Abstract
The opportunistic pathogen Pseudomonas aeruginosa can cause acute or chronic infections in humans. Little is known about the initial adaptation of P. aeruginosa to host tissues and the factors that determine whether a P. aeruginosa-epithelial cell interaction will manifest as an acute or a chronic infection. To gain insights into the initial phases of P. aeruginosa infections and to identify P. aeruginosa genes regulated in response to respiratory epithelia we exposed P. aeruginosa to cultured primary differentiated human airway epithelia. We used a P. aeruginosa strain that causes acute damage to the epithelia and a mutant with defects in Type III secretion and in rhamnolipid synthesis. The mutant did not cause rapid damage to epithelia as did the wildtype. We compared the transcriptomes of the P. aeruginosa wildtype and the mutant to each other and to P. aeruginosa grown under other conditions, and we discovered overlapping sets of differentially expressed genes in the wildtype and mutant exposed to epithelia. A recent study reported that exposure of P. aeruginosa to epithelia is characterized by a repression of the bacterial iron-responsive genes. These findings were suggestive of ample iron availability during infection. In contrast, we found that P. aeruginosa shows an iron-starvation response upon exposure to epithelial cells. This observation highlights the importance of the iron starvation response in both acute and chronic infections and suggests opportunities for therapy.
Keywords: Acute infection, chronic infection, transcriptome, regulatory network
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that can cause clinically diverse diseases in humans with impaired immune defenses. For example, P. aeruginosa can cause acute infections such as pneumonia or bacteremia in patients with severe burns, and it can cause chronic infections in the lungs of patients with the genetic disease cystic fibrosis (CF) [1].
The adaptation of P. aeruginosa to the human host involves among other things, the expression of numerous virulence determinants. Furthermore, it appears that P. aeruginosa expresses distinct sets of virulence factors in acute and chronic infections. For example, experiments with diverse animal models implicate the Type III secretion system (TTSS) as a key virulence determinant in acute infections [2–4]. In contrast, P. aeruginosa strains isolated from chronic infections are often variants that express decreased levels of the TTSS or lack a functional TTSS [5]. Chronic P. aeruginosa infections are often biofilm infections, which are recalcitrant and rarely resolved [6]. The cues that determine bacterial commitment toward an acute or a chronic infection are not known.
Information about the factors involved in the initial adaptation of P. aeruginosa to host tissues is scarce, and currently available animal infection models do not permit a comparison of bacterial gene expression patterns in the initial phases of acute and chronic P. aeruginosa infections. Previously, Frisk et al examined the transcriptional profile of P. aeruginosa after exposure to primary normal human airway epithelial cells and found that iron-regulated genes were repressed and that the number of repressed iron-regulated genes increased over the time of exposure [7]. These findings contradict the long-held view that in the host, pathogens exist in an environment where there is intense limitation for iron [8,9]. To help resolve the conflicting reports and to identify genes regulated in P. aeruginosa upon exposure to cultured primary respiratory epithelia, we performed a transcriptome analysis using experimental conditions that closely controlled for epithelial damage. We found that the expression profiles in strains initiating chronic vs acute infections are overlapping but distinct from one another. Furthermore, the set of overlapping genes includes virtually all of the known and putative P. aeruginosa iron-responsive genes and the expression pattern of these genes is indicative of a strong iron-starvation response.
2. Results
2.1. A TTSS-rhamnolipid mutant shows attenuated epithelial cytotoxicity
Previous studies indicate the TTSS and rhamnolipids are key factors in causing damage to polarized epithelia following infection [10,11]. We first examined the ability of the wildtype and a TTSS, rhamnolipid synthesis double mutant to damage epithelial integrity. To assess epithelial integrity we measured trans-epithelial resistance (Rt). With wildtype P. aeruginosa the Rt begins to decrease within about two h of co-incubation. In comparison, epithelia infected with the mutant remain undamaged for at least 18 h (Fig. 1). Epithelia retain their integrity and there is not detectable leakage of medium from the basolateral to the apical surface until the resistance drops well below 50% of the starting resistance [33]. By using standard plate count procedures we determined that both wildtype and mutant bacteria undergo about one doubling during the initial 5 h of incubation with epithelia (data not shown). Thus exposure of epithelia to the wildtype resulted in a more acute injury then exposure to the mutant, which did not injure the epithelium rapidly. In this way the infection of epithelia with the mutant P. aeruginosa resembled a chronic infection.
