Abstract
Cubitus interruptus (Ci) is a transcriptional factor that is positively regulated by the hedgehog (hh) signaling pathway. Recent work has shown that a 75-kDa proteolytic product of the full-length CI protein translocates to the nucleus and represses the transcription of CI target genes. In cells that receive the hh signal, the proteolysis of CI is inhibited and the full-length protein can activate the hh target genes. Because protein kinase A (PKA) inhibits the expression of the hh target genes in developing embryos and discs and the loss of PKA activity results in elevated levels of full-length CI protein, PKA might be involved directly in the regulation of CI proteolysis. Here we demonstrate that the PKA pathway antagonizes the hh pathway by phosphorylating CI. We show that the PKA-mediated phosphorylation of CI promotes its proteolysis from the full-length active form to the 75-kDa repressor form. The PKA catalytic subunit increases the proteolytic processing of CI and the PKA inhibitor, PKI, blocks the processing. In addition, cells do not process the CI protein to the 75-kDa repressor when all of the PKA sites in CI are mutated. Mutant CI proteins that cannot be phosphorylated by PKA have increased transcriptional activity compared with wild-type CI. In addition, exogenous PKA can increase further the transcriptional activity of the CI mutant, suggesting that PKA has a second positive, indirect effect on CI activity. In summary, we show that the modulation of the hh signaling pathway by PKA occurs directly at the level of CI phosphorylation.
Although several signaling cascades have been characterized in detail in multiple organisms, the hedgehog (hh) signaling pathway is of special interest because it plays an important role in the developmental process. The hh gene was first identified by a genetic screen for mutations affecting segment polarity in the Drosophila embryo (1). The hh gene encodes a secreted protein (2), HH, that is involved in pattern formation in both embryogenesis and disc development (3, 4). HH is synthesized in cells located in the posterior compartment of the imaginal discs. The expression of HH is maintained by another secreted protein, Wingless (WG) (2, 5–7), which regulates the expression of a homeodomain transcriptional factor engrailed (en) during early stages of embryo development (8–10). Upon secretion, HH diffuses anteriorly to function in a distance- and concentration-dependent manner to stimulate the transcription of the hh target genes, which include decapentaplegic (dpp), wingless (wg), and patched (ptc) (3, 4, 11, 12). At the membrane level, HH binds to its postulated receptor (ptc), which has at least seven putative transmembrane domains, relieving the inhibition of ptc on smoothened (smo) (13–17). Consequently, smo activates cubitus interruptus (ci), which encodes a transcriptional factor involved in mediating the hh signal from the membrane to the nucleus (18–20). Additional regulators of the hh pathway include costal2 (cos2), which is believed to tether CI in the cytoplasm (21, 22), and two protein kinases, fused (fu) and protein kinase A (PKA), whose specific targets are unknown. fu is believed to activate and PKA to inhibit hh signaling pathway, although it is not clear how these protein kinases regulate the expression of hh target genes (21–23).
Domain exchange experiments in flies and yeast have demonstrated that CI protein has separable domains: a zinc finger DNA binding domain and an acidic carboxyl-terminal activation domain (19, 20, 24). The CI DNA binding domain consists of five tandem C2H2 zinc fingers located between aa 442 and 621 (18). Electromobility shift assay shows that this fragment of CI binds specifically to the ptc and wg promoters containing CI consensus binding sites (20, 24). In vivo cytological studies also show that CI protein binds to the promoters of ptc, wg, and en (25). Substituting the activation domain (aa 970–1,282) in ci with the repressor domain of en (aa 228–282) converts CI from an activator to a repressor in vivo (19, 24). Within its five tandem zinc fingers, CI protein shares a high degree of homology with the proteins of the vertebrate Gli gene family (26, 27), the Caenorhabditis elegans sex determination gene tra-1 (28), and the Xenopus transcriptional factor TFIIIA (29). GLI proteins are transcriptional factors involved in the Sonic hedgehog (Shh) signaling cascade that play an important role in limb patterning and in determining the cell fate in the central nervous system during vertebrate development (30–32).
