Abstract
We treated 10 children with X-linked SCID (SCID-X1) using gammaretrovirus-mediated gene transfer. Those with sufficient follow-up were found to have recovered substantial immunity in the absence of any serious adverse events up to 5 years after treatment. To determine the influence of vector integration on lymphoid reconstitution, we compared retroviral integration sites (RISs) from peripheral blood CD3+ T lymphocytes of 5 patients taken between 9 and 30 months after transplantation with transduced CD34+ progenitor cells derived from 1 further patient and 1 healthy donor. Integration occurred preferentially in gene regions on either side of transcription start sites, was clustered, and correlated with the expression level in CD34+ progenitors during transduction. In contrast to those in CD34+ cells, RISs recovered from engrafted CD3+ T cells were significantly overrepresented within or near genes encoding proteins with kinase or transferase activity or involved in phosphorus metabolism. Although gross patterns of gene expression were unchanged in transduced cells, the divergence of RIS target frequency between transduced progenitor cells and post-thymic T lymphocytes indicates that vector integration influences cell survival, engraftment, or proliferation.
Introduction
Retroviral vectors have been widely used in human HSC gene therapy trials because they stably integrate into the genome and therefore provide an opportunity for sustained clinical effect. This principle has been applied successfully to treat inherited immunodeficiencies including X-linked SCID (SCID-X1) (1–3), adenosine deaminase–deficient SCID (4–6), and, more recently, X-linked chronic granulomatous disease (7). Despite highly encouraging results, evidence has accumulated in animal and human studies that mutagenic side effects occur as a direct result of vector integration (8–12). It has therefore become of particular importance to understand the risks of harmful mutagenesis and to define the patterns of retroviral insertion that may predispose to these events.
Recent studies have shown that the distribution of retroviral integration sites (RISs) within the genome is not arbitrary and is variable in pattern depending on the nature of the virus or vector. Murine leukemia virus– (MLV-), HIV-1–, and avian sarcoma leukemia virus–based (ASLV-based) vectors exhibit quite distinct target site preferences (13). Gammaretroviral vectors and HIV-1–based lentiviral vectors both preferentially integrate into gene coding regions (14), although gammaretroviruses particularly favor a 5–kilobase pair (5-kbp) window on either side of the transcription start site (TSS) (15). In contrast, ASLV exhibits only a weak preference for genes. The mechanisms that dictate the differential integration site patterns have not been clearly elucidated, but may depend to some extent on the accessibility of euchromatin to the preintegration complex, the transcriptional activity of the locus, and binding or tethering to specific DNA sequences via host proteins at the sites of insertion (16). It is therefore likely that integration patterns may also be skewed by the nature and activation status of the target cell.
Although integration patterns are easily defined in homogeneous cell populations in vitro, the influence of integration when measured in complex in vivo situations is more relevant for our understanding of the risks of harmful mutagenesis. In HSC gene therapy, starting cell populations that are transduced ex vivo are heterogeneous, and the minority of progenitor cells among them that do engraft are subject to postengraftment influences that dictate survival, homing to appropriate microenvironmental niches, and subsequent differentiation and proliferation in vivo. As a result, substantial selection pressure may favor specific retroviral insertions if they change the expression of one or several cellular genes, thereby influencing the biological fate of a cell clone over and above (or even cooperating with) any selective advantage arising from successful expression of the vector transgene. In this study we performed high-throughput analysis to examine RISs patterns in post-thymic CD3+ T cells following successful treatment of SCID-X1 and compared them with those of freshly transduced CD34+ cells. Significant changes in RISs distribution among engrafted compared with pretransplant cell populations demonstrate that vector insertion influences the biological characteristics of a significant percentage of transplanted cells.
Results
Successful recovery of immunity following gammaretrovirus-mediated gene therapy.