Figure 1. Measurements of transepithelial electrical resistance (Rt) of airway epithelia grown in millicells and infected with the parent PAO1 or the mutant PAO-SC11.
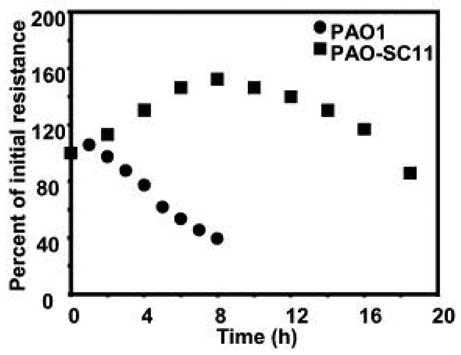
The inoculum size is ~2 × 107 CFU/millicell. Rt values are reported as averages of 5 millicells at each time point expressed as a percentage of the starting values. The range was +/−15%.
2.2. A transcriptome analysis of the P. aeruginosa response to epithelia
To identify genes regulated in the initial phase of an acute P. aeruginosa infection, we performed transcriptome analyses of wildtype PAO1 co-incubated with respiratory epithelia. For our initial comparison we used wildtype P. aeruginosa PAO1 grown in Trypticase Soy Broth (TSB) (Fig. 2). We appreciate that with these experiments the use of other media or growth conditions would alter the data, and the results would be different, but this analysis provides a basis on which to build further analyses (described below). There is not a perfect control for the experiments performed on epithelia. The bacteria on the epithelia are exposed to a thin layer of epithelium-produced fluid at an interface with the ambient atmosphere. They are not exposed to the medium for epithelial growth (Dulbecco’s Modified Eagle Medium). As described in Section 2.3 we ultimately compared our transcriptome results to many other transcriptome analyses performed under a variety of growth conditions in a variety of different media. But for the initial comparison of P. aeruginosa grown on epithelia to P. aeruginosa grown in the TSB medium used to grow bacteria used to inoculate epithelia we identified 1025 genes and 31 intergenic regions that were differentially expressed (Supplementary Table 1). Of the 1025 genes, 70% (721) were induced and 30% (304) repressed by exposure to epithelia. To identify genes regulated in the initial phase of a chronic P. aeruginosa infection (an infection that does not result in acute injury to the epithelium), transcriptome analyses were done with the TTSS, rhamnolipid synthesis mutant co-incubated with respiratory epithelia in our in vitro infection model. We identified 1570 genes and 44 intergenic regions as differentially regulated in bacteria exposed to epithelia vs bacteria grown in TSB (Supplementary Table 2). Of the 1570 genes, 58% (906) were induced and 42% (664) were repressed when co-incubated with epithelia. There were 722 genes and 23 intergenic regions common to our acute and chronic in vitro infection models (wildtype vs TTSS, rhamnolipid synthesis mutant) (Supplementary Table 3 and Fig. 3). In addition to the subset of overlapping genes, a unique set of 303 genes and 8 intergenic regions was regulated in the acute infection (Supplementary Table 4) and a unique set of 848 genes and 21 intergenic regions was regulated in the P. aeruginosa mutant (Supplementary Table 5).
Figure 2. Absolute expression profiles of genes during growth of P. aeruginosa in TSB and on epithelia.
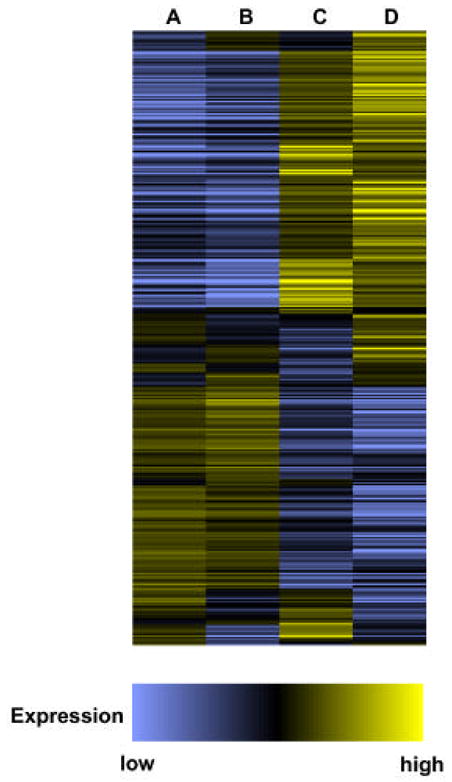
Wildtype P. aeruginosa PAO1 in TSB (A), the mutant PAO-SC11 in TSB (B), wildtype P. aeruginosa PAO1 on epithelia (C) and, the mutant PAO-SC11 on epithelia (D). The profile was generated by combining lists of genes assigned a present call in at least one of the two replicates for each growth condition. Depicted are the averaged expression values normalized to the median. Genes are displayed in the order of the hierarchical clustering of their expression profiles as determined using the Spearman corelation in GENESPRING.
