Abstract
There is widespread aberrant expression of mature and/or precursor microRNAs in cancer cells, as microRNAs are deregulated consequent to chromosomal alterations and other genomic abnormalities. The identification of such abnormalities has a clear diagnostic and prognostic significance, and there are ever increasing examples of links between certain human cancers and modifications at microRNA loci.
The cancer-microRNA link: an old disease and a newly described mechanism
Currently, one in four deaths in the United States is due to cancer (1). Therefore, every major pathogenetic, diagnostic or therapeutic advance has the potential to save many lives. With the recent discovery of thousands of members of the class of noncoding RNAs (ncRNAs, which are genes without a significant open reading frame [ORF]), it has become evident that the genomic complexity of the cancer cell is far greater than expected. At present, cancer is considered a complex genetic disease involving structural and expression abnormalities of both coding (2–4) and noncoding oncogenes (OG) and tumor suppressor genes (TSG) (5, 6). Data accumulated in the last six years clearly show that alterations in microRNA (miRNA) genes play a critical role in cancer initiation and progression (Figure 1).
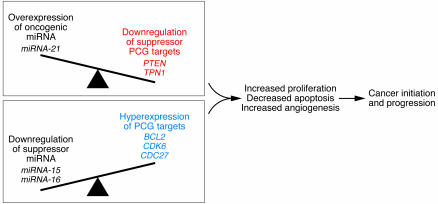
Figure 1. miRNA involvement in cancer initiation and progression.
Alterations in miRNA genes play a critical role in the pathophysiology of many, perhaps all, human cancers. Loss or amplification of miRNA genes has been reported in a variety of cancers, and altered patterns of miRNA expression may affect cell cycle and survival programs. Downregulation of suppressor miRNA is accompanied by the hyperexpression of the oncogenic PCG targets (marked in blue), while overexpression of oncogenic miRNA is followed by downregulation of suppressor PCG targets (marked in red). The two paradigms presented in this Review, miR-15a/miR-16-1 cluster and miR-21, are shown in correlation with their main identified targets.
The genetic identification of hot spots for chromosomal abnormalities showed that miRNAs, a small ncRNA class of genes, frequently reside in such genomic regions (7, 8). ncRNAs range in size from 19–24 nt for the large family of miRNAs that modulate development in several organisms including mammals, up to more than 10,000 nt for RNAs involved in gene silencing in higher eukaryotes (9). In 1993 came the first descriptions of miRNAs in Caenorhabditis elegans (10, 11). Over 4,500 miRNAs have been identified in the last six years in vertebrates, flies, worms, plants, and viruses (12–14). Naturally occurring miRNAs are 19–24 nt in length cleaved from 60- to 110-nt hairpin precursors that are produced from large primary transcripts. Many miRNAs are conserved in sequence between distantly related organisms, suggesting that these molecules participate in essential processes. The functions of miRNAs are highly varied, ranging from the control of leaf and flower development in plants to the modulation of hematopoietic lineage differentiation in mammals (15). Several groups have uncovered roles for miRNAs in the coordination of cell proliferation and cell death during development as well as in stress resistance and fat metabolism (12, 13, 16). Functionally, it was shown that miRNAs reduce the levels of many of their target transcripts and of their target translation (17).
Variations in the chromosomal numbers and structural aberrations have been systematically reported over the past five decades (18). The plethora of non-random chromosomal abnormalities consistently reported in malignant cells of hematopoietic and solid tumors (19–22) facilitates the identification of cancer-associated protein-coding OGs and TSGs (2, 23, 24). One classic paradigm is represented by the translocation t(9;22), which is identified in nearly every patient with chronic myelogenous leukemia, in which a large portion of chromosome 22 is moved to the long arm of chromosome 9 and a very tiny portion of the latter chromosome sticks to chromosome 22. This was the first-described chromosomal abnormality in human cancers (19, 25), and it results in the production of a fusion protein tyrosine kinase known as BCR-ABL (26). The findings that imatinib mesylate (Gleevec), a drug that inhibits this kinase and others, is highly effective in the treatment of chronic myelogenous leukemia clearly proved the therapeutic significance of the molecular dissection of the karyotype abnormalities in human cancers (27). Among the BCR-ABL expression–dependent genes is miR-17-92, an oncogenic miRNA polycistron; in BCR-ABL–positive cells, the expression of the six miRNAs contained in the cluster are specifically downregulated several fold by both imatinib and RNA interference against the fusion protein (28). This example further proves that there is an interplay between ncRNAs and protein-coding genes (PCGs) in the pathogenesis of human cancers (5, 8, 29).
