Abstract
Sunlight ultraviolet A (UVA) irradiation has been implicated in the etiology of human skin cancer. A genotoxic mode of action for UVA radiation has been suggested that involves photosensitization reactions giving rise to promutagenic DNA lesions. We investigated the mutagenicity of UVA in the lacI transgene in Big Blue mouse embryonic fibroblasts. UVA irradiation of these cells at a physiologically relevant dose of 18 J/cm2 caused a 2.8-fold increase in the lacI mutant frequency relative to control, i.e., 12.12 + 1.84 vs 4.39 + 1.99 × 10-5 (Mean + SD). DNA sequencing analysis showed that of one hundred UVA-induced mutant plaques and fifty-four spontaneously arisen control plaques, 97 and 51, respectively, contained a minimum of one mutation along the lacI transgene. The vast majority of both induced- and spontaneous mutations were single base substitutions, although less frequently, there were also single and multiple base deletions and insertions, and tandem base substitutions. Detailed mutation spectrometry analysis revealed that G:C→T:A transversions, the signature mutations of oxidative DNA damage, were significantly induced by UVA irradiation (P < 0.003). The absolute frequency of this type of mutations was 7.4-fold increased consequent to UVA irradiation as compared to control (3.38 vs 0.454 × 10-5; P < 0.00001). These findings are in complete agreement with those previously observed in the cII transgene of the same model system, and reaffirm the notion that intracellular photosensitization reactions causing promutagenic oxidative DNA damage are involved in UVA genotoxicity.
INTRODUCTION
Solar ultraviolet (UV) irradiation is implicated in the etiology of basal and squamous cell carcinomas and malignant melanoma of the skin in humans [1-3]. The UV components of the sunlight of relevance for these neoplasias are UVB (280-320 nm wavelength) and UVA (320-400 nm wavelength) [4,5]. The mechanistic involvement of UVB in skin carcinogenesis is primarily defined by induction of promutagenic cis-syn cyclobutane pyrimidine dimers (CPDs), pyrimidine (6-4) pyrimidone photoproducts, and Dewar valence photoisomers [6,7]. However, the underlying mechanism of UVA carcinogenicity is not fully delineated [4]. Despite the weak absorbance of UVA by DNA [5], a genotoxic mode of action for UVA has been demonstrated [6,7]. Yet, the exact process through which UVA exerts genotoxicity remains elusive [6].
An increasingly popular theory ascribes UVA genotoxicity to its ability to trigger intracellular photosensitization reactions, thereby giving rise to promutagenic DNA lesions [8,9]. In fact, UVA has been shown to induce CPDs [10-16] and oxidative DNA damage [12,16-21], as well as mutagenesis [11,20-31]. However, the correlation between UVA-induced DNA damage and -mutations has not been straightforward inasmuch as the spectrum of mutations produced by UVA has inconsistently matched the mutagenic potential of the various induced lesions [6,7]. Depending on test system, e.g., human vs rodent cells, experimental conditions, including irradiation protocol, i.e., UVA source and dose, cell type, and mutational target gene, divergent findings have been reported [6,7]. Comparatively, however, UVA mutagenicity has not been investigated in different target genes within a single test system. Such an approach could systematically account for the above-mentioned experimental variables.
Recently, we have shown distinctive oxidative-DNA damage-mediated mutagenicity of UVA alone [21] and in combination with δ-aminolevulinic acid (δ-ALA) [20], a precursor of the intracellular photosensitizer protoporphyrin IX (Pp-IX) [32], in the cII transgene in Big Blue mouse embryonic fibroblasts. In the present study, we have used a similar experimental approach to determine UVA mutagenicity in a different mutational target gene, the lacI transgene, in the same model system. The presence of two target loci within a single test system [33,34] enabled us to investigate UVA mutagenicity independently under uniform experimental conditions. Furthermore, in the Big Blue system, the availability of two chromosomally integrated target genes offers a unique opportunity to explore the effects of DNA sequence context on lesion formation and mutation induction. This advantage is significant for the present study because a variety of chemical and physical carcinogens, including solar UV are known to induce specific DNA lesions and mutations in certain DNA sequence contexts [35]. For example, sunlight UV irradiation has been shown to form photodimeric lesions at dipyrimidine sites along the p53 tumor suppressor gene, one of the most frequently mutated genes in human cancers, especially in sunlight-associated skin cancer. The same nucleotide positions are also the hotspots of single or tandem C→T transitions, the established signature mutations of photodimers, in the p53 gene of skin tumors (reviewed in ref. [35]). In the Big Blue system, the lacI and cII transgenes are 1080 and 294 base long, respectively [33,34], and represent diverse sequence contexts for DNA damage-targeted mutagenesis.