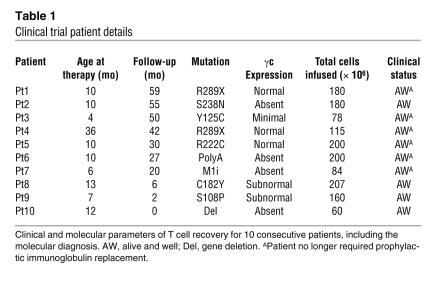
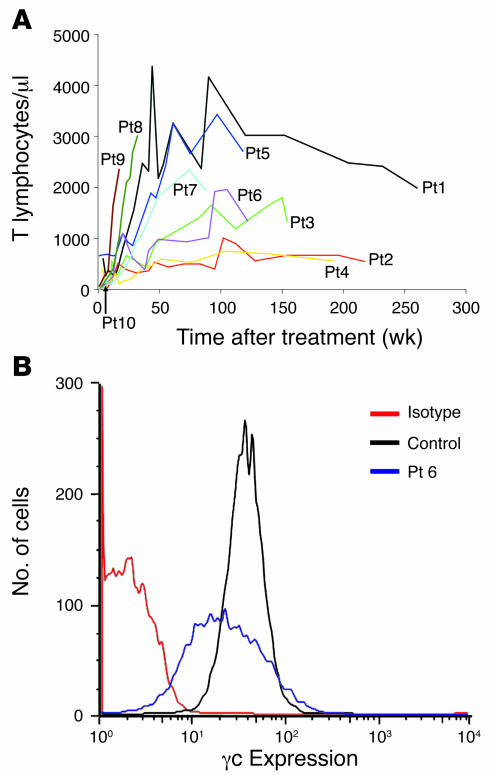
Ten patients with molecularly defined SCID-X1 were treated by retrovirus-mediated gene transfer to autologous bone marrow CD34+ progenitor cells (Table 1). Details of the gibbon ape leukemia virus–pseudotyped (GALV-pseudotyped) vector and transduction conditions have been published previously (2) and were unchanged for the duration of the study. Between 60 and 207 × 106 cells were infused in the absence of conditioning, of which 20%–60% were estimated (in patients with null mutations) to be positive for CD34 and common γ chain (γc). Where possible to evaluate, all patients experienced substantial immunological recovery, usually with normalization of T cell numbers (s 1A), TCRVβ diversity, and proliferative responses in vitro (data not shown). In most patients with sufficient follow-up, this was accompanied by recovery of humoral immunity and withdrawal of immunoglobulin supplementation. The levels of functional cell surface γc on engrafted CD3+ populations were generally less than in control cells, and there was no selection for higher expressing cells over time in any patient (Figure 1B and data not shown). The average transgene copy number in CD3+ T cells from all patients was determined by quantitative PCR on sorted populations to be 1 (data not shown). No serious adverse events were documented, and all patients are clinically well at home.
Table 1 .
Clinical trial patient details
Figure 1. Functional restoration of immunity.
(A) Lymphocyte recovery in patients after treatment in the clinical trial. CD3+ counts were obtained for each patient at regular time points after treatment. All patients demonstrated an increase in lymphocyte count, albeit varied, that was stable over time. (B) Surface expression of γc protein. Expression of γc on CD3+ T cells was determined 25 months after treatment for Pt6, who had no cell-surface γc protein before gene therapy. The isotype is a negative control, which shows levels of background fluorescence. The control, a healthy donor, demonstrates normal levels of γc expression on the cell surface.
Distribution analysis of retroviral vector insertions in transduced CD34+ cells and engrafted patient CD3+ T cells.
Linear amplification–mediated PCR (LAM-PCR) (17, 18) and high-throughput sequencing of insertion sites was performed on DNA isolated from purified peripheral blood CD3+ T cells derived from 5 patients (obtained >9 months after gene therapy) and compared with DNA from transduced CD34+ cells of patient 6 (Pt6; preengraftment sample) and of a healthy donor (transduced under identical conditions). In total, 439 unique insertion sites were isolated from postengraftment CD3+ T cell populations, of which 304 could be mapped exactly to the human genome (see Methods). Similarly, 134 and 140 unique mappable sites were isolated from transduced preengraftment CD34+ cells and transduced CD34+ cells from a healthy donor, respectively (Table 2 and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI31661DS1). The chromosomal distribution of RISs was analyzed to determine the relationships among chromosome size, gene density, and insertion frequency. For each of the human chromosomes, the number of integrations was not related to the size of the chromosome, whereas a correlation between gene content and insertion number in CD34+ progenitor cell and posttransplantation CD3+ T cells was evident (Figure 2A). This suggests that integration is dependent on the gene density of a chromosome, rather than its size. Given the number of genes in the human genome and assuming random integration, 25% of RISs would be expected to fall into or within a 10-kbp window around RefSeq genes, which account for approximately one-third of the human genome. The actual frequency of insertions within these genes (44%) and including the 10-kbp window up- and downstream (64%) was significantly higher, although similar, for all cell populations (Table 2). This indicates that in these treated cell populations, genes are preferred targets over noncoding regions for integration of MLV vectors.
Table 2 .
Genomic distribution of all mappable RISs
Figure 2. Genomic distribution of RISs.
(A) The relationship of chromosome size, number of known genes, and RIS frequency. Values for chromosome lengths are shown as a percentage of the total genome size. Values for gene density are shown as a percentage of all genes from the genome. Values of RISs are shown as a percentage of RISs from the corresponding fraction. White bars, autosome length, which was counted twice to allow for the diploid status of hematopoietic cells (X and Y chromosomes were counted once only); light gray bars, gene density of each chromosome; medium gray bars, RISs detected in CD34+ cells from a healthy donor; dark gray bars, RISs detected in pretransplant CD34+ cells from Pt6; black bars, RIS detected in patients’ engrafted cells. (B and C) RISs location related to RefSeq genes. All mappable insertions detected in different fractions are shown as a percentage of all insertions derived from the corresponding fraction. Medium gray bars, RISs derived from transduced CD34+ cells of a healthy donor; dark gray bars, RISs derived from transduced preengraftment CD34+ cells from Pt6; black bars, RISs derived from patients’ engrafted cells. RefSeq gene, RISs in gene region. (B) RISs distribution 10 kbp up- and downstream of TSSs. Up, upstream of TSSs. (C) RISs in and near gene coding regions. RISs locations inside genes are expressed as the percentage of the overall length of each individual vector targeted gene. –5 kbp, all RISs located 5 kbp upstream of TSSs; +5 kbp, all RISs located 5 kbp downstream of RefSeq genes; Down, downstream of RefSeq genes.