Causes of disturbed miRNA expression: a mix of chromosomal abnormalities, epigenomics, and processing
A substantial body of evidence supports the proposal that at least some common chromosomal fragile sites (FRAs) predispose to DNA instability in cancer cells (30, 31). Indeed, FRAs are preferential sites of sister chromatid exchange, translocation, deletion, amplification, or integration of plasmid DNA and tumor-associated viruses such as human papilloma virus. More than half of miRNA genes are located in cancer-associated genomic regions (CAGRs), including minimal regions of loss of heterozygosity (LOH) and minimal regions of amplification, described in a variety of tumors including lung, breast, ovarian, colon, gastric, and hepatocellular carcinoma as well as leukemias and lymphomas (7). In addition, looking at 113 FRAs scattered in human karyotypes, 61 miRNAs were found to be located in the same cytogenetic positions within FRAs (7).
If the location of miRNAs is relevant to tumorigenesis, then structural or functional alterations of miRNAs should be identified in various types of cancers. A growing number of reports are providing such evidence and suggest that abnormal expression of miRNAs is central to cancer pathogenesis (6, 29, 32–41). The combination of non-random chromosomal abnormalities and other types of genetic or epigenetic events could contribute to the downregulation or overexpression of miRNAs (Figure 1). It is logical to ask why this association between CAGR and miRNAs is of significant importance for tumorigenesis. The answer could be that genomic aberrations in primary tumors preferentially involve the genomic loci where miRNAs reside (7, 42). As a consequence, the clones that host genomic aberrations within miRNA loci could have biological advantages.
Several arguments suggest that the highly significant association between the location of miRNAs and chromosomal or molecular genomic aberrations is not without consequences. Expression profiles of 241 human miRNAs in normal tissues and in the NCI-60 panel of human tumor–derived cell lines showed that certain miRNAs identified as potential TSGs or OGs are located in CAGRs known to be important in various malignancies (43). Furthermore, an extensive study of high-resolution array–based comparative genomic hybridization on 227 human ovarian cancer, breast cancer, and melanoma specimens clearly proved that regions hosting miRNAs exhibit high-frequency genomic alterations in human cancer (42). Strengthening the importance of these findings is the fact that miRNA copy changes correlate with miRNA expression. The analyses of the same histotype, breast ductal carcinoma, performed by two independent groups using distinct techniques revealed overlapping sets of miRNAs differentially expressed (44) and of DNA copy number gains or losses (42) compared with those found in normal breast tissue.
Recent data proved the existence of epigenetic regulation of miRNA expression. DNA hypomethylation, CpG island hypermethylation, and histone modifications represent epigenetic markers of malignant transformation (45). It has been demonstrated that miR-127, which is embedded in a CpG island exhibiting imprinting in mice, is physiologically expressed in normal fibroblasts but strongly silenced and/or downregulated in cancer cells. It was further shown that this silencing was mediated by hypermethylation of the putative miRNA promoter region and could be reversed only by the combination of a DNA demethylating agent (5-aza-2′-deoxycytidine) and a histone deacetylase inhibitor (4-phenylbutyric acid) (46). A recent study determined the miRNA expression profiling in cancer cells genetically deficient for the DNA methyltransferase enzymes (DNMTs) and for de novo (DNMT3B) and maintenance (DNMT1) gene-promoter methylation and identified miR-124a as being silenced by hypermethylation of the promoter. The epigenetic loss of this miRNA was functionally linked to the activation of cyclin D kinase 6 (Cdk6), a bona fide oncogenic factor, and to the phosphorylation of the TSG retinoblastoma (47).
miRNA processing involves a large number of genes and proteins, and only some of them have been identified. The long primary transcripts transcribed by the RNA polymerase II are processed by two ribonuclease IIIs. One is located in the nucleus and called “Drosha” and the other, called “Dicer,” is located in the cytoplasm. The final miRNA duplex is incorporated into a large protein complex called “RNA-induced silencing complex”, the core of which includes components of the argonaute protein family (48). Recently it was found that the impaired miRNA processing enhances cellular transformation and tumorigenesis (49). In accordance with this, conditional deletion of Dicer1 enhances tumor development in a K-Ras–induced mouse model of lung cancer (49). A large study of non–small cell lung cancer patients found that Dicer, but not Drosha, expression levels were reduced in a fraction of lung cancers and correlated with shortened postoperative survival and tumoral undifferentiated state, which is a mark of poor prognosis (50, 51). As a possible explanation of these results, Dicer expression is limited to prognostically favorable, noninvasive bronchioloalveolar carcinoma (52).