Aza-Blanc et al. (25) have shown that CI exists in two cellular forms, a 155-kDa full-length transcriptionally active form and a 75-kDa N-terminal truncated repressor form. Most of the 155-kDa full-length CI exists in the cytosol in a multiprotein complex, including COS2 and FU tethered to the microtubules (21, 22). The domain responsible for keeping CI from entering the nucleus is mapped from aa 703 to 850, C terminal to the CI nuclear localization signal (aa 611–614) (25). In the absence of hh–smo signaling, the 155-kDa CI protein is either sequestered in the cytosol by its tethering domain or proteolyzed to generate the 75-kDa repressor, thereby exposing the nuclear localization signal. Consequently, this repressor form translocates to the nucleus, binds to the promoters of the hh target genes, and inhibits their expression. When cells receive the hh signal, the full-length active form is released from the microtubule complex through unknown mechanisms and the proteolytic process is inhibited. Therefore, more active CI translocates into the nucleus, less of the repressor form is generated, and the hh target genes are activated.
PKA participates in Drosophila embryo and disc development by antagonizing the hh signaling cascade. In the absence of PKA, the level of full-length CI increases and the expression of dpp, wg, and ptc is induced (33–36). In addition to its negative regulation of wg and ptc, PKA can also potentiate the activation of these genes by the hh pathway, suggesting a dual role of PKA in signaling (37). However, the details of the mechanism for PKA modulation of the hh target gene expression still remain unclear. In this study, we demonstrate that the PKA pathway acts directly on CI to regulate its activity by controlling the degree of proteolysis. In addition, we show that the proteolysis of the CI protein requires the phosphorylation of CI by PKA.
MATERIALS AND METHODS
Plasmids.
We used pPac5c (kindly provided by M. Krasnow, Stanford University), which has the actin 5C proximal promoter to drive protein expression, as our expression vector in Kc cells. A luciferase cDNA containing the simian virus 40 poly(A) signal was excised from pGL3-Basic (Promega) and cloned into pPac5c at the BamHI site to generate pPac-luciferase. The rat PKA catalytic subunit (α) cDNA was excised from RCABSSK (provided by R. Maurer, Oregon Health Sciences University) by an NdeI/BamHI digestion and cloned into pPac5c to generate pPac-PKA. pPac-PKI was constructed by inserting the 182-bp HindIII/BglII fragment, which contains the cDNA encoding PKI (inhibitor of cAMP-dependent protein kinase) into pPac5c. pPac-CI (wild type) and the rest of the CI mutants were made by ligating a BamHI linker to the ends of the Ci cDNA and inserting the fragment into the BamHI site of pPac5c. The CI PKA mutants were generated by mutating the serine codon in each PKA site to alanine by using the Promega site-directed mutagenesis kits and following their protocol. The PKA sites mutated are Ser-838, Ser-856, Ser-892, and Thr-1006, and we designate each of the alanine substitutions of these sites as m1, m2, m3, and m4, respectively. Hemagglutinin (HA)-tagged CI constructs were made by inserting oligos encoding HA sequences into the MluI site at the fifth amino acid in CI. The ADHCAT/GLI6BS (ADHCAT with six GLI binding sites fused upstream of ADH promoter) reporter vector has been described elsewhere (38).
Maintenance and Transfection of Kc Cells.
Kc cells (kindly provided by L. Cherbas, Indiana University) were maintained in Schneider medium (GIBCO/BRL) supplemented with 5% heat-inactivated fetal calf serum (HyClone), 100 units/ml penicillin G, and 100 mg/ml streptomycin sulfate in a humidified incubator at 25°C. Transfections were carried out by the standard calcium phosphate method (GIBCO/BRL). Approximately 3 × 106 cells were seeded in 6-well dishes. Fifteen micrograms of DNA (100 ng of pPac-luciferase/5 μg of ADHCAT/GLI6BS reporter gene/2 μg of pPac-CI construct/4 μg of pPac-PKA or PKI plus 4 μg of pPac) was transfected into Kc cells the following day and allowed to express for 2 days. The cells were washed with PBS and processed for luciferase and chloramphenicol acetyltransferase (CAT) assays as described (39). The CAT activities were normalized to the luciferase activities. All of the transfections were repeated at least three times with different plasmid preparations and were carried out in triplicate.