Recent studies in cell lines and primary cells have demonstrated a preferred integration of MLV-based gammaretroviral vectors near the region of the TSS (13, 15, 19, 20). Here, we observed a similar preference for transduced CD34+ cells from a healthy donor, transduced preengraftment CD34+ progenitors, and postengraftment CD3+ T cells (Figure 2B and Table 2): 29%, 25%, and 23% of RISs, respectively, were located within 5 kbp of the TSS. When the entire region of the targeted RefSeq gene was examined, the frequency of integration decreased with distance from the TSS (Figure 2C). As previously shown by Wu et al. for HeLa cells in vitro (15), CpG islands and a surrounding 1-kbp genomic region also represented preferred targets of vector integration, harboring 24% and 13% of all integrants derived from the patient pre- and posttransplantation samples, respectively, and 14% in transduced healthy donor CD34+ cells.
Insertions are clustered in common integration sites.
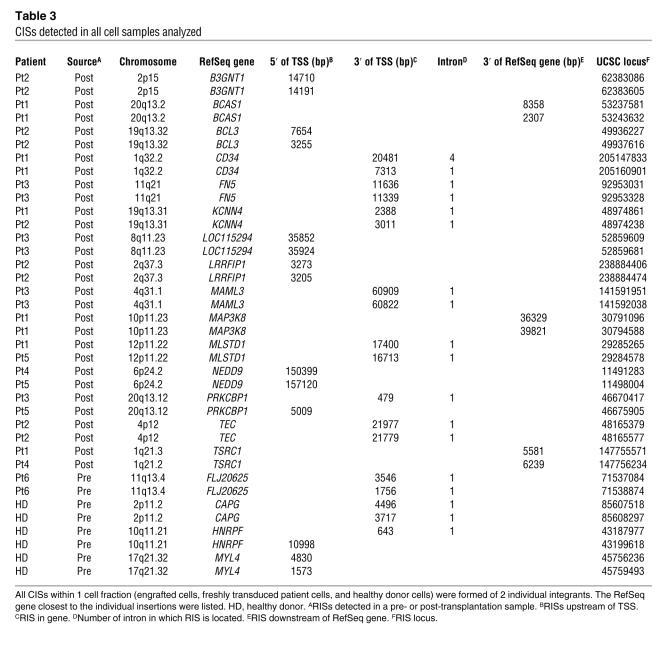
In order to define whether there was preferential insertion or selection of insertions at specific genomic loci, mapped insertions were examined for common integration sites (CISs) (Table 3). CISs have been defined as integrations into the same intergenic locus in 2 different cells or samples that are not more than 30 kbp apart from each other (21). To maintain a high degree of stringency, we followed this definition, but counted 2 integrants as a CIS including intragenic location of integrants. Accordingly, an average of 38 of the 578 (6.6%) exactly mappable RISs found in all cell populations were located in CISs (Table 3). Within the same cell fraction (engrafted CD3+ T cells, preengraftment CD34+ cells from Pt6, and healthy donor CD34+ cells), each CIS was composed of 2 RISs: 30 of 304 RISs (9.9%) from CD3+ T cells, 6 of 140 RISs (4.3%) from transduced normal CD34+ cells, and 2 of 134 RISs (1.5%) from preengraftment CD34+ cells. These findings were analyzed by performing computer simulations to compare the likelihood of random CIS occurrence based on the size of the human genome with the number of CISs detected in real samples (Supplemental Table 2). By this analysis, the number of CISs in engrafted CD3+ cells was significantly greater than expected (P < 0.0001). Also, in CD3+ T cells, 16 RISs located in CISs were intergenic, 12 RISs were intragenic, and 2 RISs were located in and near a RefSeq gene. Of the intragenic CISs, 12 RISs were detected in intron 1 of the affected genes, and 1 RIS was detected in intron 4. In normal CD34+ progenitor cells, 2 intergenic RISs, 2 intragenic RISs, and 2 intergenic/intragenic RISs were located in CISs, of which 3 RISs were located in the first intron of the genes involved. In preengraftment CD34+ cells from Pt6, only 1 CIS consisting of 2 RISs was detected in the first intron of a gene.
Table 3 .
CISs detected in all cell samples analyzed
RISs in relation to gene expression levels.