MATERIALS AND METHODS
Cell culture and UVA irradiation
Early passage Big Blue mouse embryonic fibroblasts (prepared from 13.5-day old embryos) were grown to monolayer ∼70% confluence in Dulbecco′s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum. Prior to irradiation, the media were removed form the culture dishes and the cells were washed thoroughly with phosphate buffered saline (PBS). The dishes were filled with a 1-cm layer of PBS, placed on ice, and irradiated with 18 J/cm2 of UVA. The UVA source was a Sellas Sunlight System (Medizinische Geräte GmbH; Gevelsberg, Germany) with an average fluence rate of 60 mW/cm2 emitting exclusively wavelengths between 340-400 nm (see fig. 1 of ref. [21]). For homogeneous irradiation of the cells, the culture dishes were placed directly under the source at a distance of 1 cm, and were rotated during the course of irradiation, i.e., 5 min. No excessive heat was generated throughout. Following the irradiation, the cells were washed with PBS, and cultured in complete growth medium for 8 days. The 8-day growing period is essential for the fixation of all mutations into the genome. To prevent the cultures from reaching full confluence, all cell cultures were passed once (1 to 3 split) during the 8-day growing period. At the time of harvesting, both the irradiated and control cell cultures had undergone 2-3 rounds of replications, reaching a maximum of 70-80% confluence. All cell culture experiments were performed under conventional O2 and CO2 tensions (21% and 5%, respectively) using regular incubators (Forma Scientific, Inc., Marietta, OH). All experiments were conducted in triplicate.
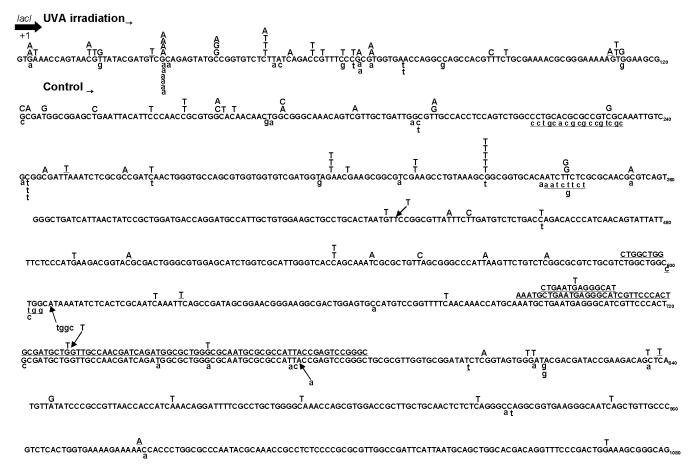
Figure 1.
Detailed mutation spectra of the lacI transgene in Big Blue mouse embryonic fibroblasts irradiated with 18 J/cm2 of UVA or control. Mutations were quantified 8 days after irradiation using the Lambda Transgenic Shuttle Vector Recovery kit for Big Blue rodents (Stratagene). Verified mutant plaques were subsequently subjected to DNA sequencing analysis. Of one hundred UVA-induced- and fifty-four spontaneously arisen control mutant plaques, 95 and 51, respectively, contained a minimum of one mutation along the lacI transgene. UVA-induced mutations are typed in capital letters above the reference lacI sequence, whereas control mutations are shown in small letters below the reference lacI sequence. Substituted bases are in bold. Deleted bases are underlined; multiple deletions are continuously underlined. Inserted bases are shown with an arrow. Numbers below the bases are the nucleotide positions.
Genomic DNA isolation
Genomic DNA was isolated using a standard phenol and chloroform extraction and ethanol precipitation protocol [36]. The DNA was dissolved in TE buffer (1 mM EDTA, 10 mM Tris-HCl, pH 7.5), and preserved at -80°C until further analysis.