For immunoprecipitation and western blots, 10 μg of pPac-HACI (WT) or pPac-HACI (null) (mutant CI with all the PKA sites mutated), and 10 μg of pPac vector, pPac-PKA, or pPac-PKI were transfected into Kc cells (≈10 × 106 cells) seeded on 100-mm dishes. Transfection mix was washed from the cells 24 hr after transfection and cells were cultured for 2 more days before harvesting for immunoprecipitation.
Immunoprecipitation and Western Blots.
Cells were washed briefly with cold PBS, scraped off the plates, and collected by centrifugation. The cell pellet was resuspended in Harlow buffer (50 mM Hepes, pH 7.5/0.2 mM EDTA/10 μM NaF/0.5% Nonidet P-40/1 mM DTT) containing 250 mM NaCl supplemented with protease inhibitors [10 μg/ml leupeptin/10 μg/ml pepstatin A/10 μg/ml antipain/10 μg/ml trans-epoxysuccinyl-leucylamido (4-guanidino)-butane (E64)/1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF)] and lysed at 4°C with rocking for 30 min. The cell extract was collected by saving the supernatant after centrifuging the lysate for 5 min at 4°C. The cell extract was precleared with protein G agarose beads for 30 min at 4°C, and the precleared extract was recovered by centrifugation followed by incubation with rat monoclonal anti-HA antibody (1:200 dilution, Boehringer Mannheim) on ice for 2 hr. The HACI–HA antibody complex was precipitated by incubating the extract with protein G agarose beads at 4°C for 1 hr. The immunocomplex was centrifuged, resuspended in protein loading buffer, boiled, and loaded onto an SDS/6% PAGE gel. Fractionated cell extracts were then electronically transferred to Immobilon-P membrane (Millipore) by using a semi-dry transfer apparatus and reacted to antibody (mouse monoclonal anti-HA antibody, 1:100 dilution; Boehringer Mannheim) following the protocol provided with enhanced chemiluminescence kits (Amersham).
Blot stripping was performed in solution containing 62.5 mM Tris⋅HCl (pH 6.7), 2% SDS, and 100 mM 2-mercaptoethanol at 50°C for 30 min. Next the blot was reacted with primary CI antibody [made in rat against the carboxyl terminus of CI; provided by R. Holmgren (Northwestern University), 1:1 dilution] and proceeded as described above.
RESULTS AND DISCUSSION
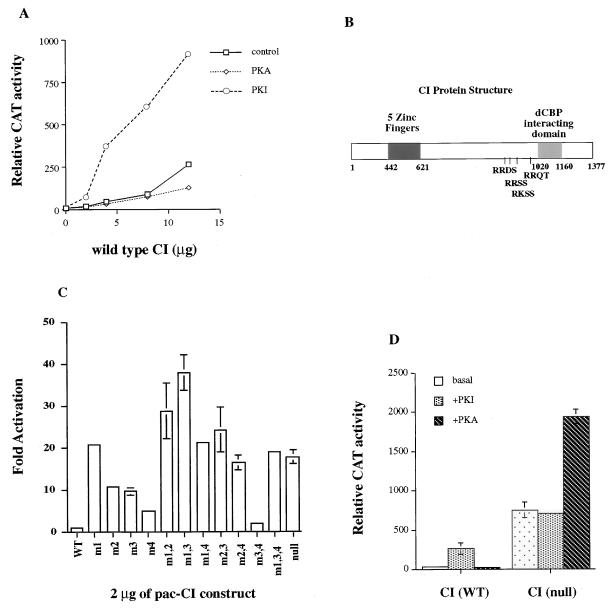
We have recently described the interaction between CI and the transcriptional coactivator Drosophila CREB-binding protein (dCBP) (38). In an effort to understand the role of PKA in the regulation of this interaction, we examined the effect of PKA phosphorylation on CI-mediated transcriptional activation in Drosophila Kc cells. Our previous results showed that CI activates the ADHCAT/GLI6BS reporter gene (a CAT reporter gene whose expression is driven by six tandem Gli binding sites fused upstream of the ADH promoter) in a dose-dependent manner in Schneider S2 cells (38). CI has the same dose-response curve in Kc cells as in S2 cells, with an EC20 of 2 μg (data not shown). When the catalytic subunit of PKA, pPac-PKA, is cotransfected with pPac-CI, we observe a slight decrease in CAT activity (Fig. 1A). To suppress any endogenous PKA that might affect CI activity, we examined the effects of a PKA inhibitor, pPac-PKI, in similar cotransfection experiments. In these experiments, we observe a 5- to 8-fold increase in CAT activity (Fig. 1A) suggesting that PKA negatively regulates CI-mediated transactivation.