To determine whether gene expression was globally altered in postengraftment CD4+ T cells (which by default contain 1 RIS each) as a result of vector integration, expression profiles were compared with an untransduced age-matched control using an Affymetrix U133A microarray. The sensitivity of the assay was low because of the polyclonal nature of the sample, but no gross disturbance of gene expression was observed (Figure 3A). To assess whether the expression of the 96 probesets exceeding the log2 fold change >2 threshold could be caused by RISs, the distance of the RIS to the significantly expressed genes was calculated. For 77 of 96 probesets, a closest RIS could be determined. The smallest distance observed was 112 kbp, and in only 6 cases was the distance smaller than 1 Mbp. Because the largest distance reported for a RIS influencing gene expression is 90 kbp (22), we conclude that significant differences in expressed probesets in the patients likely represent individual variation rather than differential expression caused by the RIS (Figure 3B).
Figure 3. Comparative analysis of vector integration and gene expression.
(A and B) MvA plots for all probesets and probesets closest to RISs in Pt1 and healthy donor. (A) RNA expression determined by Affymetrix U133A microarray of CD3/CD28-stimulated CD4+ cells of Pt1. All 22,283 probesets on the array are shown in blue. Of these, 3,173 were significantly different in Pt1 versus control (P < 0.05, adjusted Sidak step-up; red), corresponding to 1,549 upregulated and 1,624 downregulated genes. 96 probesets (65 upregulated and 16 downregulated) genes exceeded log2 fold change of 2. None of these were associated with RISs. (B) MvA plots for 200 probesets (blue) describing 134 genes closest to RISs in Pt1. Expression in 48 probesets was significantly different (red), corresponding to 17 upregulated and 19 downregulated genes. Most differences were marginal; only 5 of these probesets — describing FLJ10986 and SPTLC2 (upregulated), and ITGAL, PDCD4, and DPH5 (downregulated) — had log2 fold changes between 1.5 and 2. (C) Comparative analysis of gene expression in CD34+ cells stimulated under transduction conditions and RISs retrieved from engrafted CD3+ cells in 5 patients and (D) comparison of gene expression in engrafted CD4+ T cells and RISs retrieved from corresponding CD3+ population of Pt1. There was a significant correlation between gene expression and the number of integration events, as expected, although less pronounced. All genes on the array were organized into 10 bins according to expression levels, and the number of integrations was calculated for each category. The number of genes in each expression level category assuming uniform random distribution is shown by horizontal line.
As MLV-based vectors are known to favor highly expressed genes for integration, we hypothesized that a correlation might exist between the gene expression profile obtained in transduced CD34+ progenitors and the genes discovered in the RIS analysis on engrafted CD3+ T cells. For a statistical analysis of this relationship, genes were organized into 10 bins according to their relative expression levels on Affymetrix U133 Plus 2.0 microarray. The number of integrations in or closest to each gene of the bins was calculated. The integration sites observed in the peripheral CD3+ T cells of 5 patients were found at a higher frequency in genes that were more highly expressed in representative purified CD34+ progenitor cell populations during transduction (P = 8.5 × 10–13 for CD34+ PBMCs; Figure 3C). Interestingly, the distribution of RISs in CD3+ T cells from a single patient, Pt1, also demonstrated a clear correlation (P = 5.7 × 10–4) with gene expression patterns in a matching CD4+ T cell population from the same patient (Figure 3D), presumably a reflection of the shared portion of the gene expression pattern of mature T cells and their immature progenitors.
Oncogenes and tumor suppressor genes.
A total of 475 genes annotated with “tumor suppressor gene” and 390 genes annotated with “oncogene” were identified from Entrez Gene and were searched for in the RISs data set, and 11 oncogenes and 14 tumor suppressor genes were found (Supplemental Table 1). STAT3, RUNX1, BCL2, and HIF1A are present in both oncogene and tumor suppressor gene categories. The T cell acute lymphocytic leukemia (T-ALL) oncogenes LMO2, TAL1, TAN1, LCK, LMO1, HOX11, HOX11L2, LYL1, TAL2, and C-MYC were not present in the RIS data sets.
Gene ontology annotation.