Chromosomal translocations target miRNA loci
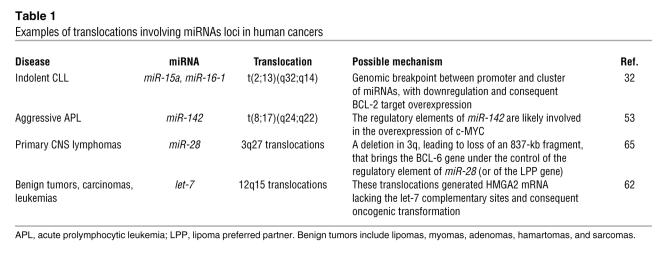
The first report linking a chromosomal breakpoint with the genomic location of miRNAs was published a couple of decades ago (53) (Table 1). A masked t(8;17) translocation resulted in a high activation of the MYC OG: MYC from chromosome 8 was truncated at the end of the first exon (which is noncoding), and the coding region joined the regulatory elements of a gene located on chromosome 17, called BCL3 (B cell leukemia/lymphoma 3). Despite extensive genomic search, BCL3 remained an elusive entity until the identification of the human miRNAs. Fifteen years after the initial discovery, the miR-142 gene was found to be located 50 nt from the t(8;17) break involving chromosome 17 and MYC, meaning that the regulatory elements of this miRNA are likely involved in the overexpression of MYC (32). The clinical consequences were dramatic for the patient, leading to aggressive acute prolymphocytic leukemia (53). Apart from the involvement in the t(8;17) breakpoint of B cell acute leukemia, miR-142-3p and miR-142-5p are also within the 17q23 minimal amplicon described in breast cancer (54) and near the FRA17B site, a target for HPV16 integration in cervical tumors (7).
Table 1 .
Examples of translocations involving miRNAs loci in human cancers
Despite careful examination of the genome, some chromosomal abnormalities had unknown targets, and the mechanisms by which these changes affected certain specific disorders remained unrevealed. For example, overexpression of BCL-2 protein was reported in many types of human cancers including leukemias, lymphomas, and carcinomas (55). In follicular lymphomas and in a fraction of diffuse B cell lymphomas, the mechanism of BCL2 activation was found to be the translocation t(14;18)(q32;q21), which places BCL2 gene under the control of immunoglobulin heavy-chain enhancers, resulting in deregulated expression of the gene (56, 57). In the most frequent adult leukemia in the Western world, the B cell chronic lymphocytic leukemia (CLL) (58), the malignant, mostly nondividing B cells of CLL overexpress BCL-2 protein (59). However, with the exception of less than 5% of cases in which BCL2 is juxtaposed to immunoglobulin loci (60), no mechanism was discovered to explain BCL-2 deregulation in CLL. The answer to this unsolved puzzle came from a decade of studies of the most frequent deleted genomic region in CLL at chromosomal band 13q14.3. Among these chromosomal aberrations, a t(2;13)(q32;q14) translocation in a CLL patient and a small deletion in a patient with CLL, bilateral retinoblastoma, and ulcerative colitis pinpointed the location of the genes involved in CLL (32) (Figure 2 and Table 1). By analyzing these patients, a 30-kb region of deletion between exons 2 and 5 of the LEU2 gene was identified and a cluster of two noncoding miRNA genes, miR-15a and miR-16-1, was found to be located precisely in this region (Figure 2) (32). Thus the miR-15/miR-16 family was proven to negatively regulate BCL-2 expression and promote apoptosis contributing to malignant transformation by upregulating BCL-2 in CLL cells (61).
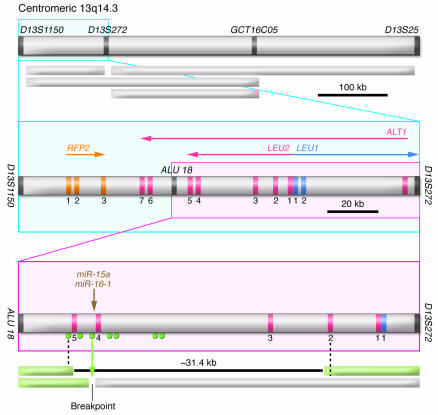
Figure 2. The t(2;13)(q32;q14) translocation involving miR-15a and miR-16-1 in indolent CLL.
The genomic map of the 13q14.3 locus between ALU 18 and D13S272 markers is shown. Exons for LEU2/ALT1 and LEU1 are indicated by numbers. The brown arrow marks the position of miR-15a and miR-16-1 genes. Green circles mark the positions of PCR primers used to screen somatic cell hybrid clones derived from a fusion of two independent leukemia cases. The green arrow represents the position of the breakpoint in CLL carrying a t(2;13)(q32;q14) translocation. Green boxes represent portions of chromosome 13 present in the hybrids. The 31.4-kb deletion was present in a clone derived from a patient with CLL, bilateral retinoblastoma, and ulcerative colitis. Figure modified from Proceedings of the National Academy of Sciences of the United States of America (32).