LacI mutation detection system
The mutation assay requires rescuing of the coliphage λLIZ shuttle vector containing the lacI reporter gene from the genomic DNA of Big Blue mouse embryonic fibroblasts. The λLIZ shuttle vector is present in approximately 40 copies, and integrated into the genome at a single locus in a head-to-tail arrangement. The rescued vector is packaged into viable bacteriophages, and the infective phage particles are introduced into an appropriate host Escherichia coli (E.coli). Inactivating mutations in the lacI repressor lead to the transcription of lacZ gene, which encodes β-galactosidase. The mutated lacI are phenotypically scored using 5-bromo-4-chloro-3-indolyl-β—D-galactopyranoside (X-gal) as the chromogenic substrate in the bacterial lawn. Thus, phages bearing a mutated lacI possess β-galactosidase activity, thereby cleaving the X-gal and forming blue plaques, whereas wild type lacI -containing phages give rise to colorless plaques. The lacI mutant plaques are isolated and subjected to DNA sequencing, thereafter (reviewed in ref. [37]).
LacI mutant frequency and mutation spectrum analyses
The lacI mutant frequency was determined using the Lambda Transgenic Shuttle Vector Recovery kit according to the manufacturer’s recommended protocol (Stratagene, La Jolla, CA). Briefly, the λLIZ shuttle vectors were rescued from the genomic DNA (∼5 μg) and packaged into viable phage particles using the Transpack Packaging Extract (Stratagene). The phages were pre-adsorbed to an E. coli host, strain SCS-8, and the bacterial culture was plated on 25-cm NZY agar trays in the presence of X-gal. The trays were incubated overnight at 37°C, and subsequently screened for blue plaque formation. Titration trays were prepared under the same conditions to compute the total number of plaques plated. Putative mutant plaques were verified by replating at low density. The lacI mutant frequency was calculated as the ratio of the number of verified circular blue plaques-not sectored or pinpoint ones - to the total number of plaques plated.
For mutation spectrometry, the verified blue plaques were cored, and subjected to polymerase chain reaction (PCR) using primer 1: 5′-GTACCCGACACCATCGAATG-3′(positions -55 to -36) and primer 2: 5′-GAGTCACGACGTTGTA-3′ (positions+1270 to +1283) as described previously [38]. The amplified 1338-bp fragment containing the lacI and flanking regions was purified using the QIA quick PCR purification kit (QIAGEN GmbH, Hilden, Germany). The entire length of the lacI gene was sequenced by a Big Dye terminator cycle sequencing kit on an ABI-377 DNA Sequencer (ABI Prism, PE Applied BioSystems, Foster City, CA) using the following primers: P3: 5′-CCCGACACCATCGAA-3′(position -53 to -37), P5: 5′-TGTAAAGCGGCGGTGCA-3′ (position +347 to +363), and P : 5′-ATTACCGAGTCCGGGCT-3′ 6 (position +797 to +813).
Statistical analysis
Results are expressed as means + SD. The entire mutation spectra and the specific types of mutation produced by UVA irradiation vs control were compared by the hypergeometric test of Adams and Skopek [39] and chi-square test, respectively. Values of P ≤0.05 were considered statistically significant.
RESULTS
UVA irradiation of mouse embryonic fibroblasts at a physiologically relevant dose of 18 J/cm2 of UVA [22] resulted in approximately 25% cytotoxicity as determined by trypan blue dye exclusion assay. The mutagenicity of UVA was confirmed by a 2.8-fold increase in the relative frequency of lacI mutants in the genomic DNA of UVA-irradiated cells, i.e., 12.76 + 1.77 vs 4.63 + 2.18 × 10-5. For verification purposes, all replated mutant lacI plaques including one hundred UVA-induced plaques and fifty-four spontaneously arisen control plaques were subjected to DNA sequencing. Of these, 97 and 51 plaques, respectively, contained a minimum of one mutation along the lacI transgene (Table 1). Thus, we adjusted the mutant frequency data on the basis of the above-mentioned DNA sequencing results. As shown in Table 1, the adjusted fold-increase in the relative frequency of lacI mutants induced by UVA irradiation remained unchanged, i.e., 2.8-fold increase relative to background (12.12 + 1.84 vs 4.39 + 1.99 × 10-5).
TABLE 1.
