Abstract
Gene transfer into HSCs is an effective treatment for SCID, although potentially limited by the risk of insertional mutagenesis. We performed a genome-wide analysis of retroviral vector integrations in genetically corrected HSCs and their multilineage progeny before and up to 47 months after transplantation into 5 patients with adenosine deaminase–deficient SCID. Gene-dense regions, promoters, and transcriptionally active genes were preferred retroviral integrations sites (RISs) both in preinfusion transduced CD34+ cells and in vivo after gene therapy. The occurrence of insertion sites proximal to protooncogenes or genes controlling cell growth and self renewal, including LMO2, was not associated with clonal selection or expansion in vivo. Clonal analysis of long-term repopulating cell progeny in vivo revealed highly polyclonal T cell populations and shared RISs among multiple lineages, demonstrating the engraftment of multipotent HSCs. These data have important implications for the biology of retroviral vectors, the dynamics of genetically modified HSCs, and the safety of gene therapy.
Introduction
Transplantation of genetically modified HSCs is an effective treatment for inherited blood disorders, such as SCID resulting from lack of common γ chain (γc) receptor (X-linked SCID; SCID-X1) (1, 2), adenosine deaminase–deficient SCID (ADA-SCID) (3), and chronic granulomatous disease (CGD) (4). Gammaretroviral vectors derived from the murine leukemia virus (MLV) are widely used to deliver therapeutic genes into human HSCs (5) because of their capacity to stably integrate into the cell genome. In addition to their therapeutic function, stably integrated proviruses represent a unique biomarker to study the biology, dynamics, and clonality of HSCs and their progeny after transplantation. Studies in human cell lines (6–8) and animal models (9–11) have indicated that MLV-derived vectors integrate in a nonrandom fashion into the host genome, favoring transcriptionally active genes, CpG islands, and transcription start sites (TSSs) (12). Recent studies have associated retroviral integrations at sensitive genomic sites to clonal expansion of hematopoietic progenitors. Retroviral insertions leading to activation of the MDS1-EVI1 locus have been described in murine models (13, 14) and in dominant hematopoietic clones after gene therapy (GT) for CGD (4). The occurrence of leukemia-like lymphoproliferative disorders in 3 patients with SCID-X1 treated by GT (15) has been associated with insertional activation of the protooncogene LMO2 (16), although there is increasing evidence that the γc gene product might have contributed to the establishment or progression of the malignancies (17, 18).
We showed previously that transplantation of autologous, MLV-transduced CD34+ HSCs combined with low-dose busulphan conditioning results in immunological and metabolic reconstitution in patients with ADA-SCID, without requirement for enzyme replacement therapy (3). ADA is a housekeeping enzyme of the purine metabolic pathway (19), and its constitutive expression provides an appropriate model for assessing the role of vector-mediated insertional oncogenesis in human HSCs. On the other hand, the selective growth advantage of ADA+ lymphocytes in an ADA-deficient context (20) might favor retrovirally transduced cells containing insertions that allow sustained transgene expression.
Here, we provide a genome-wide analysis of retroviral integration sites (RISs) in BM-derived, ADA-deficient CD34+ cells before transplantation and in their myeloid and lymphoid progenies 1.5–47 months after transplantation into 5 patients with ADA-SCID. Our data revealed similar patterns of integration into the human genome before and after transplantation, with a preference for gene-dense regions, TSSs, and highly expressed genes. In addition, they allowed us to gain information on the clonality and dynamics of the transplanted cells that ultimately affect the safety and long-term efficacy of GT for ADA-SCID.
Results
Analysis of RISs in hematopoietic cells from patients with ADA-SCID.
A genome-wide analysis of RISs was carried out in BM-derived CD34+ cells before transplantation and in their myeloid and lymphoid progenies 1.5 to 47 months after autologous transplantation into 5 patients with ADA-SCID (Pt1–Pt5). The patients received 0.9 to 9.4 × 106 BM CD34+ cells/kg transduced with an ADA-expressing MLV vector after low-dose conditioning with busulphan, as previously described (3). All patients showed multilineage engraftment of ADA-transduced hematopoietic cells, which resulted in immunological and metabolic correction with marked clinical improvement. The proportion of vector-containing cells ranged from 70% to 100% in T cells and from 0.7% to 16% in granulocytes, with the exception of Pt2, who displayed low myeloid engraftment (<0.1%; ref. 3 and A. Aiuti, unpublished observations). No adverse events related to gene transfer were observed in a follow-up with a median of 3.1 years (A. Aiuti, C. Bordignon, and M.G. Roncarolo, unpublished observations).
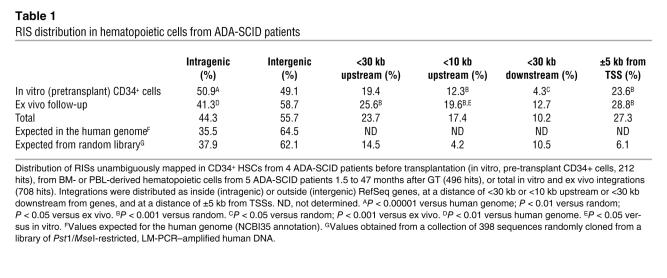
Vector-genome junctions were cloned from pretransplant transduced CD34+ cells (in vitro) or hematopoietic cells after GT (in vivo) by inverse or linker-mediated PCR (LM-PCR), sequenced and mapped onto the human genome. Overall, 212 in vitro and 496 in vivo RISs could be unambiguously assigned to a chromosomal position (for the complete list of RISs, see Supplemental Data Files 1–3; supplemental material available online with this article; doi:10.1172/JCI31666DS1). The distribution of RISs with respect to RefSeq genes is shown in Table 1. RISs within the transcribed portion of genes were more represented in the in vitro than in the in vivo sample (50.9% versus 41.3%, P = 0.03), at a frequency not significantly different than that observed in a collection of 398 control sequences randomly cloned by LM-PCR (37.9%). Exons were rarely hit, both in vivo (2.0%) and in vitro (0.5%), in agreement with the overall occupancy of coding sequences in the human genome (1.8%). RISs less than 30 kb upstream of a gene were overrepresented with respect to those less than 30 kb downstream and were more frequent in vivo than in vitro in the 10-kb region upstream of the TSS (Table 1). In addition, both samples contained a high proportion of RISs landing within a 5-kb window on either side of the nearest TSS (Table 1 and Figure 1A) and showed a distribution significantly different from random after normalization for gene length (Supplemental Figure 1A).