Figure 3. Venn diagram showing overlaps between genes regulated in the parent PAO1 and the mutant PAO-SC11 upon exposure to epithelia.
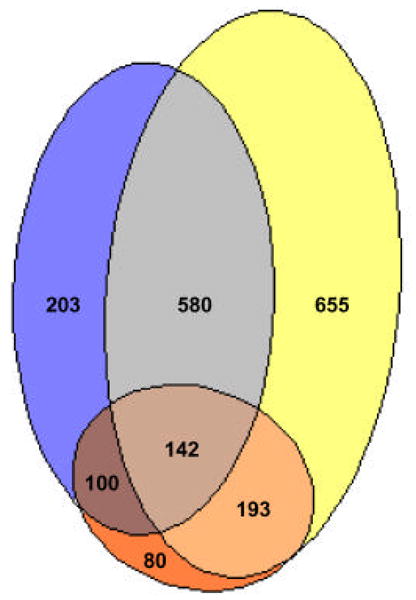
Blue, genes regulated in wildtype PAO1 exposed to epithelia versus grown in TSB. Yellow, genes regulated in the mutant PAO-SC11 exposed to epithelia versus grown in TSB. Red, genes regulated in a comparison of wildtype PAO1 exposed to epithelia and the mutant PAO-SC11 exposed to epithelia.
When we used the wildtype exposed to epithelia and the mutant exposed to epithelia for comparison we found 515 genes and 8 intergenic regions that were differentially regulated (Supplementary Table 6 and Fig. 3). Of these 40% (210) were induced and 60% (305) were repressed in the mutant. When grown in TSB we found only 48 genes that were differentially expressed in mutant vs wildtype. Of these, 28 were either known or putative members of the TTSS. Thus, with the exception of genes we expected to be influenced by the mutations only a small number were affected when cells were grown in TSB rather than exposed to epithelia. Taken together, it is apparent from the transcript profiles that compared to the parent, the mutant strain which is unable to initiate an acute infection shows significant differences in transcriptional regulation upon exposure to epithelia but not during growth in broth culture (Fig. 2).
One would expect that all genes in an operon be regulated in a similar manner. In fact, genes in known operons appear to be coregulated. For example, all genes in the hcnABC operon were repressed by epithelia in the mutant. Another observation that validates our analyses is that exoS, a TTSS gene whose expression is induced upon contact with eukaryotic host cells, was induced in the acute infection (wildtype infection) and not in the chronic infection (mutant infection).
2.3. Relationship between epithelia-regulated P. aeruginosa genes and regulons defined in previous transcriptome analyses
To gain an insight into the physiology of P. aeruginosa growing on epithelial cells we compared the sets of genes identified in our acute and chronic infection models with sets of genes identified as differentially regulated in previous microarray analyses of various P. aeruginosa strains grown under a variety of conditions. We obtained from published articles lists of quorum-regulated genes [12], genes belonging to the RpoS regulon [13], RetS-regulated genes [14], iron-responsive genes [15], SadARS-regulated genes [16], and the transcript profile of P. aeruginosa grown within the rat peritoneum in a dialysis membrane [17]. Additionally, we analyzed experiments available through the GeNet database (http://cfgenomics.unc.edu), and chose for comparison, lists of genes that met our filtering criteria as defined in the Materials and methods. Thus, we were able to compare our lists of differentially expressed genes with regulons defined using P. aeruginosa strains that were mutants in different genes; these include regulators of alginate biosynthesis (algU and mucA) (GeNet, Deretic_May03) [18], regulators of flagellar motility (fleQ, fleR, and fliA) (GeNet, Ramphal_Jun02) [19], several components of the TTSS (exsA, vfr, and cyaAB) (GeNet, Lory_Nov02) [20], and mutants in mvfR (GeNet, Rahme_Mar03) [21], and algR (GeNet, Schurr_May02) [22]. We also compared our lists with previously reported transcript profiles of P. aeruginosa grown under experimental conditions designed to identify genes regulated in response to iron availability (GeNet, Vasil_Jul02) [23], and genes regulated under nitrosative stress conditions (GeNet, Deretic_May03) [24].