Mutant frequency of the lacI transgene in Big Blue mouse embryonic fibroblasts irradiated with 18 J/cm2 of UVA or control
| Treatment | Sample No. | Total number of plaques (pfu*) | Verified mutant plaques | Mutant frequency (× 10-5) | Average mutant frequency (× 10-5)† | Adjusted average mutant frequency (× 10-5)†, ¶ |
|---|---|---|---|---|---|---|
| UVA | 1 | 301,972 | 44 | 14.57 | 12.76±1.77 | 12.12±1.84 |
| 2 | 217,566 | 24 | 11.03 | |||
| 3 | 252,618 | 32 | 12.67 | |||
| Control | 4 | 299,234 | 15 | 5.01 | 4.63±2.18 | 4.39±1.99 |
| 5 | 219,300 | 5 | 2.28 | |||
| 6 | 514,849 | 34 | 6.6 |
Plaque forming unit
Results are expressed as Mean ± SD
Mutant frequency data were adjusted after the verified lacI mutant plaques were analyzed by DNA sequencing. Of 100 UVAinduced- and 54 spontaneously arisen control mutant plaques, 95 and 51, respectively, contained a minimum of one mutation along the lacI transgene. Each sample is assayed three times and the average results are presented.
Mutation spectrometry analysis showed that the vast majority of both induced- and spontaneous mutations were single base substitutions (90.5 vs 90.2%, respectively), although less frequently, there were also single and multiple base deletions and insertions, and tandem base substitutions (see Table 2). Detailed spectra of mutation produced by UVA irradiation or derived spontaneously are presented in Figure 1. To underscore the difference(s) between UVA-induced and control mutation spectra, we compared the frequency of each type of mutation, e.g., transitions, transversions, etc. between the respective mutation spectra. Because the lacI transgene in the Big Blue system is a non-transcribed gene [37], the strand bias of mutagenesis, a phenomenon caused by transcription-coupled DNA repair [40,41], is unlikely to affect the spectrum of mutations in this transgene [37]. Therefore, it is justified to combine the strand mirror counterparts of all transitions (e.g., G→A + C→T) and transversions (e.g., G→T + C→A and G→C + C→G) when comparing the specific types of mutation between different treatment groups.
TABLE 2.
Types of UVA-induced and spontaneously derived mutations in the lacI transgene in Big Blue mouse embryonic fibroblasts irradiated with 18 J/cm2 of UVA or control
| Mutation type | UVA | Control | ||
|---|---|---|---|---|
| Single mutation | 86 (90.5%) | 46 (90.2%) | ||
| Multiple mutations | 9 (9.5%) | 5 (9.8%) | ||
| Base substitution | Single | 93 (89.4%) | 48 (85.7%) | |
| Tandem | (CC→AT) | 0 | 1 (1.8%) | |
| (TT→AC) | 0 | 1 (1.8%) | ||
| Deletion | Single | 6 (5.8%) | 1 (1.8%) | |
| Multiple | 3 (2.9%) | 3 (5.4%) | ||
| Insertion | Single | 2 (1.9%) | 1 (1.8%) | |
| Multiple | 0 | 1 (1.8%) | ||
One hundred UVA-induced- and fifty-four spontaneously arisen control lacI mutant plaques were analyzed by DNA sequencing.
As shown in Table 3, the frequency of G:C→T:A transversions was significantly elevated in the UVA-induced mutation spectrum as compared to control (P < 0.003). The absolute frequency of this type of mutations was 7.4-fold increased consequent to UVA irradiation relative to control (3.38 vs 0.454 × 10-5; P < 0.00001). In addition, the UV-induced mutation spectrum showed a 3.8-fold increase in the absolute frequency of A:T→G:C transitions relative to control; however, the difference did not reach a statistically significant level (P = 0.28). The absolute frequencies of other types of mutation were variably and less extensively elevated as compared to control (see Table 3). The predominant rise in the absolute frequency of G:C→T:A transversions accounted for approximately 38% of the observed induction in the mutant frequency consequent to UVA irradiation. The combined increase in the absolute frequencies of other types of UVA-induced mutation contributed to the remainder of the observed induction (see Table 3).
TABLE 3.