Table 1 .
RIS distribution in hematopoietic cells from ADA-SCID patients
Figure 1. RISs favor TSSs and gene-dense regions.
(A) Distribution of RISs within a 30-kb window around TSSs. Each bar corresponds to the frequency of integration sites retrieved by LM-PCR technique in pretransplant CD34+ cells (in vitro, n = 212) or posttransplant hematopoietic cells (in vivo, n = 251) at 5-kb intervals from the TSS of the nearest gene. (B) Correlation between RISs and gene density in the human genome within a 1-Mb window. The number of RefSeq genes was determined in a region of 500 kb on either side of the RIS for both in vitro and in vivo samples. The data are compared with the average gene density of the human genome.
A strong preference of RISs for gene-dense chromosomal regions was observed in both pre- and posttransplant samples, with 66.1% and 70.6% of RISs, respectively, landing in regions containing greater than 10 genes/Mb compared with the expected frequency of 24.7% (P < 0.001; Figure 1B). Accordingly, RISs retrieved from both samples were unevenly distributed in human chromosomes, and their frequencies directly correlated with the number of genes per chromosome (Supplemental Figure 1B).
LMO2 is a recurrent RIS in CD34+ cells, but is not associated with clonal selection in vivo.
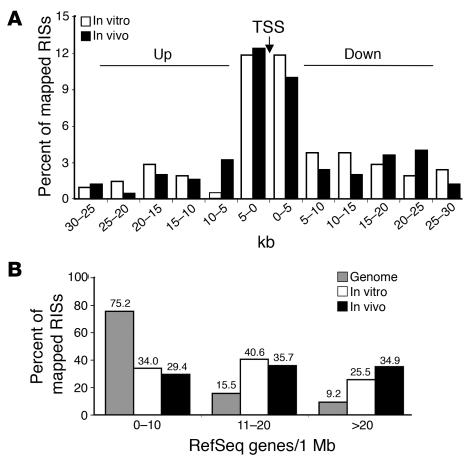
Overall, 18 of the 295 RefSeq genes targeted by an RIS were hit twice within their transcribed portion (Supplemental Table 1A). Thirty-two additional hot spots were identified by the occurrence of at least 2 independent RISs within arbitrarily chosen windows of 30 kb (2 hits), 50 kb (3 hits), or 100 kb (≥4 hits) (ref. 20 and Supplemental Table 1B). Overall, 14.6% (31 of 212) of in vitro RISs and 15.9% (79 of 496) of in vivo RISs were classified as hot spots. The most frequently hit genes (≥3 times) encoded a protein kinase (DYRK1A), a putative RNA-binding protein (RNPC1), and a cell-cycle regulator (CCND2) or were involved in tumor-associated chromosomal translocations (BCL2, BLM, or LMO2; Figure 2A).
Figure 2. RIS hot spots in patients with ADA-SCID without clonal expansion or overexpression.
(A) Schematic representation of RISs in the most frequently hit genes. Additional RISs outside these intervals in the proximity of CCND2 and BCL2 are also indicated. In vitro RISs are shown as green arrows and in vivo RISs as blue arrows; the direction of the arrow denotes orientation of the retroviral vector. The upper or lower position of the arrow refers to an LTR-genomic junction sequence mapping to the positive (above line) or negative (below line) chromosomal strand, respectively. Genes are depicted in red, with filled boxes and red arrows indicating exons and TSSs, respectively. RISs in the LMO2 locus were detected in T cells (Pt1, S1_049, 27 months after GT; Pt3, S3_042, 18 months after GT; Pt4, S4_048, 26 months after GT), granulocytes (Pt5, S5_144 and S5_163, 6 and 12 months after GT, respectively), and in Pt5 preinfusion CD34+ cells (S5_P048). (B) Quantification of the LMO2-containing clones within the total T cell population. The frequency of insertions was measured by sequence-specific real-time PCR in purified CD3+ T cells isolated at different time points after GT. Dotted lines show the frequencies of nonrecurrent (neutral) RISs retrieved from Pt3 as a control of physiologic clonal fluctuations. (C) Expression of LMO2 in peripheral blood T cells and granulocytes. Total RNA was isolated from the indicated cell subsets of GT-treated patients, patients with ADA-SCID undergoing polyethylene glycol-modified bovine ADA treatment (UT, not treated with GT), or age-matched healthy controls (ND). LMO2 gene expression was measured by quantitative RT-PCR using the standard curve of a reference sample and normalized for the expression of the housekeeping gene HPRT.