Mechanisms of miRNA alterations by translocations
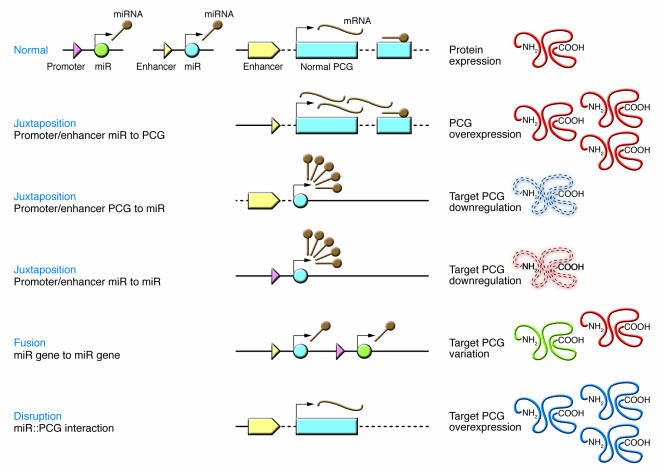
Chromosomal translocations alter PCG loci through two main mechanisms (23). The first is the juxtaposition of promoter/enhancer elements from one gene to the intact coding region of another gene, while the second is the recombination of the coding regions of two different genes. The former is more frequently found in B and T cell lymphomas and leukemias and the latter in human myeloid leukemias and soft-tissue sarcomas. The translocations that alter miRNA loci can be classified by analogy with these mechanisms (Figure 3). At least five different situations can be postulated, the last three of which have yet to be identified in human cancers: (a) juxtaposition of promoter/enhancer elements from miRNA genes to a PCG ORF with overexpression of the protein [e.g., t(8;17)(q24;q22)]; (b) disruption of the region of interaction between the target PCG and the interactor miRNA with the disruption of the repression and the overexpression of the protein (e.g., 12q15 translocations involving HMGA2 gene); (c) juxtaposition of promoter/enhancer elements from PCG to a miRNA gene with overexpression of the noncoding gene; (d) juxtaposition of promoter/enhancer elements from miRNA to another miRNA gene with overexpression of the noncoding gene (termed “promoter swapping”); and (e) miRNA gene–to–miRNA gene fusion with the consequent production of a “new” cluster of coexpressed or independently expressed miRNAs.
Figure 3. miRNA deregulation by chromosomal translocations.
The main mechanisms (some identified in patient samples [refs. 32, 53, 62, 64, 65], others only shown in the case of PCGs altered by translocations) of miRNA deregulation are schematically presented. On the left side, the primary genomic alteration is presented, while on the right side, the correspondent consequence at proteomic levels is shown. miR, miRNA gene.
One interesting mechanism of oncogenic transformation was recently proved in the cases of chromosomal translocations involving HMGA2 (62, 63). It was reported that the TSG let-7 represses the HMGA2 OG, a high mobility group protein with oncogenic properties in benign mesenchymal tumors and lung cancers. The effect was dependent on multiple target sites in the 3′ untranslated region (3′UTR), and the growth-suppressive effect of let-7 on lung cancer cells was rescued by overexpression of HMGA2 ORF without a 3′UTR (63). Another group showed that disruption of the pairing between let-7 and HMGA2 by translocations enhanced oncogenic transformation (62). Chromosomal translocations previously shown to generate a truncated version of HMGA2 mRNA lacking the let-7 complementary sites of the wild-type mRNA were described in benign tumors, carcinomas, and leukemias. This disrupted repression of HMGA2 promoted anchorage-independent growth, conferring support for a new mechanism of oncogenesis by translocations: the loss of miRNA-directed repression of a protein-coding OG (Figure 3).
Other miRNA genes are located near breakpoint regions. miR-180 is only 1 kb from the MN1 gene involved in a t(4;22) chromosomal translocation in meningioma that inactivates MN1 and possibly the miRNA gene located in the same position. Also, in a patient with precursor B cell acute lymphoblastic leukemia, an insertion of miR-125b-1 into a rearranged immunoglobulin heavy-chain locus was described, possibly as an early step in leukemogenesis (64). Chromosomal translocations fusing the BCL-6 OG to the regulatory elements of miR-28 or to the lipoma preferred partner were described in primary central nervous system lymphomas and may be associated with aberrant somatic hypermutation or defective class switch recombination (65).