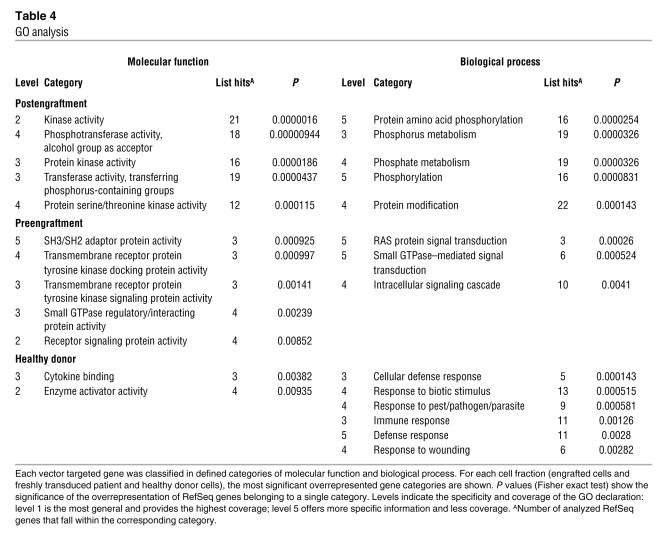
To determine the biological characteristics of RefSeq genes containing a RIS either within the gene or within the neighboring 10-kbp window, these were analyzed according to gene ontology (GO) terms. Characterization of 175 RefSeq genes derived from postengraftment CD3+ T cells revealed a significant overrepresentation of genes that encode proteins with kinase or transferase function and phosphorylation activity compared with that expected by random integration over the whole genome (Table 4). In contrast, analysis of 92 RefSeq genes from transduced normal CD34+ cells revealed that genes encoding proteins with cytokine binding or enzyme activator activity and genes involved in defense responses harbored significantly more RISs (Table 4). Analysis of 86 RefSeq genes from preengraftment CD34+ cells showed an overrepresentation of genes encoding proteins with SH3/SH2 adaptor protein activity, receptor signaling proteins, or proteins controlling GTPases as well as genes whose products are involved in biological processes like RAS protein– and small GTPase–mediated signal transduction (Table 4).
Table 4 .
GO analysis
Discussion
The RISs profile in peripheral CD3+ T cells derived from patients successfully treated by gene therapy for SCID-X1 showed interesting differences compared with transduced cells prior to transplantation. To date, most large-scale RISs analyses have been conducted in cell lines and primary cells in vitro (13–15). Although large amounts of information concerning the preferred sites of viral integration have been amassed, the data may be skewed by the use of aneuploid cells and cell lines with atypical gene expression patterns. Likewise, it is possible that the distribution of RISs determined from in vivo samples may have been strongly influenced by expansion or deletion of cells as a result of vector-specific or host contextual influences. Overall, the distribution of RISs in patient materials is consistent with earlier descriptions of preferential integration around TSSs of RefSeq genes both in vitro and in myeloid cells and transduced T cells recovered from patients in other human clinical trials (7, 23). Remarkably, the distribution of RISs in CD3+ T cells correlated well with that of transduced CD34+ progenitors, in which integration is clearly favored for highly expressed genes. This pattern therefore persists even though it is likely that the majority of pretransplantation cells have no long-term engraftment potential. Furthermore, developing T cells have undergone extensive selection within the thymus as well as post-thymic antigen-mediated clonal expansion. Gene expression patterns were also essentially identical to those of normal CD3+ T cells. It therefore appears that retroviral insertion does not cause major global disturbances, although perturbation of gene expression at a single locus would not be measurable in this setting. However, this does indicate that there are no major disturbances of T cell development or function as a result of nonphysiological regulation of transgene expression from the vector promoter and enhancer sequences.
On a more subtle level, however, GO analysis of RefSeq genes carrying insertions within a 10-kbp window revealed divergence from what would be expected from a semirandom distribution and differences between transduced CD34+ progenitor cells and postengraftment CD3+ T cells. The frequency of CISs was specifically increased in the engrafted T lymphocyte population. It therefore seems likely that within a generally conserved pattern of vector insertion, there is skewing as a result of either host- or vector-specific influences. For example, RISs that result in preferential cell survival (through homing, engraftment, or proliferative advantage) may be overrepresented and may favor the appearance of CISs. This may arise as a result of inadvertent gene activation, as has been noted in murine and nonhuman primate recipients of gammaretrovirus-transduced stem and progenitor cell populations (10, 24). In a recent clinically successful trial of gene therapy for chronic granulomatous disease, it was demonstrated by high-throughput sequencing of RISs and expression analyses that insertions in PRDM16, MDS1/EVI1, and SETBP1 led to an upregulation of these genes and conferred a selective growth advantage to affected cells without signs of malignancy (7). In this study we did not observe clonal outgrowth in vivo (over and above normal T cell receptor clonal diversity), although the occurrence of increased numbers of CISs in CD3+ T cells may be an indicator of similar effects. The profound growth advantage conferred by successful expression of γc to immature CD3– thymocytes and to all mature CD3+ T cells may, however, obscure other, more subtle influences. Consideration should also be given to the possibility that RIS patterns may be skewed in a negative way through transgene silencing, as has been previously noted for gammaretroviral vectors (25–27), although there is of course no direct evidence for this phenomenon in this study as it would preclude T cell development and compromise survival. Finally, it is possible that antigen-mediated skewing of the T cell repertoire would alter the representation of RISs, although this is unlikely because TCR diversity is highly polyclonal.