LMO2 was overall the most frequently hit locus, with 1 RIS cloned from pretransplant CD34+ cells (Pt5), 2 from granulocytes (Pt5), and 3 from T cells (Pt1, Pt3, Pt4; Figure 2A). Three RISs clustered in a 39-bp region 39.2 kb upstream of the TSS (S3_042, S5_144, and S5_P048), while the others were located 15 kb upstream (S4_048), 1.3 kb upstream (S1_049), and inside the second intron (S5_163, Pt5; Figure 2A). To assess the relative contribution of clones carrying LMO2 insertions, we measured the frequency of 4 RISs by sequence-specific real-time PCR. Three RISs were detected in T cells from Pt1 (S1_049), Pt3 (S3_049), and Pt4 (S4_048) from 6 to 60 months after GT at levels fluctuating between 0.03% and 0.7% of the total CD3+ populations (Figure 2B). These frequencies are comparable to those observed for 2 nonrecurrent insertions in T cells from Pt3 (Figure 2B). Of notice, RIS S1_049 became undetectable at the latest time of observation, 6 years after GT. RIS S5_144 was undetectable in T cells and at the limit of PCR sensitivity in granulocytes (0.01%) from Pt5 (Figure 2B). To assess whether the presence of LMO2 insertions could result in gross abnormalities in LMO2 gene expression, we measured mRNA levels in purified cell populations from both untreated and treated patients with ADA-SCID as well as from healthy controls. We observed no difference in LMO2 expression in purified bulk T cell subsets compared with normal controls (Figure 2C). LMO2 expression in granulocytes was higher in untreated patients with ADA-SCID than in healthy controls, and did not increase in granulocyte samples from treated patients.
RISs favor genes active in CD34+ and T cells.
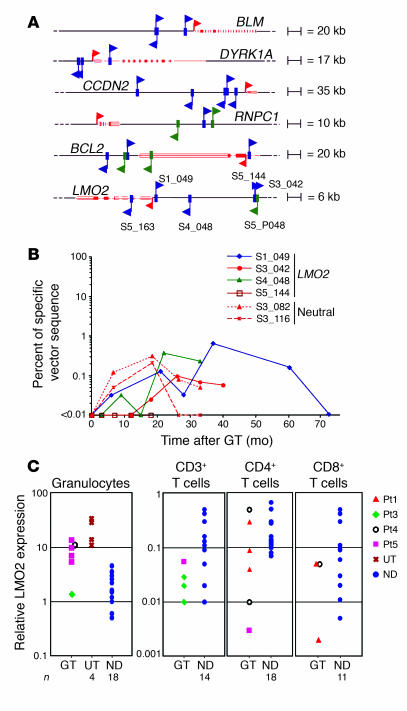
To correlate retroviral integration preferences with transcriptional activity at the time of gene transfer, we analyzed the gene expression profiles of cytokine-exposed BM CD34+ cells by Affymetrix microarray analysis. The expression of genes hit by insertions was compared with a normalized data distribution of all probesets in the microarray that was divided into 4 arbitrary expression classes. The proportion of expressed genes in the array (42.8%) was significantly increased when the analysis was restricted to genes hit by RISs inside (60.9%) or within 10 kb of TSSs (67.4%) in pretransplant CD34+ cells (P < 0.005; Figure 3A). One-fourth of the RISs upstream of the TSS hit genes in the highest gene expression class, suggesting that highly active promoters are preferentially targeted by retroviral vectors in CD34+ cells. A similar tendency, although less pronounced, was observed in the posttransplant sample (Figure 3A). Of interest, hot spot genes were on average more expressed compared with the whole-chip distribution (63.3% versus 42.8%), with a strong overrepresentation of highly expressed genes (21.1% versus 10.7%, P < 0.005; Figure 3A).
Figure 3. Gene expression profiling of genes hit by retroviral vectors.
(A) Correlation between insertion sites and gene expression profile of CD34+ cells at the time of transduction. The expression level profile of all probesets present on an Affymetrix HG-U133A microarray was determined in CD34+ cells and compared with the expression profile of probesets corresponding to targeted genes (inside or <10 kb upstream) by retroviral vector insertion in preinfusion CD34+ cells (in vitro) and cells after GT (in vivo), respectively, or in both samples. Bars show the percentage distribution of expression levels classified as present and absent by Affymetrix MAS 5.0 software. The present probesets were categorized in 3 subgroups according to 2 cutoffs identified by first (0–25) and third (75–100) quartiles of expression levels. (B) Correlation between insertion sites and genes expressed in T cells. Comparison of expression level of all probesets versus probesets of genes hit (inside or <10 kb upstream) by retroviral vector insertion and found in T cells of patients with ADA-SCID after GT. Analysis was performed as in A.
We also investigated whether the expression characteristics of a hit gene could influence the in vivo behavior of transduced T cells. The gene expression profile of purified T cells was determined and divided into arbitrary expression categories as described above (Figure 3B). Hit genes were expressed at a significantly higher frequency (53.1% versus 43.3%, P < 0.01) and enriched in the high expression category (intragenic hits, 14.0% versus 10.8%, P < 0.05; hits <10 kb from TSS, 22.7% versus 10.8%, P < 0.005). These data suggest that the selective pressure for vector-derived ADA expression favors the survival of T cells carrying insertions in the proximity of transcriptionally active regions.
Functional clustering analysis of hit genes before and after transplantation.
To understand whether the bias observed in the genomic distribution of RISs was associated to in vivo selection of specific integration events, we carried out a functional clustering of genes targeted by retroviral integration based on their gene ontology (GO) classification (21). The expected distribution of genes among the different GO families was compared with the observed distribution of genes hit by RISs (within or ±30 kb from genes) in the in vitro and in vivo samples. The analysis showed a tendency for a modest, but not statistically significant, overrepresentation of several gene categories in the pretransplant transduced CD34+ cell sample (Figure 4). The posttransplant data set displayed a similar profile, with the exception of a slight but significant increase in the frequency of genes encoding protein-binding factors (Figure 4).
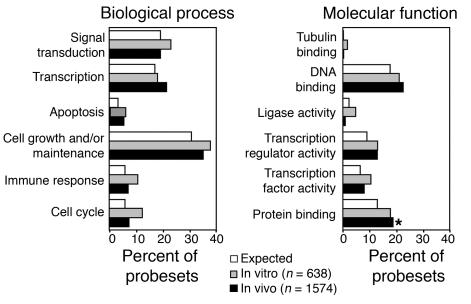
Figure 4. Functional clustering of targeted genes by GO.