The cluster miR-15a/miR-16-1 as a paradigm for suppressor miRNAs
Several groups have used positional cloning in order to identify the gene or genes targeted by the 13q14.3 deletions in CLL. A region of more than 1 Mb has been fully sequenced and characterized in detail (66) and various and relatively large (between 130 and 550 kb) minimal regions of LOH have been described (Figure 2). Until 2001, a total of 11 genes had been identified and screened for alterations at the DNA and/or RNA level in sporadic and familial cases of CLL. However, detailed genetic analysis including extensive LOH, mutation, and expression studies failed to demonstrate the consistent involvement of any of the genes located in the deleted region (32, 67). Further detailed investigation of the 13q14.3 deletions showed that both members of an miRNA cluster, miR-15a and miR-16-1, are deleted or downregulated in approximately 68% of CLL cases as compared with healthy donors (32). Furthermore, a rare mutation lowering the expression of these genes was identified in two CLL patients including one from a family with individuals having CLL and breast cancer, and was found to be associated with the loss of the normal allele in the leukemic cells (68). It was shown that the levels of both miR-15 and miR-16 inversely correlate with the BCL-2 protein expression and that BCL-2 repression by these miRNAs induces apoptosis in leukemia cells (61). Strongly supporting the involvement of miR-16 mutations in cancer predisposition, a recent study showed that in NZB mice (a model for human CLL), the region of synteny with mouse D14mit160 is the human 13q14 region, which contains miR-15a/miR-16-1. DNA sequencing of multiple NZB tissues identified a point mutation in the same 3′ flanking sequence of miR-16-1 as identified in human CLLs that was not present in other strains, including the nearest neighbor, NZW. Levels of miR-16 were decreased in NZB lymphoid tissue, and exogenous miR-16 delivered to an NZB malignant B-1 cell line resulted in cell cycle alterations and increased apoptosis. Linkage of the miR-15a/miR-16-1 complex to the development of CLL in this spontaneous mouse model suggests that the altered expression of these genes is the molecular lesion in CLL (69).
Expanding the spectrum of tumors in which these genes act as TSGs, the same cluster of genes was found to be downregulated in breast cancer (70). Functional studies performed in solid cancer cell lines showed that miR-16 negatively regulated cellular growth and cell cycle progression. miR-16–downregulated transcripts were enriched with genes whose silencing by small interfering RNAs causes an accumulation of cells in G0/G1. Simultaneous silencing of these genes was more effective at blocking cell cycle progression than was disruption of the individual genes. Thus, miR-16 coordinately regulates targets that may act in concert to control cell cycle progression (71) (Figure 1).
miR-21 and miR-155 as paradigms for oncogenic miRNAs
The only miRNA found to be overexpressed in any type of solid tumor analyzed (breast, colon, lung, prostate, stomach, and endocrine pancreas tumors, glioblastomas, and uterine leiomyomas) is miR-21 (70, 72–74). This gene is located in the 3′UTR of the vacuole membrane protein 1 (VMP1) gene at chromosome 17q23.2, a region frequently found amplified in neuroblastomas and breast, colon, and lung cancers. Knockdown of miR-21 in glioblastoma cell lines induces a caspase-mediated apoptosis, further supporting the oncogenic role of this miRNA (75). miR-21 exerts its carcinogenetic function at least in part by targeting a phosphatase and tensin homolog (PTEN; mutated in multiple advanced cancers), a TSG that normally inhibits the survival and growth promoting activity of PI3K signaling (76) (Figure 1).
To better evaluate the role of miR-21 in tumorigenesis, studies in MCF-7 breast cancer cells transfected with anti–miR-21 oligonucleotides showed that both cell growth in vitro and tumor growth in the xenograft mouse model were suppressed. Furthermore, this anti–miR-21–mediated cell growth inhibition is associated with increased apoptosis and decreased cell proliferation, which could be in part due to downregulation of the antiapoptotic BCL-2 in anti–miR-21–treated tumor cells (77). Further strengthening the oncogenic role of this miRNA in breast cancer, miR-21 was shown to target the TSG tropomyosin 1 (TPM1), and overexpression of TPM1 in breast cancer MCF-7 cells suppressed the anchorage-independent growth. All these data taken together suggest that downregulation of TPM1 by miR-21 may explain, at least in part, why suppression of miR-21 can inhibit tumor growth (78).
Clear proof that deregulation of a single miRNA gene can lead to cancer was provided with the identification of pre–B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in miR-155 transgenic mice (79). This miRNA is frequently overexpressed in several hematopoietic and solid cancers (70, 80–82). This widespread cancer deregulation suggests a critical role for this miRNA in mammalian cells, and in fact, by generating knockout mice, miR-155 was shown to be essential for normal immune function (83) and for the regulation of the germinal center response (84).
miRNAs and cancer predisposition: the missed link?
Cancer predisposition has been studied by thousands of scientific groups, but we remain ignorant about the genetic basis in the majority of familial cases. miRNAs represent ideal candidates for cancer predisposition genes, since they work in a very particular way: a small variation in expression of a specific miRNA has effects on tens or hundreds of target mRNAs, with possible functional consequences. Therefore, inherited variations in miRNA expression could represent the basis for the predisposition in various types of familial cancers with unknown pathogenesis, such as familial CLL or familial prostate cancers (5). Few hints in favor of this hypothesis have been found in the short time since the link between miRNAs and cancers was discovered (32). For example, a mutation in the primary transcript of miR-16 was found in a patient with familial CLL and familial breast cancer and was shown to induce a lower expression of the gene (68). Linkage of the miR-15a/miR-16-1 mutations and the development of a CLL-like disease in a spontaneous mouse model suggest that the altered expression of miR-15a/miR-16-1 is the likely the main molecular lesion in CLL (69).