Figure 1.
Effect of PKA phosphorylation on CI-mediated transcriptional activation. (A) One hundred nanograms of pPac-luciferase, 5 μg of the ADHCAT/GLI6BS reporter gene, 4 μg pPac-PKA or pPac-PKI, and an increasing amount (0–12 μg) of pPac-CI (wild type) were transfected into Kc cells. Four micrograms of PKA and PKI represent the amount of DNA that gives maximal activities in the dose response curves (data not shown). (B) Schematic diagram showing the structure of CI protein. (C) Transfections were carried out by using 100 ng of pPac-luciferase, 5 μg of the ADHCAT/GLI6BS reporter gene, and 2 μg of the different PKA mutants. (D) Transfections were performed as in A by using 2 μg of pPac-CI (wild type) and 2 μg of pPac-CI (null). CAT activities were normalized to the corresponding luciferase activities. Data represent means ± SEM.
A search for the RRXS/T consensus PKA phosphorylation motif (40) yields four consensus PKA phosphorylation sites clustered in the C-terminal transactivation domain (Fig. 1B). To determine whether PKA affected CI transcriptional activity directly, we mutated the serines in the PKA sites to alanines either singly [m1 (Ser to Ala at aa 838), m2 (Ser to Ala at aa 856), m3 (Ser to Ala at aa 892), m4 (Thr to Ala at aa 1,006)] or in various combinations including the mutation of all four sites (null). We cotransfected the PKA mutants with the reporter construct as before and compared the CAT activities with that of wild-type CI. As shown in Fig. 1C, m1, m2, m3, and the mutation that is null for all four PKA sites greatly stimulate CI activity, whereas m4 has less of an effect. This result suggests that the inhibition of phosphorylation of any one of the first three CI PKA sites is sufficient to stimulate CI transcriptional activity. We concentrated the remainder of our study on CI (null) to study the effects that PKA phosphorylation might have on CI activity or processing.
To ensure that we had eliminated all of the PKA sites required for the PKA regulation of CI transcriptional activity, CI (null) was cotransfected with PKI into Kc cells. As expected, we do not observe any further increase in CAT activity when CI (null) was cotransfected with PKI (Fig. 1D). However, when PKA and CI (null) were cotransfected, we observed a further increase in CI-mediated transcriptional activation (Fig. 1D). Although the increase (2- to 3-fold) is not as large as the increase of CI (null) versus wild-type CI (10-fold), it was found to be consistent. Consequently, although the major effect of PKA activity in our system is negative, when CI is no longer the target for PKA phosphorylation, a minor positive effect of PKA is unmasked. PKA might phosphorylate other factors crucial for CI-mediated transactivation, such as dCBP, to activate CI-mediated transcription. Our observation of this dual role played by PKA is in agreement with the genetic result by Ohlmeyer and Kalderon (37) that overexpression of the PKA catalytic subunit increases the hh target gene expression in a ci- and smo-dependent manner.