The probesets of genes targeted (inside and within 30 kb) by retroviral insertion were classified in accordance with GO criteria. Each bar refers to the percentage of all targeted probesets falling within a specific gene category. The difference between groups was tested by both Fisher exact test and EASE score, corrected for multiple comparisons with the Bonferroni-Holm correction. The only difference was revealed for in vivo sample versus expected in “protein binding” category (*P = 0.022, Fisher exact test; P = 0.034, EASE score). No major differences were observed in RIS distribution and hierarchical clustering between lymphoid and myeloid cells.
RISs shared among different hematopoietic progenies indicate transduction of multipotent stem cells.
Among the 496 RISs isolated from in vivo samples from 5 patients, 124 came from highly purified granulocytes, 399 from T cells (per-patient range, 42–121), and 10 from other cell types (Supplemental Table 2). Using RISs as a marker of cell clonality, we found that T cell populations remained polyclonal throughout the follow-up (Figure 5, A and B, and Supplemental Figure 2), in agreement with the polyclonal TCR repertoire observed in the same samples (data not shown). This heterogeneity was confirmed by the analysis of 49 T cell clones generated ex vivo from Pt2 and Pt3 18 months after GT, 38 of which had distinct RISs. T cell clones displayed high TCR diversity and carried either 1 (84%) or 2 (16%) vector copies per cell. In contrast, granulocytes showed a more oligoclonal pattern by gel electrophoresis analysis (Figure 5A), with fewer but stable RISs retrieved by random cloning in 4 patients (range, 6–84; Supplemental Table 2).
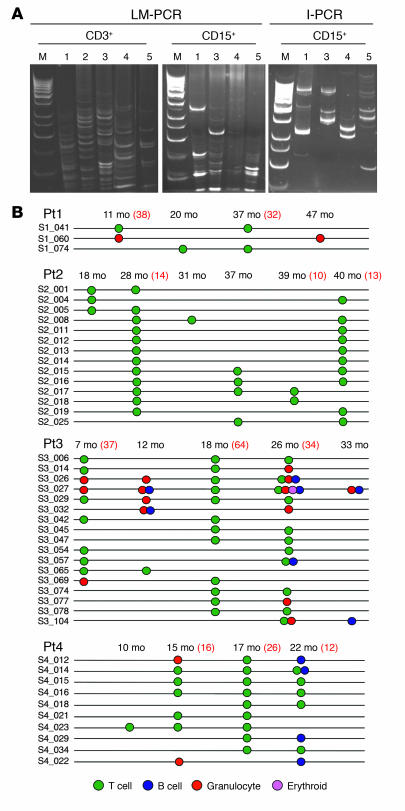
Figure 5. Vector clonal composition in distinct hematopoietic lineages after GT.
(A) Clonal analysis by LM-PCR and I-PCR in purified CD3+ T cells and CD15+ granulocytes from 5 patients with ADA-SCID treated with GT. Nested PCR products were analyzed by electrophoresis on acrilamide gels with a 100-bp ladder as a marker (M). In Pt2, who received a low CD34+ cell dose and displayed poor myeloid engraftment (<0.1%), no RISs could be isolated from multiple samples of granulocytes, thus confirming the lack of potential contamination from T cells. The following representative time points are shown for LM-PCR in T cells: Pt1, 20 months; Pt2, 39 months; Pt3, 26 months; Pt4, 22 months; Pt5, 9 months. The following representative time points are shown for LM-PCR in granulocytes: Pt1, 47 months; Pt3, 33 months; Pt4, 22 months; Pt5, 12 months. The following representative time points are shown for I-PCR in granulocytes: Pt1, 5 months, Pt3, 26 months; Pt4, 4 months; Pt5, 3 months. (B) Evidence for common integrants identified in different cell subpopulations during long-term follow-up. Each RIS from Pt1, Pt2, Pt3, or Pt4 is identified by a unique number. The circles indicate the presence of a specific RIS detected after 6 months of follow-up at 2 or more different time points by random cloning in the indicated population. Numbers in red text refer to the number of independent RISs retrieved by random cloning for the time points in which ≥30 colonies were analyzed. The random cloning analysis in Pt1 (47 months) and Pt3 (33 months) T cells was not carried out because the material was not available. In Pt5, 1 common integration site was detected in T cells at 6 and 8 months after GT. The presence of common integrants was confirmed in selected cases by sequence-specific PCR, which allowed us to unequivocally track integrants in different lineages.
Furthermore, the analysis performed in various hematopoietic lineages revealed the presence of shared integrants between myeloid (granulocytes) and lymphoid (T and/or B) cells in Pt3 (n = 9) and Pt4 (n = 4), with RISs retrieved more than once during the follow-up (Figure 5B). In Pt5 we compared in vitro RISs at early (day 45) and late (day 180 or later) time points after GT and observed a progressive restriction in the complexity of the integration pattern, as expected, due to a progressive loss of transduced committed progenitors after GT. In addition, later clones tended to contain more promoter-proximal and less intragenic integration events, as observed for the bulk of RISs after GT (Supplemental Table 3).
