Abstract
The possibility of performing fast and small-volume nucleic acid amplification and analysis on a single chip has attracted great interest. Devices based on this idea, referred to as micro total analysis, microfluidic analysis, or simply ‘Lab on a chip’ systems, have witnessed steady advances over the last several years. Here, we summarize recent research on chip substrates, surface treatments, PCR reaction volume and speed, architecture, approaches to eliminating cross-contamination and control and measurement of temperature and liquid flow. We also discuss product-detection methods, integration of functional components, biological samples used in PCR chips, potential applications and other practical issues related to implementation of lab-on-a-chip technologies.
INTRODUCTION
In many cases, amplification is indispensable for the analysis of nucleic acids. Currently, nucleic acid amplification methods include but are not limited to polymerase chain reaction (PCR) (1), strand-displacement amplification (SDA), nucleic acid sequence-based amplification (NASBA), rolling-circle amplification (RCA) and the Qβ replicase reaction. Among these methods, PCR has been the most popular due to its simplicity; however Peltier effect or metal-block-based PCR system are characterized by high thermal mass, large reaction volume and thus slow heating/cooling rates. The PCR speed can be improved by increasing the heat transfer rate or decreasing the thermal mass. With the advent of micro-electro-mechanical-systems (MEMS) technology, the development of miniaturized PCR chips becomes possible (2,3). The miniaturization of PCR devices offers several advantages such as short assay time, low reagent consumption and rapid heating/cooling rates, as well as great potential of integrating multiple processing modules to reduce size and power consumption. The number of publications on PCR chips has grown rapidly recently, and the articles are spread over a large number of journals. The development of PCR microchips has been discussed in recent reviews (2–4). In this article, we will review the latest advances and future trends based on literature published since January 2005. In addition, we will also discuss some practical issues related to the development of PCR chips. As a supplement to this review, the reader may wish to refer to several reviews of general microfluidic technologies (5–9).
The organization of this article is as follows. First, several important topics on the microfluidic PCR chips will be presented. Those topics, which are crucial in the development of PCR chips, include chip substrates and surface treatments, PCR chip architecture, on-chip PCR reaction volume and reaction speed and approaches to eliminating cross-contamination. Then, the temperature and fluidic controls and measurements in PCR chips are discussed, which include thermal insulation, evaporation and gas-bubble formation and measures to counteract these phenomena, semi-invasive or noninvasive temperature and fluidic measurements and numerical simulation of temperature and fluid fields in PCR chips. Finally, product detection methods used in PCR chips, e.g. off-line and on-line detection, are covered, followed by integration of functional components in PCR chips, biological samples used in PCR chips and potential applications of PCR chips, as well as practical issues related to the development of PCR chips.
SUBSTRATES AND SURFACE TREATMENTS TO REDUCE BIOMOLECULE ADSORPTION
Substrates
Most PCR microchambers or microchannels are fabricated from silicon (10–25) or glass (26–36) substrate. Polymers, such as polydimethylsiloxane (PDMS) (37–55), polycarbonate (PC) (56–63) and polymethylmethacrylate (PMMA) (64–68) have increasingly been utilized as alternative substrates. New substrates, such as SU-8 (69), cyclic olefin copolymer (COC) (65), Gene Frame® (70), perfluoroalkoxy-modified polytetrafluoroethylene (PFA) (13,71–76), LiNbO3 (77) and 317 stainless steel (78), have also been used in PCR microfluidic devices. Each substrate has different properties and therefore different advantages and disadvantages. The superior thermal conductivity of silicon makes rapid PCR cycling possible. Silicon fabrication processes are well developed, and thus precise and complex chip structures can be achieved (4). However, silicon can be problematic: bare silicon inhibits PCR; its high thermal conductivity requires thermal insulation and therefore results in structural complexity (14,23,25,36,79–81); its opacity limits optical detection; and its electrical conductivity makes it difficult to combine micro PCR with micro capillary electrophoresis (CE) (26,27,31,35,40) on a single silicon chip. Transparent glass is suitable for optical detection. The electro-osmotic-flow (EOF) property of glass allows the integration of PCR and CE on a monolithic chip (26,27,31,35,40). However, the PCR chips made from silicon or glass cannot be disposed due to the high cost of fabrication. The use of polymers as substrates may overcome these disadvantages. PDMS, an inexpensive elastomeric polymer, has emerged as a promising substrate. It exhibits high flexibility, better optical transparency, lower cost of fabrication and better biocompatibility than silicon. The flexibility of the polymers gives rise to the highly integrated PCR chips incorporating PDMS micropumps and/or microvalves (19,20,26,27,31,35,40,46,48,52,53). PDMS adsorbs less PCR sample than other materials and thus can be used to coat glass surfaces to avoid the possible inhibitory effect of glass on PCR (39,50,54). However, the permeability of PDMS can result in diffusional loss of biological sample, and implantation of polyethylene (PE) vapor barrier in the PDMS has been proposed to prevent this fluid loss (53). Air bubbles can be formed on PDMS surface during sample loading due to its hydrophobicity (51).