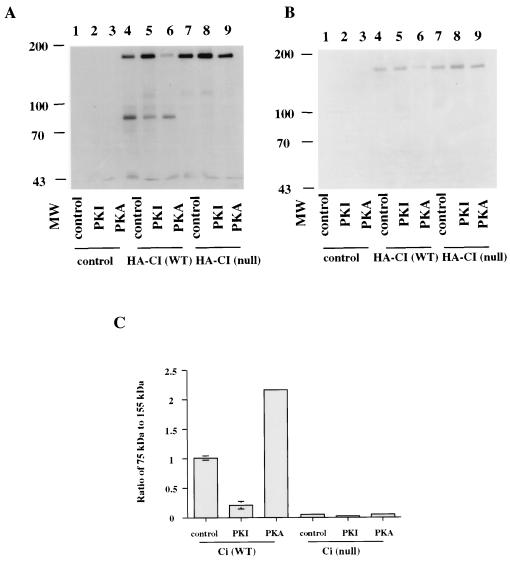
Recent studies show that the hh signal transduction pathway activates CI by inhibiting the proteolytic process that generates a transcriptional repressor form (25). The evidence shows that CI exists in two forms: (i) a full-length protein that is tethered in the cytosolic fraction by a domain near the carboxyl terminus and (ii) a 75-kDa N-terminal proteolytic product that translocates to the nucleus and inhibits CI target gene expression (25). When cells are stimulated by hh, the proteolysis is inhibited and the transcriptional block imposed by the 75-kDa CI protein is relieved. Because our results indicate that the phosphorylation of CI by PKA is involved in the regulation of CI activity, we determined whether the PKA phosphorylation of CI regulates the proteolytic processing as well. Kc cells were transfected with HA-CI (WT) or HA-CI (null) (Ci cDNA fused to an N-terminal HA tag) and the exogenous HA-CI protein was precipitated from whole-cell extracts by a rat monoclonal anti-HA antibody. The immunoprecipitated proteins were separated by PAGE, transferred to nitrocellulose, and probed with a mouse monoclonal anti-HA antibody or CI antibody made against the CI carboxyl terminus (18). As shown in Fig. 2A, the HA antibody detected both the 155-kDa full-length form and the 75-kDa form when HA-CI wild type was transfected into Kc cells. When HA-CI (null), which has all four PKA sites mutated, was transfected into cells, we could not detect the 75-kDa CI protein (Fig. 2A), indicating that PKA phosphorylation of CI is required for the proteolysis of CI. Kc cells were also cotransfected with HA-CI (WT) or HA-CI (null) and either PKI or PKA. Cotransfection of PKI with HA-CI (WT) inhibited the formation of the 75-kDa CI protein, whereas the cotransfection of HA-CI (WT) and PKA stimulated the formation of the 75-kDa CI product (Fig. 2A). (The ratio of the 75- and 155-kDa proteins is quantitated in Fig. 2C.) This result suggests that the phosphorylation of CI by PKA stimulates the proteolytic processing of CI in Kc cells. When we coexpressed PKA or PKI with HA-CI (null) we could not detect the 75-kDa CI protein (Fig. 2 A and C). To ensure that the 75- and 155-kDa bands represented the N terminus and full-length CI, respectively, the filter that was probed with the HA antibody was stripped and probed with a rat anti-CI antibody made against the CI carboxyl terminus. As shown in Fig. 2B, the CI antibody detected the full-length CI and failed to detect the 75-kDa protein, indicating that the 75-kDa band detected with HA antibody is the N terminus of CI.
Figure 2.