Discussion
Our study provides a comprehensive analysis of the profile of RISs in hematopoietic cells before and after transplantation into patients with ADA-SCID. Clonal analysis in the T cell compartment demonstrated a high number of distinct insertion sites, in agreement with the complexity of the T cell repertoire following immune reconstitution in all patients. Fewer RISs were retrieved in the myeloid compartment compared with T lymphocytes, reflecting the 1–2 log difference in hematopoietic cells versus lymphoid engraftment observed in these patients (ref. 3 and A. Aiuti, unpublished observations). The presence of shared integrants between myeloid and lymphoid cells in 2 patients confirmed that the gene transfer protocol combined with low-dose chemotherapy is adequate to achieve engraftment of genetically corrected, multipotent hematopoietic cells. Overall, 13% of the RISs identified in granulocytes more than 6 months after GT were detected also in lymphoid cells by random cloning. A systematic approach of clone-specific tracking would be useful to determine the actual frequency of multipotent hematopoietic cells versus long-lived lineage-specific progenitors. Our results differ from those obtained in nonmyeloconditioned patients with ADA-SCID (23, 24), which showed poor engraftment of transduced hematopoietic cells and as few as 1 progenitor cell clone contributing to long-term T lymphopoiesis (24). On the other hand, preconditioning is not an absolute requirement for hematopoietic cell engraftment, because progenitor cell clones with myeloid and lymphoid potential have been detected at low frequencies in the SCID-X1 GT trial (25).
Our analysis revealed a nonrandom distribution of in vitro–integrated proviruses, with a strong preference for TSSs and gene-dense regions and a tendency to hit genes that are highly expressed in CD34+ cells at the time of transduction. The bias toward highly expressed genes observed in previous studies (9, 12, 26) may be related to the preferential interaction of the viral integrating machinery with factors associated with transcriptionally active genes (8). A similar profile was detected in long-term engrafted hematopoietic cells analyzed in vivo with an additional tendency to favor integrations near TSSs. This may reflect a growth advantage for clones expressing adequate ADA levels, in which RISs near promoters could increase the chance of productive interaction between the vector and the cell transcriptional machinery. Indeed, RISs retrieved from T cells were enriched for highly expressed genes, suggesting the occurrence of in vivo selective pressure for vector-derived ADA expression. Alternatively, retroviral insertions might have influenced the expression of genes involved in cell expansion or clonal dominance. This explanation appears unlikely because, in contrast to previous reports in animal models (11, 14), we observed no in vivo skewing toward RISs in genes controlling crucial steps in self renewal or survival of HSCs, such as cell cycling, proliferation, or signal transduction. Thus, the 3- to 4-fold potential increase in the genotoxic risk predicted by the nonrandom genomic distribution of RISs appears to have a limited impact in patients with ADA-SCID, confirming that the actual risk of insertional oncogenesis is several orders of magnitude lower than the simple risk of hitting a potential oncogene (10–3 to 10–2) (27).
A comparison of our RIS collection with that of a recently reported CGD GT trial (4) reveals that 9.7% of the RISs we detected in vitro (n = 24) or in vivo (n = 46) are in and/or near the same genes. Interestingly, we detected only 1 RIS inside the MDS1-EVI1 locus (S3_067; see Supplemental Data File 1), which became undetectable at later time points by sequence-specific PCR. This is in contrast with the observation that the MDS1-EVI1 locus was overrepresented (1.9% of the total events) in a preclinical study in primates (28) and associated with clonal dominance in the CGD trial (4). Two factors might have contributed to this difference in our trial: (a) the use of BM-derived CD34+ cells, which have different properties (expression of retroviral receptors, phenotype, cell-cycle status) compared with the mobilized peripheral blood CD34+ cells used in the studies of patients with CGD and primates; and (b) the use of a long-terminal repeat (LTR) with substantially less enhancer activity in HSCs and myeloid cells compared with that of the spleen focus-forming virus (4) used in the CGD trial.
In addition, 34 RISs (4.8%) from our samples are in common with a collection of 322 RISs retrieved from peripheral blood T lymphocytes transduced with an MLV-derived vector (26). Overall, these results reveal the existence of a common fingerprint of vector insertions in human cells, which should be further defined in larger collections of RISs and in multiple GT trials.
A striking finding of our study is the overrepresentation of RISs in the proximity of CCND2 and LMO2 genes. In both cases, 2 RISs were found in granulocytes (of 124 total independent RISs) and 3 in T cells (of 399 RISs) in vivo after transplantation. Of interest, CCND2 and LMO2 insertions were found at relatively high frequencies also in vivo in T cells isolated from the French SCID-X1 trial (CCND2, 9 RISs; LMO2, 5 RISs; ref. 29). LMO2 insertions were also identified in the peripheral blood cells (2 of 765) of GT-treated patients with CGD (4). These findings may reflect a potential growth advantage associated with the clones carrying such RISs. However, in our study, CCND2 insertions were detected by random sequencing only in the first 2 years of follow-up and not later, suggesting the lack of selective pressure for the expansion of these clones. Moreover, RISs in the proximity of LMO2 were also overrepresented in the pretransplant CD34+ cell sample (1 of 212) and in a separate collection of RISs from cord blood–derived CD34+ cells (2 of 595; ref. 30). These data indicate that the LMO2 gene is a hot spot for retroviral integration in human CD34+ cells. Because LMO2 is highly expressed in CD34+ cells and lies in a gene-dense region, this bias may just reflect the general preferences of gammaretroviral vectors. However, specific properties of the LMO2 locus, such as binding of transcription factors or other chromatin characteristics, may further increase its targeting by the retroviral vector integration machinery. Indeed, the LMO2 locus (11p13) is a fragile chromosomal site (FRA11E) (31) and is a major site of aberrant trans-V(D)J recombination between immune loci in thymocytes (32), a typical marker of genomic instability.