Recent data further support the role of miRNAs as candidate genes for cancer risk and suggest that miRNAs could be responsible for susceptibility to solid tumors (85). By comparing the genome positions of mouse tumor susceptibility loci with those of mouse miRNAs, a statistically significant association was found between the chromosomal location of miRNA and that of the mouse cancer susceptibility loci that influence the development of solid tumors. Furthermore, distinct patterns of flanking DNA sequences for several miRNAs located at or near susceptibility loci were identified in inbred strains with different tumor susceptibilities exist (85). Certainly, further cloning of ncRNAs located at chromosomal breakpoints will identify new ncRNA genes involved in cancers, while linkage analyses in large families affected by cancer will offer the final proof of miRNA involvement as cancer-predisposing genes.
Abnormal miRNA expression in cancer can be clinically exploited
The identification of chromosomal translocations and other genomic abnormalities has a clear diagnostic and prognostic significance (5, 8). In the near future more examples of links between certain human cancers and alterations at miRNA loci are likely to be described. The identification of alterations that possibly affect miRNA loci by various cytogenetic techniques could be of help for diagnosis or prognosis. For example, deletions or translocations involving the miR-15a/miR-16-1 locus at 13q14.3 in CLL patients (easily detected by cytogenetics) could signal a good prognosis, as 13q14 deletions are associated with the longest treatment-free interval (86). On the other hand, profiling of CLL samples could identify down-regulation of the members of miR-29 and miR-181 families, recently shown to target the TCL1 OG whose overexpression is associated with a poor prognosis (87). In this way, miRNA profiling could represent a good indicator of discrimination between patients with a promising prognosis versus those with a poor prognosis. Further strengthening this point, a common signature of only 13 miRNAs out of about two hundred could discriminate the two distinct types of CLL (68). The development of new miRNA markers in the near future will represent one of the main goals in molecular medicine as miRNA expression profiles better classify poorly differentiated tumors as compared with the mRNA-based (expressed sequence tag–based) classifiers (88). Furthermore, cancer-specific miRNA fingerprints were identified in every type of analyzed tumor, benign or malign, including breast carcinoma (44), primary glioblastoma (72), hepatocellular carcinoma (89), papillary thyroid carcinoma (90, 91), lung cancer (70, 92, 93), gastric carcinoma, colon carcinoma (94, 95), endocrine pancreatic tumors (70), pituitary adenomas (96), and uterine leiomyomas (74).
miRNAs as new targets of gene therapy
The potential for using miRNAs and/or their antisense inhibitors in cancer treatment has only recently been envisioned; no clinical trials have begun or even have been scheduled. Patient-specific therapeutic “drugs” could be designed in the future in individuals who harbor abnormalities in the miRNA genes in their tumors. miRNAs might be targets for therapy, as knocking down overexpressed miRNAs or expressing silenced miRNAs in cancer cells might contribute to tumor killing.
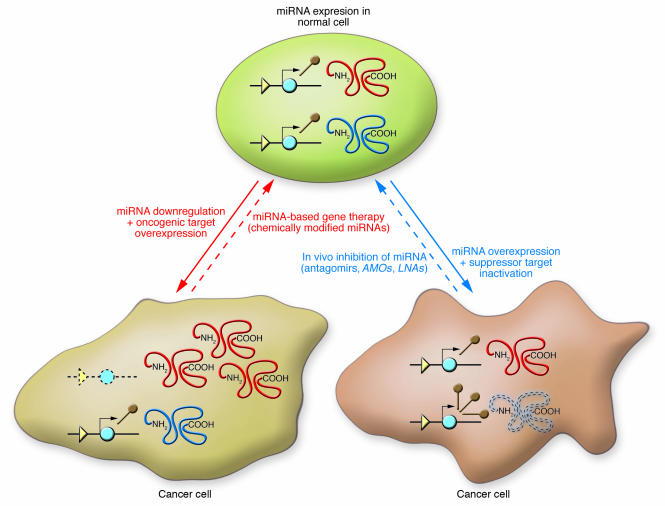
Several new therapeutic agents are under investigation at present (Figure 4). Among these are the nucleic acids that are antisense to the miRNA sequence named as anti-mRNA oligonucleotides (AMOs). By base pairing with the miRNA, these agents impaired the interaction between an overexpressed miRNA and the target mRNAs (97). Different modifications of the RNA backbone into 2′-O-methyl group or 2′-O-methoxyethyl group prevented nuclease degradation in the culture media and, importantly, also prevented endonucleolytic cleavage by the RNA-induced silencing complex nuclease, leading to irreversible inhibition of the miRNA. Locked nucleic acids (LNAs) represent nucleic acid analogs containing one or more LNA nucleotide monomers with a bicyclic furanose unit locked in an RNA-mimicking sugar conformation (98). LNA is thought to increase the functional half-life of miRNAs in vivo by enhancing the resistance to degradation and by stabilizing the miRNA-target duplex structure that is crucial for silencing activity (99). Finally, efficient specific and effective silencers of miRNA expression in mice were recently developed (100). The chemically engineered oligonucleotides, named “antagomirs,” represent single-stranded 23-nucleotide RNA molecules complementary to the targeted miRNAs modified to increase the stability of the RNA and protect it from degradation (101). Development of antagomirs against oncogenic miRNAs such as miR-17-92, miR-21, or miR-155 and future in vivo experiments of miRNA transgenics and knockout mice will offer valuable information regarding safety and efficiency. Although exciting, the use of miRNAs/anti-miRNAs in human cancer therapy should still meet the most difficult criteria: high efficiency of target inhibition with a significant increase in patient survival and reduced toxic effects.