Comparative mutation spectra of the lacI transgene in Big Blue mouse embryonic fibroblasts irradiated with 18 J/cm2 of UVA or control
| Mutation type | Number of Mutations | % Mutations | Absolute Mutant Frequency (× 10-6) | |||||
|---|---|---|---|---|---|---|---|---|
| UVA | Control | UVA | Control | P value | UVA | Control | Fold increase | |
| G:C→C:G | 6 | 6 | 5.8 | 10.3 | 0.4 | 6.99 | 4.54 | 1.5 |
| G:C→T:A | 29 | 6 | 27.9 | 10.3 | 0.003 | 33.80 | 4.54 | 7.4* |
| G:C→A:T | 41 | 25 | 39.4 | 43.1 | 0.7 | 47.78 | 18.92 | 2.5 |
| A:T→T:A | 6 | 6 | 5.8 | 10.3 | 0.4 | 6.99 | 4.54 | 1.5 |
| A:T→G:C | 5 | 2 | 4.8 | 3.4 | 0.9 | 5.83 | 1.51 | 3.8 |
| A:T→C:G | 6 | 7 | 5.8 | 12.1 | 0.2 | 6.99 | 5.30 | 1.3 |
| Del/Ins | 11 | 6 | 10.6 | 10.3 | 0.9 | 12.82 | 4.54 | 2.8 |
One hundred UVA-induced- and fifty-four spontaneously arisen control lacI mutant plaques were analyzed by DNA sequencing.
Del = deletion; Ins = insertion.
As compared with control; P < 0.00001
For comparison purposes, we have also determined the spontaneous and UVA-induced cII mutant frequencies in the genomic DNA of UVA-irradiated and control cells, respectively. The UVA-induced and control cII mutant frequencies were 27.2 + 0.38 and 6.0 + 0.42 × 10-5, respectively. The respective mutant frequencies reported in our previous study were 28.6 + 0.43 and 6.2 + 0.4 × 10-5 [21]. Furthermore, we have quantified the UVA-induced and control lacI mutant frequencies in the leftover genomic DNA from our above-mentioned previous study [21]. The respective mutant frequencies were 13.1 + 1.9 and 4.6 + 2.1 × 10-5, which are very comparable to the respective values obtained in the present study. The slightly higher lacI mutant frequencies quantified in the leftover genomic DNA are presumably due to the adventitious generation of DNA strand breaks during freeze-thaw process of the long-term preserved samples. Taken together, these findings are reassuring in that they clearly show that under standardized experimental conditions, the induced and spontaneous mutations quantified in both the lacI and cII transgenes (even in independent sets of experiment) are perfectly reproducible and can be used for comparative analysis.
DISCUSSION
Sunlight UV irradiation is a prime etiologic factor for human skin cancer [1-3]. The predominant UV component of the sunlight is UVA radiation [4,5]. As a known genotoxic agent, UVA radiation has been shown to induce both DNA damage and mutation [6,7]. However, the mechanism of UVA-induced mutagenesis is obscure [6,7]. A widely recognized theory raises the possibility that UVA triggers photosensitization reactions, thereby giving rise to promutagenic oxidative DNA damage, especially 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) [6,7]. Because the mutagenic potential of 8-oxo-dG is well-established, i.e., induction of G→T transversions [6], one can easily test the above-mentioned theory by investigating UVA-induced DNA damage-targeted mutagenesis.
Recently, we have shown that UVA irradiation of Big Blue mouse embryonic fibroblasts at a physiologically relevant dose of 18 J/cm2 results in a significant generation of 8-oxo-dG [21]. We have also demonstrated that the extent of 8-oxo-dG formation can be pronounced by combining the UVA irradiation with a treatment with δ-ALA [20], a precursor of the intracellular photosensitizer Pp-IX [32]. In both cases, we have observed a unique spectrum of mutations induced by UVA irradiation in this transgenic system [20,21]. The spectrum of UVA-induced mutations was characterized by a significant increase in the relative frequency of G:C→T:A transversions in the cII transgene [20,21]. In the present study, we have used a similar experimental approach to investigate the mutagenicity of UVA in a different mutational target gene, the lacI transgene, in the same model system.
Analysis of mutant frequency data revealed a 2.8-fold increase in the relative frequency of lacI mutants consequent to UVA irradiation. The elevation of relative mutant frequency in the lacI transgene is slightly lower than that previously observed in the cII transgene after irradiation of the same cells under similar conditions [21]. Our mutation spectrometry analysis revealed that G:C→T:A transversions were significantly induced by UVA irradiation (P < 0.003). The absolute frequency of this type of mutations was 7.4-fold increased consequent to UVA irradiation as compared to control (3.38 vs 0.454 × 10-5; P < 0.00001). These findings are of importance because they underline a similar mutagenicity of UVA in different mutational target genes in two separate experiments. Our overall findings obtained independently in the lacI and cII transgenes support the theory that intracellular photosensitization reactions causing promutagenic oxidative DNA damage are involved in UVA genotoxicity.
