Abstract
Transcription factor-7–like 2 (TCF7L2) is the most important type 2 diabetes susceptibility gene identified to date, with common intronic variants strongly associated with diabetes in all major racial groups. This ubiquitous transcription factor in the Wnt signaling pathway was not previously known to be involved in glucose homeostasis, so defining the underlying mechanism(s) will provide new insights into diabetes. In this issue of the JCI, Lyssenko and colleagues report on their human and isolated islet studies and suggest that the risk allele increases TCF7L2 expression in the pancreatic β cell, reducing insulin secretion and hence predisposing the individual to diabetes (see the related article beginning on page 2155).
Over 170 million people in the world can blame their type 2 diabetes, at least in part, on their genes. It has been hoped for over 2 decades that identifying the “guilty” genes would help us to understand the fundamental pathophysiology of this common and important disorder. Now, at last, not only are common gene variants being reproducibly associated with type 2 diabetes, but work such as that of Lyssenko and colleagues, reported in this issue of JCI, is turning this genetic information into novel biological insights (1).
Early genetic studies in type 2 diabetes
Early attempts to identify the genes responsible for type 2 diabetes were slow and unsuccessful: faced with 30,000 suspects, geneticists were only able to examine less than 5% and, in most cases, the coverage of the gene and sample size were too small to detect modest effects. The choice of genes studied was primarily based on evidence that these genes played biologically important roles in glucose homeostasis. By the end of 2005, despite considerable research throughout the world, only 2 polymorphisms were considered guilty beyond a reasonable doubt of predisposing to type 2 diabetes: P12A in PPARG (2) and E23K in KCNJ11 (3). One advantage of using biological candidacy to choose genes for further study was that we already knew the critical science of the proteins encoded by these genes — the nuclear transcription factor PPARγ and the potassium inward-rectifying 6.2 subunit (Kir6.2) of the potassium ATP channel. Both genes were diabetes drug targets, and mutations in both could cause monogenic diabetes (4, 5). This meant that once the association with disease was established, understanding the associated pathophysiology was relatively straightforward. However, the very reasoning that led to the genes being chosen also meant there was not a lot of new scientific insights to be learned from identifying these two genes.
TCF7L2: the most important type 2 diabetes gene
At the start of 2006, transcription factor-7–like 2 (TCF7L2) was revealed as an unexpected suspect for a type 2 diabetes gene by the DECODE group in Iceland (6). This gene had initially drawn attention during follow-up research on a small linkage signal on chromosome 10, but it turned out that, despite not explaining this linkage, multiple polymorphisms within the gene showed strong association with diabetes in multiple cohorts. The initial study was rapidly followed by widespread replication not only in white Europeans (7) but also in Indian and Japanese people (8–10), Mexican Americans (11), and West Africans (12) — representing the major racial groups with a high prevalence of type 2 diabetes. In all populations, TCF7L2 showed strong association, with the odds of developing type 2 diabetes being increased by 30%–50% for each allele inherited — approximately double the odds ratio seen with most other diabetes susceptibility polymorphisms.
The tracking of criminals and the tracking of genes have both been greatly helped by new technologies. Because of technological advances, the majority of common genetic variations can be assessed on a single chip at a very reasonable cost. This directly led to a whole new series of large-scale genome-wide genetic studies. As with many other polygenic conditions, this approach has been dramatically successful in studying type 2 diabetes, and within a few months, the number of established associated polymorphisms increased from 3 to 9 (13–17). A key result was that TCF7L2 polymorphisms have been most strongly associated with type 2 diabetes in the initial scan in 4 of the 5 recently published genome-wide scans (13–17).
Defining the mechanism by which TCF7L2 alleles predispose to diabetes
Defining the biological functions of polygenes found through genetic approaches can be very hard. Calpain 10 was the first type 2 diabetes susceptibility gene to be defined through linkage rather than a candidate gene route (18). Calpain 10 had not been previously thought to be involved in the pathogenesis of diabetes; it showed initial association with intronic SNPs, and replication required large studies (19, 20). We now recognize that these three characteristics are typical of the majority of type 2 diabetes susceptibility genes, and this may mean that the biological function of such genes will be difficult to define. In the case of Calpain 10, it was 5 years before the gene was shown to play a role in apoptosis in pancreatic islets (21).