In our comparisons with other analyses, we found overlaps of varying extents with genes identified in our experiments. Because our experimental protocol exposed the bacterial cells to an environment that mimics the apical surface of a lung we believe that the overlaps with responses to known stimuli are a reflection of how P. aeruginosa perceives the epithelium. The most overlap was seen with a comprehensive list of iron-responsive genes generated using a set of experiments from the GeNet database (Vasil_Jul02). Among the list of 375 genes identified as iron-responsive, 26% (98) were induced and 74% (277) were repressed in iron-depleted conditions. The subset of 98 induced genes encodes most known or putative iron acquisition and receptor systems. A comparison revealed that of this set of 98 induced genes, 82% were induced in the wildtype and 78% were induced in the mutant after 5 hours on epithelia. The iron-responsive genes induced in both the parent and mutant include genes required for synthesis of the iron binding siderophores pyoverdine and pyochlelin, membrane receptors, and transcriptional regulators. Among the 277 genes repressed in iron-depleted conditions were several genes involved in energy metabolism and iron storage. Of the 277 repressed genes, 18% were shared with genes repressed by epithelia in the wildtype and 30% were shared with genes repressed in the mutant.
3. Discussion
In this analysis we employed microarrays to examine P. aeruginosa transcript levels on a genome-wide basis in a strain incapable of initiating rapid epithelial destruction and its parent, which does cause rapid injury to the epithelium (Fig. 1). We have used primary differentiated human epithelial cultures. It is clear that the interaction between P. aeruginosa and the host is complex, involves cells other than epithelial cells, and evolves over time. The system we employed can be viewed as a model for the initial response of P. aeruginosa to the epithelial surface of the respiratory tract. We have not attempted to simulate aspects of a chronic infection that require more time to develop (for example, the organization of bacterial cells into structured biofilms). Cystic fibrosis lung infections are chronic but again, our model only reflects what might happen early in the colonization of any given region of a CF lung.
Our experiments show that there is a common response to epithelia regardless of whether we examine the strain causing acute injury or the more benign strain. There is also transcript regulation unique to one strain or the other. It is reasonable to assume some of the responses unique to one strain or the other are important in the process of bifurcation towards acute vs chronic infection. In this regard it is of interest that extracellular virulence factors like the PA-1 galactophilic lectin were induced in the wildtype strain, whereas alginate regulatory genes and gacA were induced in the mutant strain. The PA-1 galactophilic lectin is involved in acute P. aeruginosa infections and it has been reported to be induced by host factors, which are released in response to injury [25]. Alginate synthesis is a hallmark of chronic P. aeruginosa lung infections in CF [26,27]. Additionally, several gene clusters associated with motility were repressed by epithelia in the mutant P. aeruginosa. These findings correlate with the nonpiliated and nonflagellated phenotypes typical of isolates from chronic CF infections [28].
A recent study described a multicomponent signaling network that functions to regulate contrasting sets of virulence factors during acute and chronic infections [14]. This study indicates that the transcriptional regulators RetS and Vfr activate factors that promote an acute infection whereas the alternate activation of the GacS-GacA pathway promotes persistence. It is interesting to note that vfr was repressed and gacA was induced in the mutant P. aeruginosa, our chronic in vitro infection model. Several genes controlled by Vfr and GacA in vitro showed a response when exposed to the epithelia.
A significant finding was that in either the wildtype or the mutant P. aeruginosa a battery of genes previously shown to be activated by iron starvation were activated upon exposure to epithelia. This indicates that P. aeruginosa on epithelia rapidly sense an iron limitation, and it is of some importance for several reasons: As described in the Introduction, although it has long been known that supplies of iron are limiting in the host and that the success of pathogens hinges on their ability to effectively employ iron acquisition mechanisms, a recent analysis of P. aeruginosa on epithelia has brought this into question [7]. One explanation, suggested by the authors for their findings, was that iron is made available during interaction with primary normal human airway epithelial cells. We believe that a more likely explanation, and one that the authors invoke to explain the increasing number of repressed iron-responsive genes over time, is that the response of the bacteria was measured in experimental conditions where the epithelium had been breached and iron was available from the culture medium or from lysed epithelial cells. We believe that the contrasting findings between our study and that of Frisk et al might have to do with our attempt to closely control epithelial damage. We believe our experiments reflect a situation where there is minimal damage to the epithelium whereas this was likely not the case in the previous study. Either situation can occur during P. aeruginosa infections, but the initial interaction with many strains of P. aeruginosa likely does not involve significant epithelial damage. There is considerable evidence in the literature highlighting the importance of iron scavenging mechanisms of pathogens to overcome the low iron availability in the host [9]. There is also accumulating evidence that iron acquisition and utilization might be a target for therapeutic intervention. The envisioned therapies, which involve inert metals like zinc or gallium, and iron chelators, show maximal effects under iron limiting conditions.