The spectrum of mutations produced by UVA irradiation in different studies has not always been compatible with the mutagenic potential of the various induced lesions [6,7]. The disparity has been more noticeable in studies where the two endpoints, i.e., DNA damage and mutation, have not been quantified simultaneously and/or within a single test system [6]. We have shown that cells of various species and types are differently resistant toward cytotoxic and genotoxic effects of UVA radiation [16,20,21]. Such differences might have arisen from the varying DNA repair capacity and diverse content of intracellular photosensitizers, specific for each species and cell type [6,7]. Additionally, experimental variables such as UVA source and dose may have contributed to some discrepancies [6]. For example, irradiation sources, which emit contaminating UV wavelengths, e.g., in the UVB range, could cause distorted induced mutation spectra as a result of UVB-specific DNA lesion formation. This is of significance because per joule basis, UVB radiation is up to 50,000 times more genotoxic than UVA radiation [42]. Ikehata et al. [26] have used an irradiation source emitting a small, yet, appreciable fraction of UVB radiation, and reported an induced mutation spectrum predominated by single and tandem C→T transitions at dipyrimidine sites in the lacZ transgene in mouse skin epidermis. Woollons et al. [43] have shown that the 0.8% UVB component of a UVA sunlamp accounted for 75% of the CPDs induced in human keratinocytes irradiated in vitro.
The relatively high detection limit of most available assays for quantification of photo-induced DNA damages - especially at the nucleotide resolution level-has necessitated intense UVA irradiation, enabling sufficient production of DNA lesions [12,14,15]. Such high doses of UVA irradiation could cause severe cytotoxicity, thereby, precluding the DNA damage to be translated into mutation through cell division [6]. We have found that the type of induced DNA lesions is determined by UVA irradiation dose, i.e., at low irradiation dose (of relevance for mutagenesis), oxidative DNA damage is predominantly formed [20,21], whereas at extremely high irradiation dose, both CPDs and oxidative DNA damage are induced [16]. It is imperative that UVA-induced DNA damage and mutagenesis be investigated simultaneously and at a biologically relevant dose. Our studies are unique in that they compare DNA damage-targeted mutagenicity of UVA in independent target genes and within a single test system, thus, controlling for the potential confounding variables.
In conclusion, we have confirmed a similar mutagenicity of UVA irradiation in the lacI and cII transgenes in Big Blue mouse embryonic fibroblasts. The characteristic DNA damage-targeted mutagenicity of UVA reaffirms the notion that intracellular photosensitization reactions causing promutagenic oxidative DNA damage are responsible for UVA genotoxicity.
ACKNOWLEDGEMENT
This work was supported by a grant from the National Institute of Environmental Health Sciences (ES06070) to G.P.P.
ABBREVIATIONS
- X-gal
5-bromo-4-chloro-3-indolyl-β—D-galactopyranoside
- 8-oxo-dGs
8-oxo-7,8-dihydro-2′-deoxyguanosines
- CPDs
cis-syn cyclobutane pyrimidine-dimers
- δ-ALA
δ-aminolevulinic acid
- DMEM
Dulbecco’s Modified Eagle’s Medium
- E. coli
Escherichia coli
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- Pp-IX
protoporphyrin IX
- UV
ultraviolet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Woodhead AD, Setlow RB, Tanaka M. Environmental factors in nonmelanoma and melanoma skin cancer. J Epidemiol. 1999;9:S102–114. doi: 10.2188/jea.9.6sup_102. [DOI] [PubMed] [Google Scholar]
- [2].de Gruijl FR. Skin cancer and solar UV radiation. Eur J Cancer. 1999;35:2003–2009. doi: 10.1016/s0959-8049(99)00283-x. [DOI] [PubMed] [Google Scholar]
- [3].Jhappan C, Noonan FP, Merlino G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene. 2003;22:3099–3112. doi: 10.1038/sj.onc.1206450. [DOI] [PubMed] [Google Scholar]