Considerable attention has been focused on the development of malignant lymphoproliferation in 3 patients treated using a similar gene therapy protocol similar to that for SCID-X1 (8). Inadvertent activation of the T cell protooncogene LMO2 contributes, at least in part, to the clonal expansion through insertional mutagenesis. We have screened a large number of randomly cloned RISs from CD3+ T cells and tracked LMO2 integrations in a total of 500 ng sample DNA derived from 3 patients corresponding to approximately 75,000 transduced cells, but have been unable to detect CISs or RISs at the LMO2 locus, which suggests that there may be true differences in the frequency of LMO2-related insertions between trials. However, the overall genomic integration site distribution (chromosomal distribution, proportion of targeting events in RefSeq genes, and preference for TSS) in both studies is very similar (28). GO analysis of targeted genes in engrafted CD3+ cells revealed an overrepresentation of the same categories in both studies, namely phosphorus metabolism and kinase and transferase activity. In contrast, the frequency of CISs in CD3+ T cells and, strikingly, in preengraftment CD34+ cells was much lower in this study, suggesting that any differences may have arisen during the transduction process, for example, as a result of differences in cytokine concentrations or vector pseudotype. To address this issue at least in part, we analyzed gene expression in CD34+ cells cultured according to each transduction protocol in the absence of vector supernatant. No significant differences were detected (data not shown), indicating that divergent cell culture conditions did not lead to major differences of gene expression that could have influenced the RIS distribution. It is not clear whether patients treated in our study are at the same risk of developing lymphoproliferation, as the follow-up period was relatively short and the majority of our patients has not reached the 34-month manifestation time point typical of the previously described lymphoproliferations. However, this outcome may also be influenced by effective engrafting cell dosage, the kinetics of T cell reconstitution, and other undefined host factors. To improve our understanding of the nature of the hematopoietic reconstitution, in future trials it will be necessary to prospectively analyze clonal composition and evolution of transduced hematopoiesis in more detail. Clonality analysis and tracking of the vector is in many ways the equivalent to the pharmacodynamic studies of regular drug development.
Our data show that even without obvious side effects, there are indeed vector insertion–induced influences on engraftment, clonal proliferation, and survival. Delineation of RISs in clinical gene therapy trials such as the present study provides important information on the biology of vectors in vivo and on the way in which they interact with host genes and environment. Ultimately this has major implications for clinical efficacy and safety as well as for rationalization of vector design.
Methods
Transduction of CD34+ progenitor cells.
CD34+ cells were purified using magnetic bead sorting (CliniMACS) from bone marrow harvested under general anesthetic. Cells were preactivated for 40 hours in the presence of cytokines (300 ng/ml SCF, 100 ng/ml TPO, 20 ng/ml IL-3, and 300 ng/ml FLT3-ligand; R&D Systems) and then transduced on 3 sequential occasions over the next 56 hours in gas-permeable cell culture containers. Serum-free conditions were maintained during the entire ex vivo culture period. The clinical-grade gammaretroviral vector (containing intact Moloney MLV long-terminal repeat [LTR] sequences) was pseudotyped with a gibbon ape leukemia virus envelope and produced in PG13 cells (BioReliance).
Trial approval and patient consent.
The gene therapy protocol was approved by the Gene Therapy Advisory Committee, London, United Kingdom; the Medicine and Healthcare products Regulatory Agency, London, United Kingdom; and the local institutional research ethics committee, Great Ormond Street Hospital for Children, NHS Research Ethics Committee, Institute of Child Health. Written informed consent was obtained from each family.
Flow cytometry for the detection of γc expression.
Of 200 μl whole blood collected in EDTA, 100 μl was stained with 5 μl of an anti-γc antibody or isotype control (BD Biosciences — Pharmingen). The stained cells were detected using fluorescence-activated cell sorting (FACSCalibur; BD). Analysis was performed using CellQuest software (version 3.3; BD) to determine the level of γc expression on CD3+ gated cells.
Preparation of DNA.
Peripheral blood samples were taken from patients 9–30 months after the reinfusion of autologous CD34+ cells transduced with a Moloney MLV retrovirus encoding the therapeutic gene as previously described (2). CD3+ cells were isolated by fluorescence-activated cell sorting (Epics Altra; Beckman Coulter). A preengraftment sample of CD34+ cells was withheld from the cells of Pt6 at the time of reinfusion, and a control sample of CD34+ cells was separated from healthy donor bone marrow cells using CliniMACS (Miltenyi) and transduced with the same protocol used in the clinical trial (2). Genomic DNA was isolated from all cells using a DNeasy kit (Qiagen).
LAM-PCR and sequence alignment.