Effect of PKA phosphorylation on CI proteolysis. (A) A mixture of 10 μg of pPac (lanes 1–3) or pPac-HA-CI (WT) (lanes 4–6) or pPac-HA-CI (null) (lanes 7–9), and 10 μg of pPac (lanes 1, 4, and 7), pPac-PKI (lanes 2, 5, and 8), or pPac-PKA (lanes 3, 6, and 9) were transfected into Kc cells. HA-tagged CI proteins were immunoprecipitated with a rat monoclonal anti-HA antibody and probed with a mouse anti-HA antibody. (B) The filter in A was probed with a rat anti-CI antibody. In other experiments, the level of full-length CI is higher in the PKI lane than illustrated here and we attribute the difference to the variation often associated with stripping and reprobing. (C) Intensity ratio of 75–155 kDa plotted according to A. Because enhanced chemiluminescence signals can be nonlinear, the data represent scans of the mid-range exposures from two separate experiments. They include the means ± SD.
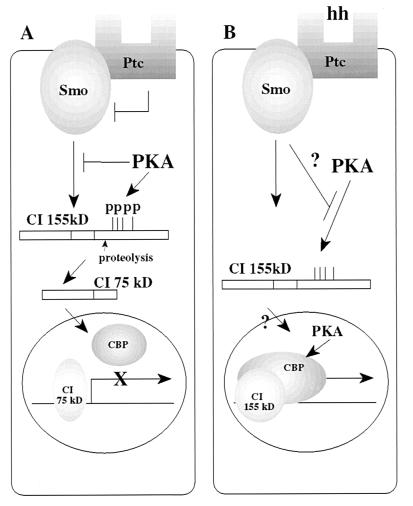
A large body of genetic evidence has demonstrated that the transcription factor ci is activated in response to the hh signal transduction cascade (20, 24, 41–43). A mechanism to account for this activation was presented by Aza-Blanc et al. (25) who showed that hh signaling suppresses the cleavage of an active CI protein into a 75-kDa repressor, which migrates into the nucleus and inhibits the activation of CI target genes. It has long been known that the PKA signaling pathway can inhibit CI-mediated transactivation (33–36); however, the direct targets of PKA and the crosstalk between the hh and PKA signaling cascades have never been elucidated. In this paper we use CI proteins, which are mutant for PKA phosphorylation sites, to show that PKA directly regulates CI activity by promoting its proteolytic processing. We provide evidence that the hh and PKA signaling cascades converge on the regulation of full-length CI protein levels (a model is summarized in Fig. 3). According to this scenario, cells located in the anterior compartment of a developing disc express an activated PKA pathway that phosphorylates the CI protein and this phospho-CI is targeted for proteolysis. The 75-kDa proteolytic product migrates into nucleus and binds to CI binding sites in the promoter of hh target genes. Because the 75-kDa repressor lacks the activation domain, which also includes the dCBP interaction domain (Fig. 1B and ref. 38), it cannot recruit dCBP to the promoter and presumably inhibits the transcription from the downstream genes (Fig. 3A). When cells located along the anterior/posterior boundary receive the hh signal, we propose that PKA activity is suppressed and CI protein phosphorylation is inhibited. As a result of this inhibition, the proteolysis of CI is suppressed. Therefore, the level of 75-kDa repressor is decreased and the level of the active, CI protein is increased. The active form of CI may be a full-length protein or an alternative proteolytic product that includes the DNA binding domain and carboxyl-terminus transactivation domain, which can act as an activator in vivo (ref. 29 and Y. Chen and S. Smolik, unpublished observation). This alternative proteolytic event is PKA independent, because PKI has no effect on the activity of the CI (null) protein. Two possible mechanisms may account for the increased CI activity. One is that the cytosolic compartment of the anterior cells can only tether a certain proportion of active CI; the other possibility is that smo signals to the CI-FU-COS2-microtubule complex through some unknown mechanism. In any case, when the level of the active form of CI increases, more CI protein migrates into the nucleus, binds to the CI binding sites, recruits the coactivator dCBP to the promoter, and activates the transcription from the hh target genes. Our immunocytochemical results support this hypothesis. When CI (null) is introduced into Kc cells, we observe an overall increase in CI full-length protein staining, including an increase in nuclear staining as compared with the CI staining observed in cells transfected with wild-type CI (data not shown).