- [4].de Gruijl FR. Photocarcinogenesis: UVA vs. UVB radiation. Skin Pharmacol Appl Skin Physiol. 2002;15:316–320. doi: 10.1159/000064535. [DOI] [PubMed] [Google Scholar]
- [5].Setlow RB. The wavelengths in sunlight effective in producing skin cancer: a theoretical analysis. Proc Natl Acad Sci U S A. 1974;71:3363–3366. doi: 10.1073/pnas.71.9.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- [7].Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- [8].Cadet J, Berger M, Douki T, Morin B, Raoul S, Ravanat JL, Spinelli S. Effects of UV and visible radiation on DNA-final base damage. Biol Chem. 1997;378:1275–1286. [PubMed] [Google Scholar]
- [9].Wang SQ, Setlow R, Berwick M, Polsky D, Marghoob AA, Kopf AW, Bart RS. Ultraviolet A and melanoma: a review. J Am Acad Dermatol. 2001;44:837–846. doi: 10.1067/mjd.2001.114594. [DOI] [PubMed] [Google Scholar]
- [10].Drobetsky EA, Turcotte J, Chateauneuf A. A role for ultraviolet A in solar mutagenesis. Proc Natl Acad Sci U S A. 1995;92:2350–2354. doi: 10.1073/pnas.92.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sage E, Lamolet B, Brulay E, Moustacchi E, Chateauneuf A, Drobetsky EA. Mutagenic specificity of solar UV light in nucleotide excision repair-deficient rodent cells. Proc Natl Acad Sci U S A. 1996;93:176–180. doi: 10.1073/pnas.93.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Douki T, Perdiz D, Grof P, Kuluncsics Z, Moustacchi E, Cadet J, Sage E. Oxidation of guanine in cellular DNA by solar UV radiation: biological role. Photochem Photobiol. 1999;70:184–190. [PubMed] [Google Scholar]
- [13].Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E. Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J Biol Chem. 2000;275:26732–26742. doi: 10.1074/jbc.M001450200. [DOI] [PubMed] [Google Scholar]
- [14].Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- [16].Besaratinia A, Synold TW, Chen HH, Chang C, Xi B, Riggs AD, Pfeifer GP. DNA lesions induced by UV A1 and B radiation in human cells: Comparative analyses in the overall genome and in the p53 tumor suppressor gene. Proc Natl Acad Sci U S A. 2005;102:10058–10063. doi: 10.1073/pnas.0502311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang X, Rosenstein BS, Wang Y, Lebwohl M, Mitchell DM, Wei H. Induction of 8-oxo-7,8-dihydro-2′-deoxyguanosine by ultraviolet radiation in calf thymus DNA and HeLa cells. Photochem Photobiol. 1997;65:119–124. doi: 10.1111/j.1751-1097.1997.tb01886.x. [DOI] [PubMed] [Google Scholar]
- [18].Kvam E, Tyrrell RM. Induction of oxidative DNA base damage in human skin cells by UV and near visible radiation. Carcinogenesis. 1997;18:2379–2384. doi: 10.1093/carcin/18.12.2379. [DOI] [PubMed] [Google Scholar]
- [19].Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- [20].Besaratinia A, Bates SE, Synold TW, Pfeifer GP. Similar mutagenicity of photoactivated porphyrins and ultraviolet A radiation in mouse embryonic fibroblasts: involvement of oxidative DNA lesions in mutagenesis. Biochemistry. 2004;43:15557–15566. doi: 10.1021/bi048717c. [DOI] [PubMed] [Google Scholar]
- [21].Besaratinia A, Synold TW, Xi B, Pfeifer GP. G-to-T transversions and small tandem base deletions are the hallmark of mutations induced by ultraviolet A radiation in mammalian cells. Biochemistry. 2004;43:8169–8177. doi: 10.1021/bi049761v. [DOI] [PubMed] [Google Scholar]
- [22].Robert C, Muel B, Benoit A, Dubertret L, Sarasin A, Stary A. Cell survival and shuttle vector mutagenesis induced by ultraviolet A and ultraviolet B radiation in a human cell line. J Invest Dermatol. 1996;106:721–728. doi: 10.1111/1523-1747.ep12345616. [DOI] [PubMed] [Google Scholar]
- [23].van Kranen HJ, de Laat A, van de Ven J, Wester PW, de Vries A, Berg RJ, van Kreijl CF, de Gruijl FR. Low incidence of p53 mutations in UVA (365-nm)-induced skin tumors in hairless mice. Cancer Res. 1997;57:1238–1240. [PubMed] [Google Scholar]