LAM-PCR was performed on 10–100 ng of DNA isolated from sorted peripheral blood leukocytes to characterize the unknown genomic DNA flanking the 5′LTR and the 3′LTR of the vector. For LAM-PCR, 5′ biotinylated LTR-specific vector primers (Carl Roth GmbH and Co. KG) were used as follows: linear PCR (5′LTR, 5′-TGCTTACCACAGATATCCTG-3′ and 5′-ATCCTGTTTGGCCCATATTC-3′; 3′LTR, 5’-TCCGATTGACTGAGTCGC-3′ and 5′-GGTACCCGTGTATCCAATA-3′), first exponential PCR (5′LTR, 5′-GCCCTTGATCTGAACTTCTC-3′; 3′LTR, 5′-TCTTGCAGTTGCATCCGACT-3′), and second exponential PCRs (5′LTR, 5′-TTCCATGCCTTGCAAAATGGC-3′; 3′LTR, 5′-GTGGTCTCGCTGTTCCTT-3′). Linear PCR, magnetic capture, hexanucleotide priming, restriction digest (Tsp509I, MseI, or HinP1I enzymes used), linker ligation, and exponential PCRs have been previously described (17, 18). Optionally, the first exponential biotinylated PCR product was magnetically captured before reamplification by the second PCR step. LAM-PCR amplicons were either isolated and cloned (Elchrom Scientific) into the TOPO TA vector (Invitrogen) or PCR purified (Qiagen), shotgun cloned, and sequenced (GATC Biotech). Sequences were aligned to the human genome (assembly July 2003) using the University of California Santa Cruz (UCSC) BLAT genome browser (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start). Relation to annotated genome features were studied using the UCSC and Ensembl database (http://www.ensembl.org).
Analysis of the LMO2 TSS region.
A total of 40 ng of CD3+ T cell DNA from each of 3 patients (Pt1, 9 months after transplant; Pt2, 9 months after transplant; Pt3, 14 months after transplant) was analyzed for potentially dangerous integration events surrounding the TSS of LMO2. To screen the region 5 kbp upstream and downstream of the TSS and possible forward and reverse orientation of integrated vector, initial midrange PCR (PeqLab) was set up 4 different ways: upstream (LMO2 forward primer 5′-TCGTCCAAACTGAGGATCAC-3′ and biotinylated LTR forward primer LTRA1, 5′-TGCTTACCACAGATATCCTG-3′, or LTR reverse primer LTRB1, 5′-TTCAAATAAGGCACAGGGTC-3′); downstream (LMO2, 5′-CTTCCCAATTCTGCTCAAGG-3′; LTR, LTRA1 or LTRB1). After a 30-cycle PCR (initial denaturation of 94°C for 1 min, followed by 94°C for 30 s, 56°C for 45 s, 68°C for 3.5 min, and a 7-min final elongation at 68°C), final PCR products were captured via magnetic beads (Dynal) and reamplified by nested PCR (Taq polymerase; Qiagen). For the TSS upstream and downstream region, each of 5 LMO2-specific primers was applied with LTR primer LTRA2 (5′-GACCTTGATCTGAACTTCTC-3′) for vector forward orientation and LTR primer LTRB2 (5′-GTGGTCTCGCTGTTCCTT-3′) for vector reverse orientation. LMO2-specific primers for nested PCR located upstream of TSS were 5′-AGCTCTCTCACACCAGATG-3′, 5′-TACATTGCTAGCTTGCAGAC-3′, 5′-ATGCAGAGTGTCAGACTATG-3′, and 5′-GCTGGCAAAGTGGAATAGTG-3′. LMO2-specific primers downstream of TSS were 5′-CAAGTCTCCACATTCTGAGT-3′, 5′-ACAGGCCGGGCACATTGGCT-3′, 5′-CAAAGAAGAGCAGAGCTCCA-3′, 5′-GAGGATCACCTGAACTCAGA-3′, and 5′-ATCCCAGCACTTTGGGAGGC-3′.
Culturing and gene expression profiling of CD34+ and CD4+ cells.
G-CSF–mobilized peripheral blood CD34+ cells from 3 donors were transduced using conditions identical to those of the gene therapy trial (2). RNA of transduced cells was isolated using Tri Reagent (Sigma-Aldrich) according to the manufacturer’s protocol. The mRNA expression levels were determined using Affymetrix U133 Plus 2.0 arrays and normalized as described previously (29). CD4 cells were isolated from Pt1 and a healthy aged-matched control using the CD4+ T Cell Isolation kit (Miltenyi). Isolated cells were expanded for 1 week in culture in RPMI containing 2% human AB serum, 100 IU/ml IL-2, and 75 μl CD3/CD28 T cell expander Dynabeads per 106 cells (Dynal) before the RNA was isolated with Tri Reagent (Sigma-Aldrich). The RNA was then double extracted with an RNA isolation kit (Qiagen) and applied to an Affymetrix U133A microarray. The normalized expression values were used to generate MvA plots, in which average log2 expression level (A; calculated as log2 √[Expressionpatient × Expressioncontrol]) and the difference in log2 expression level (M; calculated as log2 [Expressionpatient/Expressioncontrol]) for each probeset are plotted. Significant differences in probeset expression, as determined by a Sidak step-up adjusted P value, were indicated separately (30). P values less than 0.05 were considered significant. Genes described by these probesets were retrieved from the Affymetrix HG-U133a annotation file in April 2006.