Figure 4. Principles of miRNA-based gene therapy and in vivo inhibition of miRNA in cancer cells.
The major types of new miRNA drugs or manipulations toward the therapeutic goal of reverting the malignant phenotype are presented in conjunction with the main miRNA abnormalities found in human cancers. AMOs, anti-mRNA oligonucleotides; LNAs, locked nucleic acids.
Acknowledgments
C.M. Croce is supported by program project grants from the National Cancer Institute. G.A. Calin is supported by a Kimmel Scholar Award, the CLL Global Research Foundation, and an MD Anderson Trust grant. We thank Muller Fabbri for the critical reading of the manuscript. We apologize to many colleagues whose work was not cited due to space limitations.
Footnotes
Nonstandard abbreviations used: CAGR, cancer-associated genomic region; CLL, chronic lymphocytic leukemia; FRA, fragile site; LOH, loss of heterozygosity; miRNA, microRNA; ncRNA, noncoding RNA; OG, oncogene; ORF, open reading frame; PCG, protein-coding gene; TSG, tumor suppressor gene; UTR, untranslated region.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2059–2066 (2007). doi:10.1172/JCI32577.
George A. Calin’s present address is: Department of Experimental Therapeutics and Department of Cancer Genetics, University of Texas, MD Anderson Cancer Center, Houston, Texas, USA.
References
- 1.Jemal A., et al. Cancer statistics, 2007. CA Cancer J. Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Bishop J.M. Molecular themes in oncogenesis. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Cooperation between oncogenes. Cell. 1991;64:249–270. doi: 10.1016/0092-8674(91)90637-e. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg R.A. Tumor suppressor genes. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 5.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A., Slack F.J. Oncomirs — microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Calin G.A., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin G.A., Croce C.M. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 9.Storz G., Altuvia S., Wassarman K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 10.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 16.Harfe B.D. MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev. 2005;15:410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Lim L.P., et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 19.Nowell P.C., Hungerford D.A. Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 20.Rowley J.D. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 21.Croce C.M., Nowell P.C. Molecular basis of human B cell neoplasia. Blood. 1985;65:1–7. [PubMed] [Google Scholar]
- 22.Croce C.M. Role of chromosome translocations in human neoplasia. Cell. 1987;49:155–156. doi: 10.1016/0092-8674(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 23.Russo G., et al. Molecular analysis of a t(7;14)(q35;q32) chromosome translocation in a T cell leukemia of a patient with ataxia telangiectasia. Cell. 1988;53:137–144. doi: 10.1016/0092-8674(88)90495-3. [DOI] [PubMed] [Google Scholar]
- 24.Rabbits T.H. Chromosomal translocation master genes, mouse models and experimental therapeutics. Oncogene. 2001;20:5763–5777. doi: 10.1038/sj.onc.1204597. [DOI] [PubMed] [Google Scholar]
- 25.Rowley J.D. Chromosome translocations: dangerous liaisons revisited. Nat. Rev. Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 26.Perrotti D., Calabretta B. Translational regulation by the p210 BCR/ABL oncoprotein. Oncogene. 2004;23:3222–3229. doi: 10.1038/sj.onc.1207543. [DOI] [PubMed] [Google Scholar]
- 27.Kantarjian H.M., Talpaz M., Giles F., O’Brien S., Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann. Intern. Med. 2006;145:913–923. doi: 10.7326/0003-4819-145-12-200612190-00008. [DOI] [PubMed] [Google Scholar]
- 28.Venturini L., et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 29.Calin G.A., Croce C.M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 30.Arlt M.F., Casper A.M., Glover T.W. Common fragile sites. Cytogenet. Genome Res. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- 31.Richards R.I. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 32.Calin G.A., et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McManus M.T. MicroRNAs and cancer. Semin. Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 34.Berezikov E., Plasterk R.H. Camels and zebrafish, viruses and cancer: a microRNA update. Hum. Mol. Genet. 2005;14:R183–R190. doi: 10.1093/hmg/ddi271. [DOI] [PubMed] [Google Scholar]
- 35.Gregory R.I., Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 36.Croce C.M., Calin G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 37.Caldas C., Brenton J.D. Sizing up miRNAs as cancer genes. Nat. Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 38.Chen C.Z. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 39.Calin G.A., Garzon R., Cimmino A., Fabbri M., Croce C.M. MicroRNAs and leukemias: how strong is the connection? Leuk. Res. 2005;30:653–655. doi: 10.1016/j.leukres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Hwang H.W., Mendell J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L., et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaur A., et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 44.Iorio M.V., et al. microRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 45.Fraga M.F., Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:144–148. doi: 10.4161/cc.4.10.2113. [DOI] [PubMed] [Google Scholar]