A sorting of genes by comparing the transcriptomes of the wildtype PAO1 and the mutant strain PAO-SC11 growing on epithelia yielded a relatively small list of 515 differentially regulated genes, about 10% of the genome. This eliminated genes arising in comparisons of growth of either strain on epithelia to growth in laboratory media. Thus we saw few genes involved in metabolism or in starvation (including iron starvation) among the 515 genes differentially expressed in the mutant and wildtype on epithelia.
By comparing our data to the wealth of P. aeruginosa transcriptome data available from other studies some interesting patterns emerge that suggest the function of complex regulatory networks involving multiple inputs. For example, the hcnABC operon has been shown to be quorum-sensing induced [29] and repressed in iron depleted laboratory media (GeNet, Vasil_Jul02). What about hcnABC transcription on epithelia where many quorum-sensing genes are activated and where iron appears to be limiting? One would predict that hcnABC would be induced by exposure to epithelia but it was repressed in both the wildtype and mutant strain. This indicates hcnABC regulation is more complicated then previously believed. Likewise, the phz operon is induced by quorum sensing and one might anticipate that it would be induced with other quorum-sensing activated genes by epithelia. This is the case with the wildtype P. aeruginosa but not with the mutant. All of this points to the notion that there are epithelial cell modulators of gene expression in P. aeruginosa.
Overall, our findings suggest that P. aeruginosa has evolved a network to control the expression of alternative sets of coregulated genes in acute and chronic infections. A particularly interesting finding is that the pattern of changes of several sets of genes in our chronic infection model is reminiscent of phenotypic or genotypic changes associated with CF isolates. The trend for these changes may be set early in the chronic infection process. One might imagine that different host signals are emitted depending on the degree of injury caused by the infecting strain. Our work and the work of others [25] strongly suggest the existence of signals produced by the host that lead to the development of acute vs chronic P. aeruginosa infections. These hypothetical signals remain to be identified. We hope to develop reporters based on some of the genes we have discovered to be differentially regulated in the mutant vs the wildtype P. aeruginosa upon exposure to epithelia, and to use these reporters in efforts to identify important host-generated signals.
4. Materials and methods
4.1. Bacterial strains and growth conditions
A rhlA mutant was obtained from the P. aeruginosa Transposon Mutant Library at the University of Washington Genome Center (strain ID 54908) [30]. The mutation was moved into P. aeruginosa PAO1 to generate the strain PAO-SC10. To construct a PAO1 rhlA, exsA mutant, PCR-amplified DNA fragments flanking exsA were cloned into pEX18Tc [31] resulting in a deletion of codons 8 to 245 in exsA. This plasmid was digested with BamHI and ligated with a gentamicin-resistant (GmR)-GFP FRT cassette from pPS858 [31] to generate pSAC201. We mobilized pSAC201 from E. coli SM10 [32] into PAO-SC10 and isolated GmR colonies. A tetracycline-sensitive (Tcs) recombinant was identified and the GmR-GFP marker was deleted using pFLP2 [31]. Loss of pFLP2 following excision was confirmed by the carbenicillin-sensitive and sucrose-resistant phenotype of the mutant, PAO-SC11. The rhlA mutation was confirmed by PCR analysis and the exsA mutation was confirmed by Southern analysis. PAO-SC11 had no discernible growth defect compared with wildtype.