TCF7L2 polymorphisms are clearly guilty of predisposing to type 2 diabetes on the basis of strong, reproducible association in multiple populations and would appear to be the leader among a gang of susceptibility genes. The next challenge, as with all genome-wide scans, is to define how the polymorphism predisposes to disease. The associated TCF7L2 haplotype was in the noncoding region of the gene without obvious function in gene regulation, so it was uncertain how or even whether such variants alter TCF7L2 expression. What is the critical variant of the large number of polymorphisms that are coinherited as a haplotype? Is the risk variant altering the gene it is situated in, or does it have a distant regulatory function? What biological pathway are the altered gene or genes acting in and how does this predispose to diabetes? These are the fundamental questions that need to be answered if we are to move forward from the genetic association to gain new insights into diabetes.
It is the hypothesis-free results from genome-wide association studies that have the potential to create major breakthroughs in our understanding of disease, but there are intrinsic difficulties in working on these leads. Most polymorphisms are in genes on which there has been little previous work, and the leading scientists working in cell biology and rodent models already have funding for worthy work in other areas, so why should they risk time and money working on a gene whose role is not 100% certain?
The difficulty had been trying to place TCF7L2 at the scene of the crime, especially as there was some doubt regarding which organ and cell type(s) were involved in the pathophysiology. TCF7L2 is a transcription factor involved in the Wnt signaling pathway and is ubiquitously expressed. The studies from Lyssenko and colleagues (1) confirm earlier studies (22–25) showing that the predisposition to type 2 diabetes is the result of reduced insulin secretion rather than reduced insulin action, making the pancreatic β cell the most likely primary cell target of altered TCF7L2 activity. However, this was contrary to an initial, much-repeated hypothesis, suggesting that the risk genotype was altering insulin secretion indirectly by reducing intestinal TCF7L2 activity, which in turn reduced the secretion of incretins, glucagon intestinal peptide (GIP), and glucagon-like peptide 1 (GLP-1) (6). Lyssenko et al. (1), in their detailed studies, show that insulin secretion in subjects with the at-risk genotype was reduced in response to a variety of stimuli including i.v. glucose and arginine and not just oral glucose. In addition, GIP levels were not reduced, suggesting that even though GLP-1 levels were not measured, there was a reduced β cell response to incretin secretion rather than reduced incretin secretion.
The final question is how exactly the “crime” is performed within the β cell — and here there is a final twist in the story. Lyssenko et al. (1) show that TCF7L2 expression was increased 5-fold in type 2 diabetes islets, rather than being reduced. This vital and surprising observation came from studies of pancreatic islets carefully purified from type 2 diabetic and nondiabetic human cadavers. In addition, there was some suggestion that in the nondiabetic islets that the risk genotype was associated with increased TCF7L2 expression, but the numbers are small and caution must be exercised in interpreting these data until a greater number of islets are examined. Finally, in a reconstruction of the crime, overexpressing TCF7L2 in human islets using an adenovirus system reduced insulin secretion. As with much of science that has been reported in the study of type 2 diabetes, there are bits of the story that do not fit: insulin gene mRNA was positively correlated with TCF7L2 mRNA, out of keeping with the reduced insulin secretion observed, and the overexpression of TCF7L2 did not result in the increased glucagon secretion seen in the type 2 diabetic islets.
Conclusion
So the interim verdict is that TCF7L2 risk alleles predispose to type 2 diabetes by crimes against the β cell (Figure 1). The risk allele results in overexpression of TCF7L2 in the pancreatic β cell, which reduces insulin secretion in response to a variety of stimuli (1). The reduced insulin secretion in turn explains the increased hepatic glucose production observed. There are still many unanswered questions: Is the concentration of TCF7L2 protein increased in the β cell in addition to TCF7L2 RNA? How is expression increased by intronic variants? How does increased TCF7L2 expression reduce insulin secretion? This is the first in a series of steps toward understanding the associated pathophysiology. In the end, what is desired from scientific breakthroughs is improved prevention of type 2 diabetes and improved treatment of those who develop the disease. We are still a long way from this, but there is now a new β cell pathway to be further investigated to see if it can be manipulated by drugs or lifestyle changes.