Figure 3.
Schematic drawing showing the relationship of PKA and hh pathways on CI proteolysis and activity. (A) In a cell that is not stimulated by the hh signaling pathway, PKA phosphorylates cytoplasmic CI. The phospho-CI is a target for proteolysis and the 75-kDa protein enters the nucleus and represses the CI target genes, perhaps blocking the recruitment of dCBP to the promoter. (B) In a cell that receives the hh signal, the phosphorylation of CI by PKA in the cytoplasm is inhibited and CI is not proteolysed to the 75-kDa repressor. This allows the active form of CI to enter the nucleus, recruit dCBP to the promoter, and activate the CI target genes. The CI activation of transcription is potentiated by PKA.
In addition to the inhibition of CI transcriptional activity by PKA we have shown that PKA enhances CI-mediated transactivation when CI cannot serve as a PKA phosphorylation target, suggesting that PKA plays a dual role in regulating the transcriptional activity of CI protein. Although we cannot rule out the possibility that PKA activates CI directly through the phosphorylation of a cryptic PKA site, this seems unlikely because we cannot inhibit the activity of the CI (null) protein with the addition of PKI. Our interpretation of this result is that PKA activates other factors that CI recruits to the promoter, such as dCBP. Drosophila CBP has several consensus PKA sites, one of which is located between the third and fourth zinc fingers and is well conserved among p300/CBP gene family. Swope et al. (44) have shown that a GAL-CBP1678–2441 fusion protein, which includes this conserved PKA site, is a transcription activator and the activity of this chimera increases when PKA activity is stimulated in PC12 cells; however, there is no evidence as yet that PKA increases CBP activity by phosphorylation of this site. In addition, the finding that the activity of GAL-CBP1–460 is greatly stimulated by PKA treatment in PC12 cells (17-fold) further supports the idea that PKA modulates the activity of CBP. However, our results do not rule out the possibility that PKA phosphorylates and activates other transcriptional factors involved in CI-mediated gene activation as well. Whether the hh–smo pathway directly affects PKA function is unclear. Smo encodes a seven transmembrane domain protein reminiscent of G-protein coupled receptor. It also carries a consensus carboxyl-terminal PKA phosphorylation site that is characteristic of the site involved in ligand-binding induced receptor desensitization. It may be that smo couples to an effector, such as adenylate cyclase, through a G protein, such as Gi, to negatively regulate PKA activity (16, 17).
In summary, we have shown that PKA has a direct negative effect on CI-mediated transcriptional activity, and this negative effect is, at least partially, because of the stimulation of CI proteolysis upon PKA phosphorylation of CI protein. Besides the negative effect, PKA also has a indirect positive effect on ci activity, suggesting a dual role of PKA in regulating the hh signaling output. Our data are in agreement with previous genetic findings and provide a biochemical mechanism for the PKA regulation of the hh signaling pathway.
Acknowledgments
We thank Robert Holmgren for ci cDNA and CI antibody, Marc Krasnow for pPac vector, and Richard Maurer for PKA and PKI cDNAs. This work was supported by grants from the National Institutes of Health (DK44239) and Human Frontiers Foundation (RG-67193B).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PKA, protein kinase A; CAT, chloramphenicol acetyltransferase; HA, hemagglutinin.