4.2. Culturing primary respiratory epithelia and infecting the epithelia with P. aeruginosa
Human lung epithelial cells were obtained from the Iowa Donor Network and prepared as described previously [33]. All cell layers were incubated for at least 15 days before use to allow development of morphological and functional properties of airway epithelia. Four days prior to infection with bacteria, the epithelia were transferred to antibiotic-free medium and washed daily with Dulbecco’s Modified Eagle Medium to remove antibiotics. To mimic the milieu of the apical surface of respiratory epithelia, a 24-h period was allowed between the final wash and infection. For infections, P. aeruginosa strains were grown in TSB to an optical density (OD600) of 1.0, washed once, and suspended in Dulbecco’s phosphate-buffered saline (D-PBS) (Invitrogen, cat. no. 14287). Ten μl of bacterial cell suspension (2 × 109 CFU ml−1) was applied to the apical surface of epithelium at a multiplicity of infection of approximately 25. The fluid in this small application volume is absorbed within a short time leaving the bacteria in contact with the airway cells or the normal airway surface fluid. Unless otherwise specified, the bacteria and epithelia were co-incubated for 5 h. Epithelial integrity was assessed by measuring the transepithelial electrical resistance (Rt) using an Ohm meter and 250 μl D-PBS. Only bacteria washed from epithelia that had Rt measurements ≥50% of the starting values were used for transcription profiling experiments. By using standard plate counting techniques we found that over 90% of the total bacteria on an epithelium were recovered in the wash with the remaining bacteria exhibiting a tighter binding to the epithelium.
4.3. Transcript profiling
As a baseline comparison, duplicate bacterial cultures of the wildtype P. aeruginosa PAO1 or the isogenic mutant PAO-SC11 were grown in TSB from an OD600 of 0.10 to an OD600 of 0.85 and 2×109 cells were mixed with RNA Protect Bacteria reagent (Qiagen). For infection experiments, bacteria were washed from the epithelia in 250 μl of D-PBS and the suspension was mixed with RNA Protect Bacteria reagent. RNA was isolated using RNeasy mini columns (Qiagen) according to the manufacturer’s recommendations. Bacteria pooled after washing from 9 epithelial layers yielded approximately 3.5 μg RNA. The purified RNA was treated with DNase I to ensure removal of genomic DNA followed by purification using RNeasy mini columns. Chromosomal DNA contamination was monitored by PCR as described previously [12]. To enable transcriptome analysis using low amounts of RNA, we used a modification of existing protocols for cDNA synthesis (Affymetrix protocol and a protocol suggested by M. Wolfgang, Harvard University) as described below. For the primer annealing step of the cDNA synthesis reactions we used 1.2 μg of purified RNA at a final concentration of 0.04 μg/μl and semirandom hexamer primers with an average G+C content of 75% at a final concentration of 25 ng/μl. We could synthesize about 2.5 μg cDNA from 1.2 μg RNA using our modified protocol. This allowed us to adhere to the Affymetrix recommendations for the subsequent procedure, which requires about 3 μg cDNA for GeneChip analysis. Samples were hybridized to GeneChip P. aeruginosa Microarrays (Affymetrix) at the University of Iowa Bioinformatics Core Facility. To validate our modified protocol, we performed a microarray experiment to compare expression profiles of P. aeruginosa grown in broth culture under conditions inducing or non-inducing for the TTSS. Analysis using cDNA synthesized from 12 μg RNA with the recommended protocol was compared to analysis done using cDNA synthesized from 1.2 μg of the same RNA with our modified protocol. A 98% correlation coefficient between the two analyses established the validity of our modified protocol. Replicates were biological replicates (the entire biological experiment was repeated). The microarray data have been deposited at http://www.ncbi.nlm.nih.gov/geo under the accession number GSE4614.
4.4. Analysis of microarray data
Initial data analysis was done by using the Affymetrix Microarray Software Suite (MAS) version 5.0. All genes showing expression levels above the lower detection level of the chips (present calls) in MAS 5.0 in at least one of the two conditions (baseline vs experiment) were checked for differential expression using Cyber-T (http://www.visitor.ics.uci.edu/genex/cybert/). The Bayesian prior estimate was 10, the sliding window size was 100, and the β-fit iteration value was 2. We defined the cutoff value for differential gene expression as transcripts that showed a >2.5-fold change and a t-test P value of <0.001. The posterior probability of differential expression (PPDE) (< p) value for the genes identified was >0.97.