The lack of in vivo expansion of clones carrying LMO2 RISs indicates that insertions in potentially dangerous genomic sites are not sufficient per se to induce a proliferative advantage in T cells in vivo, confirming that multiple cooperating events are required to promote oncogenic transformation in humans (33). It is tempting to speculate that the difference between the ADA-SCID and the SCID-X1 studies may be related to the SCID-X1 genetic background (34) or to the role of the therapeutic transgene, where ADA is a housekeeping enzyme and γc a potentially oncogenic growth factor receptor (17, 18), although the oncogenicity of IL2RG has been recently challenged (35, 36). On the other hand, we could not provide conclusive evidence that the RISs detected in our patients with ADA-SCID influenced LMO2 expression at single-cell levels. The probability for a retroviral vector insertion to induce over-expression of a proximal gene was estimated to be 20% in the context of T cells (26). However, we were unable to isolate T cell clones containing LMO2 RISs to directly test this hypothesis in our patients.
In summary, our data show that transplantation of ADA-transduced HSCs does not result in in vivo selection of expanding or malignant cell clones, despite the occurrence of insertions near potentially oncogenic loci. These results, combined with a relatively long-term follow-up of patients, indicate that retrovirally mediated gene transfer for ADA-SCID has a favorable safety profile. GT of other genetic diseases may require the development of safer gene-transfer tools, such as self-inactivating lentiviral or retroviral vectors and the use of physiologically controlled gene expression cassettes.
Methods
Clinical trials.
Patients with ADA-SCID were enrolled in clinical trials approved by the San Raffaele Scientific Institute and Hadassah University Ethical Committees and National Regulatory authorities. GT for ADA-SCID has received Orphan Drug Status by the European Medicines Agency. Pt1 and Pt2 have been described previously (3); Pt3–Pt5 were treated between 12 and 22 months of age (A. Aiuti, C. Bordignon, and M.G. Roncarolo, unpublished observations). Patients’ numbers were assigned for the purpose of this research study. Patients received autologous BM-derived CD34+ cells prestimulated for 24 hours in the presence of cytokines (FLT3-ligand, SCF, TPO, and IL-3) and then transduced 3 times with the GIADAl MLV retroviral vector (20), for a total of 92 hours of culture (3). Nonmyeloablative conditioning with 4 mg/kg busulphan i.v. (except Pt2, taken orally) total dose was administered on days –3 and –2 relative to CD34+ cell infusion, as previously described (3).
Purification of cell subsets from peripheral blood and BM.
Blood and/or BM samples were obtained from of patients with ADA-SCID and age-matched healthy controls after parents of patients gave informed consent following standard ethical procedures and with approval of the San Raffaele Scientific Institute and Hadassah University Ethical Committee. Mononuclear cells from peripheral blood and BM samples were isolated by density gradient centrifugation on Ficoll-Hypaque. The peripheral blood granulocyte fraction was enriched by consecutive separations on dextran and Ficoll-Hypaque gradient. Thereafter, the different subpopulations were isolated by immunomagnetic technique using antibody-coated microbeads (Miltenyi Biotec) and fluorescence-activated cell sorting using fluorescine-labeled antibodies (FACS Vantage; BD Biosciences). The following monoclonal antibodies were used for positive selection: anti-CD15 (granulocytes), anti-CD19 (B cells), anti-CD61 (megakaryocytes), anti-glycophorin A (erythroid cells), anti-CD56 (NK cells), anti-CD3 (T cells), anti-CD4 (CD4+ T cells), anti-CD8 (CD8+ T cells). Two steps of purification were performed to increase the purity to greater than 98%–99%.
DNA purification and quantitative PCR for vector-positive cells.
Genomic DNA was extracted from purified cells using QIAamp DNA Blood Mini kit (Qiagen) or after proteinase K digestion for fewer than 105 cells. Quantitative PCR analysis for vector positivity was performed as previously described (3). Briefly, 2 sets of primers and probes specific for the NeoR gene and GAPDH were used to detect transduced cells and standardize for DNA content, respectively. The frequency of transduced cells was calculated using a standard curve and expressed as the proportion of cells containing the NeoR reporter gene.
Cloning and analysis of RISs.
In order to avoid potential biases related to the use of restriction enzyme or amplification steps, 2 different techniques were used for analysis of RISs: inverse PCR (I-PCR) and LM-PCR. I-PCR was performed by a modification of previously described primers and PCR conditions (37). Briefly, 50 ng of genomic DNA was digested with TaqI, and the LTR-containing fragments were self-ligated and subjected to 2 rounds of amplification using the following LTR-specific primers: forward, 5′-CTGTTCCTTGGGAGGGT-3′; reverse, 5′-AGGAACTGCTTACCACA-3′; nested forward, 5′-GCGTTACTTAAGCTAGCTTG-3′; nested reverse, 5′-GATTGACTACCCACGACGGG-3′.
Reaction conditions were identical for the first and second rounds of PCR: denaturation at 94° for 1 minute, 50° annealing for 1 minute, and 72° extension for 1.5 minutes for a total of 29 cycles. For LM-PCR, 200 ng of genomic DNA were digested with PstI and MseI, to prevent amplification of an internal viral fragment from the 5′LTR, and ligated to an MseI double-strand linker. Nested PCR was performed with LTR- and linker-specific primers as previously described (6). The resultant PCR products were shotgun cloned without purification with the PCR 2.1 TOPO TA cloning kit (Invitrogen) and transformed into libraries of vector-integration junctions. Alternatively, nested PCR products were separated either on 2% agarose or on Spreedex gels (Elchrom Scientific), excised, and directly sequenced. Proviral insertion site sequences were aligned to a downloaded version of the human genome (NCBI Entrez Genome, version 35) using the Ensembl database (http://www.ensembl.org/). Mapped RISs were selected for analysis only if they contained the correct LTR- and primer-specific sequences and yielded a unique best hit with at least 95% identity with the human genome. We sequenced 653 RISs from samples obtained after GT, 496 of which were unambiguously assigned to a chromosomal position (in vivo samples). The remaining 157 sequences could not be mapped because of their short length or the presence of repetitive sequences. In parallel, we cloned 361 RISs from transduced BM CD34+ cells of 4 patients with ADA-SCID (Pt2, Pt3, Pt4, and Pt5) before infusion and identified 212 unique RISs (in vitro samples). Random genomic sequences originated by LM-PCR (genomic MseI-MseI, PstI-MseI fragments) were mapped as well and used as controls. The average length of generated fragment was 187 bp for I-PCR (range, 17–893 bp) and 82 bp for LM-PCR (range, 15–474 bp).