- [24].Palmer CM, Serafini DM, Schellhorn HE. Near ultraviolet radiation (UVA and UVB) causes a formamidopyrimidine glycosylase-dependent increase in G to T transversions. Photochem Photobiol. 1997;65:543–549. doi: 10.1111/j.1751-1097.1997.tb08602.x. [DOI] [PubMed] [Google Scholar]
- [25].Izumizawa Y, Yang SJ, Negishi T, Negishi K. DNA lesion and mutagenesis induced in phageM13mp2 by UVA, UVB and UVC irradiation. Nucleic Acids Symp Ser. 2000:73–74. doi: 10.1093/nass/44.1.73. [DOI] [PubMed] [Google Scholar]
- [26].Ikehata H, Kudo H, Masuda T, Ono T. UVA induces C-->T transitions at methyl-CpG-associated dipyri midine sites in mouse skin epidermis more frequently than UVB. Mutagenesis. 2003;18:511–519. doi: 10.1093/mutage/geg030. [DOI] [PubMed] [Google Scholar]
- [27].Agar NS, Halliday GM, Barnetson RS, Ananthaswamy HN, Wheeler M, Jones AM. The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations: a role for UVA in human skin carcinogenesis. Proc Natl Acad Sci U S A. 2004;101:4954–4959. doi: 10.1073/pnas.0401141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ikehata H, Nakamura S, Asamura T, Ono T. Mutation spectrum in sunlight-exposed mouse skin epidermis: small but appreciable contribution of oxidative stress-mediated mutagenesis. Mutat Res. 2004;556:11–24. doi: 10.1016/j.mrfmmm.2004.06.038. [DOI] [PubMed] [Google Scholar]
- [29].Kappes UP, Runger TM. No major role for 7,8-dihydro-8-oxoguanine in ultraviolet light-induced mutagenesis. Radiat Res. 2005;164:440–445. doi: 10.1667/rr3434.1. [DOI] [PubMed] [Google Scholar]
- [30].Kozmin S, Slezak G, Reynaud-Angelin A, Elie C, de Rycke Y, Boiteux S, Sage E. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006;126:667–675. doi: 10.1038/sj.jid.5700093. [DOI] [PubMed] [Google Scholar]
- [32].Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- [33].Kohler SW, Provost GS, Kretz PL, Dycaico MJ, Sorge JA, Short JM. Development of a short-term, in vivo mutagenesis assay: the effects of methylation on the recovery of a lambda phage shuttle vector from transgenic mice. Nucleic Acids Res. 1990;18:3007–3013. doi: 10.1093/nar/18.10.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jakubczak JL, Merlino G, French JE, Muller WJ, Paul B, Adhya S, Garges S. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci U S A. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Besaratinia A, Pfeifer GP. Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis. 2005;27:1526–1537. doi: 10.1093/carcin/bgi311. [DOI] [PubMed] [Google Scholar]
- [36].Pfeifer GP, Chen HH, Komura J, Riggs AD. Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol. 1999;304:548–571. doi: 10.1016/s0076-6879(99)04032-x. [DOI] [PubMed] [Google Scholar]
- [37].Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- [38].You YH, Li C, Pfeifer GP. Involvement of 5-methylcytosine in sunlight-induced mutagenesis. J Mol Biol. 1999;293:493–503. doi: 10.1006/jmbi.1999.3174. [DOI] [PubMed] [Google Scholar]
- [39].Adams WT, Skopek TR. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987;194:391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- [40].Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- [41].Denissenko MF, Pao A, Pfeifer GP, Tang M. Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene. 1998;16:1241–1247. doi: 10.1038/sj.onc.1201647. [DOI] [PubMed] [Google Scholar]
- [42].de Gruijl FR, Sterenborg HJ, Forbes PD, Davies RE, Cole C, Kelfkens G, van Weelden H, Slaper H, van der Leun JC. Wavelength dependence of skin cancer induction by ultraviolet irradiation of albino hairless mice. Cancer Res. 1993;53:53–60. [PubMed] [Google Scholar]
- [43].Woollons A, Kipp C, Young AR, Petit-Frere C, Arlett CF, Green MH, Clingen PH. The 0.8% ultraviolet B content of an ultraviolet A sunlamp induces 75% of cyclobutane pyrimidine dimers in human keratinocytes in vitro. Br J Dermatol. 1999;140:1023–1030. doi: 10.1046/j.1365-2133.1999.02899.x. [DOI] [PubMed] [Google Scholar]