References
- 1.Nusslein-Volhard C, Wieschaus E. Nature (London) 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee J J, von Kessler D P, Parks S, Beachy P A. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 3.Heemskerk J, DiNardo S. Cell. 1994;76:449–460. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 4.Kojima T, Michiue T, Orihara M, Saigo K. Gene. 1994;148:221–217. doi: 10.1016/0378-1119(94)90691-2. [DOI] [PubMed] [Google Scholar]
- 5.Mohler J, Vani K. Development (Cambridge, UK) 1992;115:957–971. doi: 10.1242/dev.115.4.957. [DOI] [PubMed] [Google Scholar]
- 6.Tabata T, Eaton S, Kornberg T B. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Arias A, Baker N E, Ingham P W. Development (Cambridge, UK) 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- 8.DiNardo S, Sher E, Heemskerk-Jongens J, Kassis J A, O’Farrell P H. Nature (London) 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Heuvel M, Nusse R, Johnston P, Lawrence P A. Cell. 1989;59:739–749. doi: 10.1016/0092-8674(89)90020-2. [DOI] [PubMed] [Google Scholar]
- 10.Heemskerk J, DiNardo S, Kostriken R, O’Farrell P H. Nature (London) 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabata T, Kornberg T B. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 12.Basler K, Struhl G. Nature (London) 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- 13.Nakano Y, Guerrero I, Hidalgo A, Taylor A, Whittle J R S, Ingham P W. Nature (London) 1989;341:508–513. doi: 10.1038/341508a0. [DOI] [PubMed] [Google Scholar]
- 14.Hooper J E, Scott M P. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- 15.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 16.Alcedo J, Ayzenzon M, Ohlen T V, Noll M, Hooper J E. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 17.van den Heuvel M, Ingham P W. Nature (London) 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 18.Orenic T V, Slusarski D C, Kroll K L, Holmgren R A. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 19.Hepker J, Wang Q, Motzny C K, Holmgren R, Orenic T V. Development (Cambridge, U K) 1997;124:549–558. doi: 10.1242/dev.124.2.549. [DOI] [PubMed] [Google Scholar]
- 20.Ohlen T V, Lessing D, Nusse R, Hooper J E. Proc Natl Acad Sci USA. 1997;94:2404–2409. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Therond P P. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 22.Sisson J C, Ho K S, Suyama K, Scott M P. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 23.Forbes, A. J., Nakano, Y., Taylor, A. M. & Ingham, P. W. (1993) Development (Cambridge, U.K.) Suppl., 115–124. [PubMed]
- 24.Alexandre C, Jacinto A, Ingham P W. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- 25.Aza-Blanc P, Ramfrez-Weber F, Laget M, Schwartz C, Kornberg T B. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 26.Ruppert J M, Vogelstein B, Arheden K, Kinzler K W. Mol Cell Biol. 1990;10:5408–5415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzler K W, Ruppert J M, Bigner S H, Vogelstein B. Nature (London) 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 28.Zarkower D, Hodgkin J. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- 29.Brown R S, Sander C, Argos P. FEBS Lett. 1985;186:271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- 30.Altaba A R. Cell. 1997;90:193–196. [Google Scholar]
- 31.Sasaki H, Hui C, Nakafuku M, Kondoh H. Development (Cambridge, UK) 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 32.Hynes M, Stone D M, Dowd M, Pitts-Meek S, Goddard A, Gurney A, Rosenthal A. Neuron. 1997;19:15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 33.Pan D, Rubin G. Cell. 1995;80:543–552. doi: 10.1016/0092-8674(95)90508-1. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Ohlmeyer J T, Lane M E, Kalderon D. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Struhl G. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- 36.Lepage T, Cohen S M, Diaz-Benjumea F J, Parkhurst S M. Nature (London) 1995;373:711–715. doi: 10.1038/373711a0. [DOI] [PubMed] [Google Scholar]
- 37.Ohlmeyer J T, Kalderon D. Genes Dev. 1997;11:2250–2258. doi: 10.1101/gad.11.17.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akimaru H, Chen Y, Dai P, Hou D, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Nature (London) 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 39.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 40.Kemp B E, Pearson R B. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 41.Slusarski D C, Motzny C K, Holmgren R. Genetics. 1995;139:229–240. doi: 10.1093/genetics/139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motzny C K, Holmgren R. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- 43.Johnston R L, Grenier J K, Scott M P. Development (Cambridge, UK) 1995;121:4161–4170. doi: 10.1242/dev.121.12.4161. [DOI] [PubMed] [Google Scholar]
- 44.Swope D L, Mueller C L, Chrivia J C. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]