There was no statistical difference in the frequency of RISs hitting genes with the 2 methods, but I-PCR retrieved RISs greater than 10 kb upstream from a gene more frequently than did LM-PCR (P = 0.036). This was not unexpected, because MseI sites are underrepresented in GC-rich promoters. In the in vivo sample, 22 RISs were retrieved by both methods.
Quantitative tracking of specific integrants.
To determine the relative contribution of LMO2-related molecular clones over time, real-time quantitative PCR was set up to specifically amplify the proviral-genomic junction of RISs within the LMO2 locus (S1_049, S4_048, S3_042, and S5_144). Two molecular clones isolated from Pt3 T cells mapping to “neutral” regions of the genome (S3_082 and S3_116) were used as control of physiological clonal fluctuations. Quantitative PCR analysis was performed on 50 ng of genomic DNA, directly isolated from sorted cells, using the specific unique genomic flanking primers in combination with common LTR primer and probe (Primm). The primers sequence were as follows: LTR primer, 5′-GTTTGCATCCGAATCGTGGT-3′; LTR probe, 6-FAM-TCTCCTCTGAGTGATTGACTACCCACGACG-TAMRA; S1_049 primer, 5′-GGAATCAGGCACCTTCTCTCTC-3′; S4_048 primer, 5′-GGTTAAGATCACAGGCTGTGGTG-3′; S3_042/S5_144 primer, 5′-TCTCTCCTATCAGCCAATAAAGGG-3′; S3_082 primer, 5′-CGGCTCGGGTGTCCTG-3′; S3_116 primer, 5′-AAGTGTCGGTGTTAGTACCC-3′.
Each set of primers allowed us to unambiguously track the corresponding clone, as assessed by the absence of amplification in samples from different patients. The frequency of each individual clone, expressed as the proportion of cells that contained the specific vector sequence, was calculated on the basis of a standard curve of the relative integrants diluted in untransduced PBLs (from 5 × 104 to 5 × 100).
Real-time PCR analysis for LMO2 expression.
Total RNA was isolated from CD3+, CD4+, and CD8+ T cells as well as CD15+ granulocytes purified either from GT-treated patients, untreated patients with ADA-SCID, or age-matched healthy donors. The RNA was reverse transcribed using the Archive Kit (Applied Biosystems). Expression analysis of LMO2 was carried out with the TaqMan system, developed as a custom Assay-On-Demand by Applied Biosystems. The data were analyzed by the ABI PRISM 7700 with the sequence detector system software (version 1.9.1; Applied Biosystems). LMO2 expression was determined by relative quantification method. Essentially, 2 standard curves of serially diluted cDNA, obtained from 1 healthy donor granulocyte sample, were set up to amplify the target gene and the HPRT housekeeping gene as an endogenous control. The LMO2 expression in the normalized samples was expressed as fold difference relative to the reference sample as calibrator.
Gene expression profiling.
RNA was isolated from BM-derived CD34+ cells (n = 2) cultured ex vivo according to the cytokine conditions used in the clinical protocol and from primary CD3+ T cells (n = 5). The transcribed biotinylated cRNA was cleaned up using RNeasy spin columns (Affymetrix), fragmented, and hybridized to an Affymetrix HG-U133A Gene Chip Array. The scanned images were analyzed using Microarray Suite (MAS) software (version 5.0; Affymetrix), and the signal intensity of detected transcripts (probesets) was determined with the MAS 5.0 absolute analysis algorithm. Following interarray normalization of overall fluorescence intensities, the differences in gene expression levels were expressed as numeric values. In the correlation between RISs and gene activity, the expression values of each probeset classified as present by the software were assigned to 3 arbitrary classes corresponding to 0–25, 25–75, and 75–100 percentiles of expression level of all present genes (probesets) in a normalized data distribution.
Functional clustering analysis.
Probesets corresponding to genes targeted by retroviral vector insertions were analyzed for functional clustering into specific categories according to GO criteria. The probability of overrepresentation of each category was determined using the EASE bioinformatics package from NIH-DAVID (http://david.abcc.ncifcrf.gov/ease/ease.jsp). The default metric used by EASE to rank categories of genes by overrepresentation provides the EASE score. Results were corrected for multiple comparisons with the Bonferroni-Holm correction (α = 0.05).
Bioinformatics.
The bioinformatics analysis was performed by dedicated Perl scripts allowing for the storage, retrieval, analysis, and summary of the data through a back-end MySQL database (http://www.mysql.com/). Human RefSeq chromosome sequence text-based FASTA files for the May 2004 human genome freeze (NCBI Entrez Genome, version 35) were downloaded from the NCBI genome project website (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=9558) and converted to be used in local BLAST search. Coordinates of RefSeq genes and other annotation tables for the same genome reference sequence freeze were accessed through the Ensembl Perl API from the Ensembl genome project. In the present study, we assumed the definition of “gene” as the genomic region between the TSS and stop boundaries of one of the 24,194 RefSeq genes mapped to the human genome as reported by Wu et al. (6).
Statistics.