To determine the relationship between expression levels and viral integration, the normalized microarray values were sorted on expression and divided into 10 equal-sized expression level categories. The presence of the gene closest to a virus integration site as identified by LAM-PCR analysis was determined in each expression category. A Cochran-Armitage test for trend was performed to test whether higher expression level categories correspond to larger numbers of insertions (31). For each unique Gene Symbol represented on the array, the highest expression value over all probesets representing it was used for analysis.
GO analysis.
GO analysis was performed on each RefSeq gene that was vector targeted in its gene coding region or the surrounding 10-kbp genomic region using EASE software from NIH-DAVID (http://david.abcc.ncifcrf.gov/ease/ease.jsp). The database classifies each gene into defined categories of “cellular compartment,” “biological process,” and “molecular function.” Overrepresented gene categories were calculated and defined by Fisher exact test. An overrepresentation was given for P values less than 0.05 compared with the whole human genome as a background.
Supplementary Material
Acknowledgments
Funding was provided by the European Commission (5th and 6th Framework Programs, Contracts QLK3-CT-2001-00427-INHERINET and LSHB-CT-2004-005242-CONSERT), by Deutsche Forschungsgemeinschaft (DFG) grants Ka976/5-3 and Ka976/6-2, by NIH grant R01 CA 112470-01, by the Primary Immunodeficiency Association, by the Jeans for Genes Appeal, by the Chronic Granulomatous Disease Research Trust, by SPARKS, and by The Netherlands Organisation for Health Research and Development ZonMW program grant 431-00-016. S.J. Howe, M.H. Brugman, and K. Pike-Overzet are supported by European Commission contracts. F. Zhang and J. Bayford are supported by the UK Department of Health. Mike Hubank and Nipurna Jina at the Institute of Child Health provided assistance and advice for microarray experiments. A.J. Thrasher is a Wellcome Trust Senior Clinical Fellow.
Footnotes
Nonstandard abbreviations used: common γc, γ chain; CIS, common integration site; GO, gene ontology; kbp, kilobase pair(s); LAM-PCR, linear amplification–mediated PCR; LTR, long-terminal repeat; MLV, murine leukemia virus; Pt, patient; RIS, retroviral integration site; SCID-X1, X-linked SCID; TSS, transcription start site.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2241–2249 (2007). doi:10.1172/JCI31661
Kerstin Schwarzwaelder, Steven J. Howe, and Manfred Schmidt are joint first authors.
Christof von Kalle and Adrian J. Thrasher are co–senior authors.
See the related Commentary beginning on page 2083.
References
- 1.Cavazzana-Calvo M., et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar H.B., et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. . 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S., et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 4.Aiuti A., et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 5.Aiuti A., et al. Immune reconstitution in ADA-SCID after PBL gene therapy and discontinuation of enzyme replacement. Nat. Med. 2002;8:423–425. doi: 10.1038/nm0502-423. [DOI] [PubMed] [Google Scholar]
- 6.Gaspar H.B., et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol. Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Ott M.G., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina S., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 9.Li Z., et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 10.Kustikova O., et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 11.Modlich U., et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- 12.Montini E., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. . 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell R.S., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder A.R., et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 15.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 16.Bushman F.D. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M., et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. . 2002;100:2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt M., et al. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. . Nat. Med. . 2003;9:463–468. doi: 10.1038/nm844. [DOI] [PubMed] [Google Scholar]
- 19.Hematti P., et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. . 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laufs S., et al. Retroviral vector integration occurs in preferred genomic targets in human bone marrow repopulating cells. Blood. 2003;101:2191–2198. doi: 10.1182/blood-2002-02-0627. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T., et al. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 22.Bartholomew C., Ihle J.N. Retroviral insertions 90 kilobases proximal to the Evi-1 myeloid transforming gene activate transcription from the normal promoter. Mol. Cell. Biol. 1991;11:1820–1828. doi: 10.1128/mcb.11.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Recchia A., et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calmels B., et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoeben R.C., Migchielsen A.A., van der Jagt R.C., van Ormondt H., van der Eb A.J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J. Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer T.D., Rosman G.J., Osborne W.R., Miller A.D. Genetically modified skin fibroblasts persist long after transplantation but gradually inactivate introduced genes. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L., Yee J.K., Wolff J.A., Friedmann T. Factors affecting long-term stability of Moloney murine leukemia virus-based vectors. Virology. 1989;171:331–341. doi: 10.1016/0042-6822(89)90600-4. [DOI] [PubMed] [Google Scholar]
- 28.Deichmann A., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dik W.A., et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J. Exp. Med. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge Y., Dudoit S., Speed T.P. Resampling-based multiple testing for microarray analysis. Test. . 2003;12:1–77. [Google Scholar]
- 31. Armitage, P., Berry, G., and Matthews, J.N.S. 2001.Statistical methods in medical research . 4th edition. Blackwell Publishing. Malden, Massachusetts, USA/Oxford, United Kingdom. 832 pp . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.