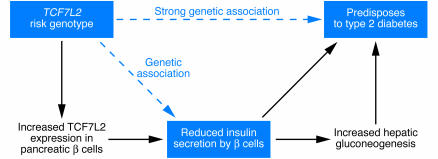
Figure 1. From genetic association to pathophysiology in TCF7L2 genotypes predisposing to type 2 diabetes.
Diagram of proposed pathophysiological pathway explaining how TCF7L2 risk genotypes predispose to type 2 diabetes. The risk genotype results in overexpression of TCF7L2 in pancreatic β cells, which in turn results in reduced insulin secretion. Reduced insulin secretion results in a predisposition to type 2 diabetes directly and also indirectly by increasing hepatic glucose output. Dotted arrows represent previous genetic associations. Solid arrows show observations reported by Lyssenko and colleagues in the current issue of the JCI (1).
Acknowledgments
Andrew T. Hattersley is a Wellcome Trust Research Leave Fellow.
Footnotes
Nonstandard abbreviations used: TCF7L2, transcription factor-7–like 2 gene.
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2077–2079 (2007). doi:10.1172/JCI33077.
See the related article beginning on page 2155.
References
- 1.Lyssenko V., et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. . J. Clin. Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altshuler D., et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 3.Gloyn A.L., et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell K-ATP channel subunits Kir6.2 (KCNJ11) and SUR1 ABCC8) confirm that the KCNJ11 E23K variant is associated with Type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 4.Barroso I., et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 5.Gloyn A.L., et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 6.Grant S.F., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 7.Zeggini E., McCarthy M.I. TCF7L2: the biggest story in diabetes genetics since HLA? Diabetologia. 2007;50:1–4. doi: 10.1007/s00125-006-0507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandak G.R., et al. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50:63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T., Iwamoto Y., Kaku K., Hirose H., Maeda S. Replication study for the association of TCF7L2 with susceptibility to type 2 diabetes in a Japanese population. Diabetologia. 2007;50:980–984. doi: 10.1007/s00125-007-0618-z. [DOI] [PubMed] [Google Scholar]
- 10.Horikoshi M., et al. A genetic variation of the transcription factor 7-like 2 gene is associated with risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50:747–751. doi: 10.1007/s00125-006-0588-6. [DOI] [PubMed] [Google Scholar]
- 11.Lehman D.M., et al. Haplotypes of transcription factor 7-like 2 (TCF7L2) gene and its upstream region are associated with type 2 diabetes and age of onset in Mexican Americans. Diabetes. 2007;56:389–393. doi: 10.2337/db06-0860. [DOI] [PubMed] [Google Scholar]
- 12.Helgason A., et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 13.Zeggini E., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sladek R., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 15.Scott L.J., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena R., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 17.Steinthorsdottir V., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 18.Horikawa Y., et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 19.Weedon M.N., et al. Meta-analysis and a large association study confirm a role for calpain-10 variation in type 2 diabetes susceptibility. Am. J. Hum. Genet. 2003;73:1208–1212. doi: 10.1086/379285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchiya T., et al. Association of the calpain-10 gene with type 2 diabetes in Europeans: results of pooled and meta-analyses. Mol. Genet. Metab. 2006;89:174–184. doi: 10.1016/j.ymgme.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J.D., et al. RyR2 and calpain-10 delineate a novel apoptosis pathway in pancreatic islets. J. Biol. Chem. 2004;279:24794–24802. doi: 10.1074/jbc.M401216200. [DOI] [PubMed] [Google Scholar]
- 22.Saxena R., et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 23.Freathy R.M., et al. Type 2 diabetes TCF7L2 risk genotypes alter birth weight: a study of 24,053 individuals. Am. J. Hum. Genet. 2007;80:1150–1161. doi: 10.1086/518517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Florez J.C., et al. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N. Engl. J. Med. 2006;355:241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loos R.J., et al. TCF7L2 polymorphisms modulate proinsulin levels and beta-cell function in a British Europid population. Diabetes. 2007;56:1943–1947.. doi: 10.2337/db07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]