Statistical analysis was performed with R statistical software (version 2.1.1; http://www.r-project.org). Overall the significance level chosen was α = 0.05. To investigate whether some regions proximal to the TSS are favored by integration, we measured the distance between each RIS and the TSS of genes immediately upstream or downstream. These distances were normalized according to gene length and orientation and they followed a β distribution (see below). This is a simple procedure to address the problem of integration occurring in areas with different gene density. The hypothesis of random distribution in probability terms is equivalent to assuming uniform distribution. Comparison between observed integration distribution and expected random integration was performed by Kolmogorov-Smirnov test based on confidence intervals of 95% built on bootstrap replications. Comparisons between in vitro and in vivo frequencies were tested with a Pearson c2 test (for details on the statistical procedure, see Supplemental Methods). In the gene expression studies, given the different sample sizes of pretransplant and posttransplant groups, an asymptotic approximation was considered via the Monte-Carlo method based on 50,000 or 100,000 replications to account for the difference in the distribution between each sample. Statistical significance was evaluated by a parametric likelihood test, distributed as a c2 test. Analysis of overrepresentation of probeset classes in the RIS data set was performed by means of both Fisher exact test and EASE score. The results were corrected for multiple testing errors within each data set and/or system combination with Bonferroni-Holm correction methods.
Supplementary Material
Acknowledgments
We are grateful to Ulrike Benninghoff and Federica Cattaneo for clinical care, Luciano Callegaro for data managing, all physicians and nurses of the Pediatric Clinical Research Unit, Claudia Cattoglio for data on the random library of human restricted fragments, Alessandro Guffanti for initial help in setting up the software for sequence analysis, Giorgio Verde for help in establishing the bioinformatic tools, Alessio Palini for cell sorting, and Eugenio Montini for critical discussion. This work was supported by grants from Italian Telethon Foundation, AFM/Telethon (GAT0205), the European Commission (CONSERT contract LSBH-CT-2004-005242 and CLINIGENE LSHB-CT2006-018933), Istituto Superiore di Sanità: Cellule Staminali, and Ministero dell’Universitá e della Ricerca–Fondo per gli Investimenti della Ricerca di Base (MIUR-FIRB).
Footnotes
Nonstandard abbreviations used: ADA, adenosine deaminase; γc, γ chain; CGD, chronic granulomatous disease; GO, gene ontology; GT, gene therapy; I-PCR, inverse PCR; LM-PCR, linker-mediated PCR; MLV, murine leukemia virus; Pt, patient; RIS, retroviral insertion site; SCID-X1, X-linked SCID; TSS, transcription start site.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2233–2240 (2007). doi:10.1172/JCI31666
See the related Commentary beginning on page 2083.
References
- 1.Cavazzana-Calvo M., et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar H.B., et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 3.Aiuti A., et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. . 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 4.Ott M.G., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 5.Bordignon C., Roncarolo M.G. Therapeutic applications for hematopoietic stem cell gene transfer. Nat. Immunol. 2002;3:318–321. doi: 10.1038/ni0402-318. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell R.S., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Palma M., et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 9.Hematti P., et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laufs S., et al. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Mol. Ther. 2004;10:874–881. doi: 10.1016/j.ymthe.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Montini E., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 12.Bushman F., et al. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 13.Du Y., Jenkins N.A., Copeland N.G. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kustikova O., et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 15.Cavazzana-Calvo M., Fischer A. Gene therapy for severe combined immunodeficiency: are we there yet? J. Clin. Invest. 2007;117:1456–1465. doi: 10.1172/JCI30953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina S., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 17.Dave U.P., Jenkins N.A., Copeland N.G. Gene therapy insertional mutagenesis insights. Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- 18.Woods N.B., Bottero V., Schmidt M., von Kalle C., Verma I.M. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 19.Hershfield M.S. Adenosine deaminase deficiency: clinical expression, molecular basis, and therapy. Semin. Hematol. 1998;35:291–298. [PubMed] [Google Scholar]
- 20.Aiuti A., et al. Immune reconstitution in ADA-SCID after PBL gene therapy and discontinuation of enzyme replacement. Nat. Med. 2002;8:423–425. doi: 10.1038/nm0502-423. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T., et al. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordignon C., et al. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA-immunodeficient patients. Science. 1995;270:470–475. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt M., et al. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. . Nat. Med. 2003;9:463–468. doi: 10.1038/nm844. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt M., et al. Clonal evidence for the transduction of CD34+ cells with lymphomyeloid differentiation potential and self-renewal capacity in the SCID-X1 gene therapy trial. . Blood. 2005;105:2699–2706. doi: 10.1182/blood-2004-07-2648. [DOI] [PubMed] [Google Scholar]
- 26.Recchia A., et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baum C., Kustikova O., Modlich U., Li Z., Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 28.Calmels B., et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deichmann A., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattoglio C., et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007 doi: 10.1182/blood-2007-01-068759. In press. [DOI] [PubMed] [Google Scholar]
- 31.Bester A.C., et al. Fragile sites are preferential targets for integrations of MLV vectors in gene therapy. Gene Ther. 2006;13:1057–1059. doi: 10.1038/sj.gt.3302752. [DOI] [PubMed] [Google Scholar]
- 32.Marculescu R., Le T., Simon P., Jaeger U., Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J. Exp. Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCormack M.P., Rabbitts T.H. Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2004;350:913–922. doi: 10.1056/NEJMra032207. [DOI] [PubMed] [Google Scholar]
- 34.Shou Y., Ma Z., Lu T., Sorrentino B.P. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pike-Overzet K., et al. Gene therapy: is IL2RG oncogenic in T-cell development? Nature. 2006;443:E5; discussion E6–E7. doi: 10.1038/nature05218. [DOI] [PubMed] [Google Scholar]
- 36.Thrasher A.J., et al. 2006Gene therapy: X-SCID transgene leukaemogenicity. Nature. 443E5–E6; discussion E6–E7. . [DOI] [PubMed] [Google Scholar]
- 37.Kim H.J., et al. 2000Many multipotential gene-marked progenitor or stem cell clones contribute to hematopoiesis in nonhuman primates. Blood. 961 – 8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.