- 46.Saito Y., et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Lujambio A., et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 48.Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 50.Karube Y., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris K.S., Zhang Z., McManus M.T., Harfe B.D., Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2005;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiosea S., et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 53.Gauwerky C.E., Huebner K., Isobe M., Nowell P.C., Croce C.M. Activation of MYC in a masked t(8;17) translocation results in an aggressive B-cell leukemia. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8867–8871. doi: 10.1073/pnas.86.22.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlund M., et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 55.Sanchez-Beato M., Sanchez-Aguilera A., Piris M.A. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101:1220–1235. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- 56.Tsujimoto Y., Finger L.R., Yunis J., Nowell P.C., Croce C.M. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 57.Tsujimoto Y., Cossman J., Jaffe E., Croce C.M. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 58.Chiorazzi N., Rai K.R., Ferrarini M. Chronic lymphocytic leukemia. N. Engl. J. Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 59.Kitada S., et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 60.Adachi M., Tefferi A., Greipp P.R., Kipps T.J., Tsujimoto Y. Preferential linkage of bcl-2 to immunoglobulin light chain gene in chronic lymphocytic leukemia. J. Exp. Med. 1990;171:559–564. doi: 10.1084/jem.171.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimmino A., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee Y.S., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonoki T., Iwana E., Mitsuya H., Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–2010. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 65.Schwindt H., et al. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J. Neuropathol. Exp. Neurol. 2006;65:776–782. doi: 10.1097/01.jnen.0000229988.48042.ae. [DOI] [PubMed] [Google Scholar]
- 66.Bullrich F., et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001;61:6640–6648. [PubMed] [Google Scholar]
- 67.Migliazza A., et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2098–2104. doi: 10.1182/blood.v97.7.2098. [DOI] [PubMed] [Google Scholar]
- 68.Calin G.A., et al. A unique microRNA signature associated with prognostic factors and disease progression in B cell chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 69.Raveche E.S., et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volinia S., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linsley P.S., et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ciafre S.A., et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 73.Krichevsky A.M., King K.S., Donahue C.P., Khrapko K., Kosik K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang T., et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- 75.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 76.Meng F., et al. Involvement of human microRNAs in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 77.Si M.L., et al. miR-21-mediated tumor growth. Oncogene. 2006;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 78.Zhu S., Si M.L., Wu H., Mo Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 79.Costinean S. Pre B cell proliferation and lymphoblastic leukemia/high grade lymphoma in Emiu miR 155 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Metzler M., Wilda M., Busch K., Viehmann S., Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 81.Eis P.S., et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kluiver J., et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 83.Rodriguez A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thai T.H., et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 85.Sevignani C., et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dohner H., et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 87.Pekarsky Y., et al. 2006Tcl1 expression in CLL is regulated by miR-29 and miR-181. Cancer Re s. 6611590–11593. [DOI] [PubMed] [Google Scholar]
- 88.Lu J., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 89.Murakami Y., et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 90.He H., et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weber F., Teresi R.E., Broelsch C.E., Frilling A., Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J. Clin. Endocrinol. Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 92.Takamizawa J., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 93.Yanaihara N., et al. microRNA signature in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 94.Michael M.Z., O’ Connor S.M., van Holst Pellekaan N.G., Young G.P., James R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 95.Cummins J.H., et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bottoni A., et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J. Cell. Physiol. 2007;210:370–377. doi: 10.1002/jcp.20832. [DOI] [PubMed] [Google Scholar]
- 97.Weiler J., Hunziker J., Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2005;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 98.Orom U.A., Kauppinen S., Lund A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 99.Naguibneva I., et al. An LNA-based loss-of-function assay for micro-RNAs. Biomed. Pharmacother. 2006;60:633–638. doi: 10.1016/j.biopha.2006.07.078. [DOI] [PubMed] [Google Scholar]
- 100.Krutzfeldt J., et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 101.Krutzfeldt J., et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892.. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]