For experiments available through the Cystic Fibrosis Foundation Therapeutics, Inc.-GeNet shared workspace we obtained the original raw data files from the Signet server (http://cfgenomics.unc.edu). Analysis was as described above except for the following modifications: For experiments with three replicates, only those genes with present calls in at least 2 out of 3 replicates in either condition (experiment vs baseline) were analyzed further using Cyber-T. The β-fit iteration value for Cyber-T analysis was 1. The cutoff value for differential gene expression was defined as transcripts that showed a >2.5-fold change and a t-test P value of <0.05. A comparison of gene expression patterns between different experiments was done by using GENESPRING (http://www.silicongenetics.com).
Supplementary Materials
Supplementary table 1: Genes regulated in wildtype PAO1 exposed to epithelial cells versus grown in TSB.
Supplementary table 2: Genes regulated in the mutant PAO-SC11 exposed to epithelial cells versus grown in TSB.19
Supplementary table 3: Genes regulated in both wildtype PAO1 and the mutant PAO-SC11 exposed to epithelial cells versus grown in TSB.
Supplementary table 4: Genes regulated in wildtype PAO1, but not in the mutant PAO-SC11, exposed to epithelial cells versus grown in TSB.
Supplementary table 5: Genes regulated in the mutant PAO-SC11, but not in wildtype PAO1, exposed to epithelial cells versus grown in TSB.
Supplementary table 6: Genes regulated in a comparison of the mutant PAO-SC11 and the wildtype PAO1 exposed to epithelial cells.
Acknowledgments
This research has been supported by grant number AI30040 from National Institutes of Allergy and Infectious Diseases. We thank Philip Karp, Tamara Nesselhauf, and Pary Weber at the University of Iowa In Vitro Models and Cell Culture Core for their generous assistance, we thank Michael Welsh for advice regarding epithelial cell biology, and we thank Yannick Lequette for advice on data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 2.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–57. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee EJ, Cowell BA, Evans DJ, Fleiszig SM. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Invest Ophthalmol Vis Sci. 2003;44:3892–8. doi: 10.1167/iovs.02-1302. [DOI] [PubMed] [Google Scholar]
- 4.Sawa T, Yahr TL, Ohara M, Kurahashi K, Gropper MA, Wiener-Kronish JP, et al. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med. 1999;5:392–8. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 5.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, et al. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol. 2004;42:5229–37. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, et al. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect Immun. 2004;72:5433–38. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 9.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–53. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 10.Holder IA, Neely AN, Frank DW. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns. 2001;27:129–30. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 11.Zulianello L, Canard C, Kohler T, Caille D, Lacroix JS, Meda P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect Immun. 2006;74:3134–47. doi: 10.1128/IAI.01772-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–79. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004;51:973–85. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 14.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–54. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Palma M, Worgall S, Quadri LE. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch Microbiol. 2003;180:374–9. doi: 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- 16.Kuchma SL, Connolly JP, O’Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187:1441–54. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–66. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firoved AM, Deretic V. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol. 2003;185:1071–81. doi: 10.1128/JB.185.3.1071-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:809–24. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 20.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–63. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 21.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M, et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 22.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J Bacteriol. 2004;186:5672–84. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–87. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 24.Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J Bacteriol. 2004;186:4046–50. doi: 10.1128/JB.186.12.4046-4050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–7. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 26.Doggett RG. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol. 1969;18:936–7. doi: 10.1128/am.18.5.936-937.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doggett RG, Harrison GM, Wallis ES. Comparison of some properties of Pseudomonas aeruginosa isolated from infections in persons with and without cystic fibrosis. J Bacteriol. 1964;87:427–31. doi: 10.1128/jb.87.2.427-431.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182:6940–9. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Proc Natl Acad Sci U S A. 2003;100:14339–44. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants . Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 32.de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 33.Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol. 2002;188:115–37. doi: 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1: Genes regulated in wildtype PAO1 exposed to epithelial cells versus grown in TSB.
Supplementary table 2: Genes regulated in the mutant PAO-SC11 exposed to epithelial cells versus grown in TSB.19
Supplementary table 3: Genes regulated in both wildtype PAO1 and the mutant PAO-SC11 exposed to epithelial cells versus grown in TSB.
Supplementary table 4: Genes regulated in wildtype PAO1, but not in the mutant PAO-SC11, exposed to epithelial cells versus grown in TSB.
Supplementary table 5: Genes regulated in the mutant PAO-SC11, but not in wildtype PAO1, exposed to epithelial cells versus grown in TSB.
Supplementary table 6: Genes regulated in a comparison of the mutant PAO-SC11 and the wildtype PAO1 exposed to epithelial cells.


