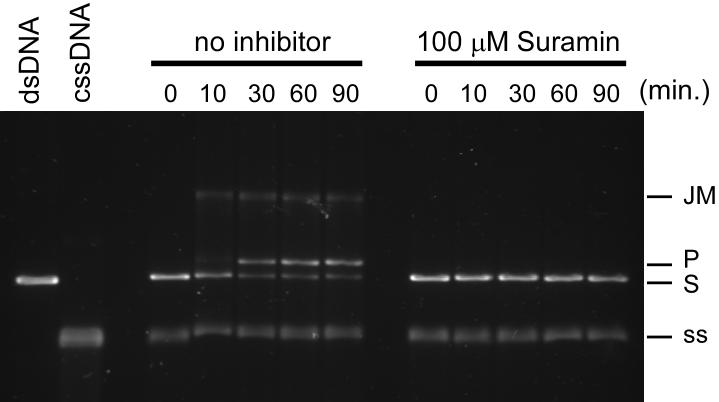
Importantly, we demonstrated that suramin interferes with the RecA-mediated DNA strand exchange reaction, an established RecA activity that serves as an in vitro model for its physiologic recombinational functions.29 It is known that RecA must hydrolyze ATP in order to carry out the strand exchange reaction between ϕχ174 cssDNA and homologous, linear dsDNA (S) to yield a nicked circular dsDNA product (P), which migrates more slowly under electrophoretic conditions than the substrate DNA molecules. The presence of suramin (100 μM) in this reaction completely abrogated the formation of nicked circular dsDNA product, even over the course of 90 min (Figure 6).
Figure 6.
Suramin inhibits the RecA-mediated DNA three strand exchange reaction.
In conclusion, we have reported two new fluorescence-based assays for screening potential inhibitors of RecA's ATPase activity. We previously developed activity assays for RecA's ATPase and filament assembly activities,6,7 and the new molecular screening assays complement and extend the previous assays by providing an observable parameter that is not influenced adversely by UV-active compounds. Moreover, the assays reported herein were optimized for use with RecA based on its production of either free phosphate or ADP. We further reported that suramin, Congo Red, and bis-ANS strongly inhibit RecA's ATPase assay and compose a new structural class of RecA inhibitors. We expect that polysulfated naphthyl compounds such as these are likely to be of little therapeutic utility due to membrane-impermeability caused by their negative charges. Nonetheless these compounds may be used in future rational modification procedures for the synthesis microbiological tools to tease apart the roles of RecA in various aspects of pathogenicity. We envision that such inhibitors may ultimately be developed into novel adjuvants for antibiotic chemotherapy that moderate the development and transmission of antibiotic resistance genes and increase the antibiotic therapeutic index.
Acknowledgments
The authors gratefully acknowledge support of this project by NIH grant GM58114.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beaber JW, Hochhut B, Waldor MK. Nature. 2004;427:72. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 2.Hastings PJ, Rosenberg SM, Slack A. Trends Microbiol. 2004;12:401. doi: 10.1016/j.tim.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Hersh MN, Ponder RG, Hastings PJ, Rosenberg SM. Res. Microbiol. 2004;155:352. doi: 10.1016/j.resmic.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Matic I, Taddei F, Radman M. Res. Microbiol. 2004;155:337. doi: 10.1016/j.resmic.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Foster PL. Mutat. Res. 2005;569:3. doi: 10.1016/j.mrfmmm.2004.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee AM, Ross CT, Zeng BB, Singleton SF. J. Med. Chem. 2005;48:5408. doi: 10.1021/jm050113z. [DOI] [PubMed] [Google Scholar]
- 7.Wigle TJ, Lee AM, Singleton SF. Biochemistry. 2006;45:4502. doi: 10.1021/bi052298h. [DOI] [PubMed] [Google Scholar]
- 8.Ohta T, Watanabe M, Shirasu Y, Inoue T. Mutat. Res. 1988;201:107. doi: 10.1016/0027-5107(88)90116-9. [DOI] [PubMed] [Google Scholar]
- 9.Shaughnessy DT, Schaaper RM, Umbach DM, Demarini DM. Mutat. Res. 2006;602:54. doi: 10.1016/j.mrfmmm.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda Y. Mutat. Res. 1995;348:67. doi: 10.1016/0165-7992(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Fix D. Mutat Res. 2006;600:193. doi: 10.1016/j.mrfmmm.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Knight KL, McEntee K. J. Biol. Chem. 1985;260:10177. [PubMed] [Google Scholar]
- 13.Scholz G, Barritt GJ, Kwok F. Eur. J. Biochem. 1992;210:461. doi: 10.1111/j.1432-1033.1992.tb17443.x. [DOI] [PubMed] [Google Scholar]
- 14.Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS. J. Am. Chem. Soc. 2006;128:15529. doi: 10.1021/ja065793p. [DOI] [PubMed] [Google Scholar]
- 15.Corbett KD, Berger JM. Nucleic Acids Res. 2006;34:4269. doi: 10.1093/nar/gkl567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Wagner MB, Kumar R, Cheng J, Joyner RW. Pflugers Arch. 2003;446:485. doi: 10.1007/s00424-003-1061-8. [DOI] [PubMed] [Google Scholar]
- 17.Lambrecht G, Braun K, Damer M, Ganso M, Hildebrandt C, Ullmann H, Kassack MU, Nickel P. Curr. Pharm. Des. 2002;8:2371. doi: 10.2174/1381612023392973. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson KA, Costanzi S, Ohno M, Joshi BV, Besada P, Xu B, Tchilibon S. Curr. Top. Med. Chem. 2004;4:805. doi: 10.2174/1568026043450961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. Anal. Biochem. 1997;253:162. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Senator D, Wilson CJ, Ng SC. Anal. Biochem. 2005;345:326. doi: 10.1016/j.ab.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. J. Biomol. Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Optimized ADP-linked assay conditions (100 μL volume, flat-bottom 96-well blackplates): 0.5 μM E. coli RecA, 5 μM-nts poly(dT) ssDNA, 10 mM Mg(OAc)2, 500μM ATP, 250 μM phosphoenolpyruvate, 100 μM amplex red, 1 U/mL pyruvate kinase, 1 U/mL pyruvate oxidase, 1 U/mL horseradish peroxidase, and 25 mM Tris·HOAc, pH 7.5. After 25-min incubation at 37 °C the reaction was excited at 485 nm and emission was observed at 595 nm in a microplate reader. Positive control reactions were performed in the absence of any inhibitor to simulate an unimpeded ATPase reaction and negative control reactions lacked RecA to simulate the complete inhibition of ATP hydrolysis.
- 23.Stopa B, Konieczny L, Piekarska B, Roterman I, Rybarska J, Skowronek M. Biochimie. 1997;79:23. doi: 10.1016/s0300-9084(97)87621-3. [DOI] [PubMed] [Google Scholar]
- 24.Konieczny L, Piekarska B, Rybarska J, Skowronek M, Stopa B, Tabor B, Dabros W, Pawlicki R, Roterman I. Folia Histochem. Cytobiol. 1997;35:203. [PubMed] [Google Scholar]
- 25.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J. Med. Chem. 2002;45:1712. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 26.McGovern SL, Helfand BT, Feng B, Shoichet BK. J. Med. Chem. 2003;46:4265. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 27.Stein CA. Cancer Res. 1993;53:2239. [PubMed] [Google Scholar]
- 28.Lee AM, Singleton SF. J. Inorg. Biochem. 2004;98:1981. doi: 10.1016/j.jinorgbio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Lusetti SL, Cox MM. Annu. Rev. Biochem. 2002;71:71. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]