Other chip substrates include PC, which has a high glass transition temperature (Tg) (∼150°C) and can withstand the sustained high temperatures associated with PCR or ligation detection reaction (LDR) (56,58–63). Biocompatible PMMA is another suitable candidate. It displays less autofluorescence than PC (59) and is suitable for conventional optical detection. Inexpensive and versatile PMMA PCR chips can be fabricated by CO2 laser ablation within minutes (64,67). In addition, the nonspecific adsorption between PMMA and DNA/protein is minimal. However, its low Tg (∼105°C) (65) may hinder widespread use.
Surface treatment
The PCR efficiency is often limited by interactions between the chip surface and the biomolecules in the PCR solution, primarily due to the increase of the surface-to-volume ratio in a micro-scale environment. In general, the hydrophilic PCR solution is not easily introduced into the hydrophobic chip (28). Therefore, a proper surface treatment is required to ensure the success of on-chip PCR. The treatment processes can be classified as static treatment and dynamic treatment.
The static treatment involves the pre-coating of chip surface, during fabrication of PCR chip or immediately before use, with one of the following substances: SiO2 (10,14,20,21,25), bovine serum albumin (BSA) (11,17,25,37,53,61,73), polyethylene glycol (PEG) (65,78,82) or silanizing agents (for example 3-glycidoxypropyl trimethoxysilane (11), dichlorodimethylsilane (23,69), Sigmacoat® (16,29) or trimethylchlorosilane (33). The SiO2 pre-coating is a reproducible and inexpensive standard MEMS process and can be accomplished in a batch fashion. BSA and other agents become popular due to their simplicity but the reproducibility is argued. It is worth noting that diacrylated PEG (DAPEG) can be grafted into PDMS polymer for polyelectrolyte multilayers (PEM) deposition (82), and that the surface characteristic of PDMS polymer shifts from hydrophobic to hydrophilic after oxygen plasma treatment (44,83). The oxygen plasma activation on the surface of PDMS can be performed at the same time with the bonding processes, and thus it is a standard MEMS process with high reproducibility and low costs. Silanization is also a commonly used process to prevent on-chip adsorption of biomolecules, but it is time-consuming and labor intensive. Moreover, its reproducibility is also problematic. Recently, Legendre et al. developed a rapid (<5 min) and simple (one-step) Sigmacoat® silanization method on a dry glass surface without extensive rinsing and cleaning steps often associated with other chip coating procedures (29). This dynamic treatment occurs during the actual operation of PCR chips because the reagents are in the PCR solution. Other widely used dynamic reagents are BSA (10,11,13,16,20,21,28,31,50,51,53,57,59,60,62–64,70,74–76,78) and polyvinylpyrrolidone (PVP) (17,21,44). Glycerol (10,78), formamide (10,78), polyethylene glycol (PEG) 8000 (21), and Tween 20 (21), have also been studied as dynamic coating agents. It is known that BSA can improve on-chip PCR efficiency because it can compete with DNA polymerase for active adsorption sites on the inner surface. However, it is not yet clear that how components such as (NH4)2SO4, MgCl2 and/or KCl in the PCR mixture affect the BSA coating. In addition, when the fluorescence detection technique is applied, possible interactions between BSA and fluorescent probe/dye may be problematic.
ARCHITECTURES FOR PCR CHIPS
Stationary chamber-based PCR amplification
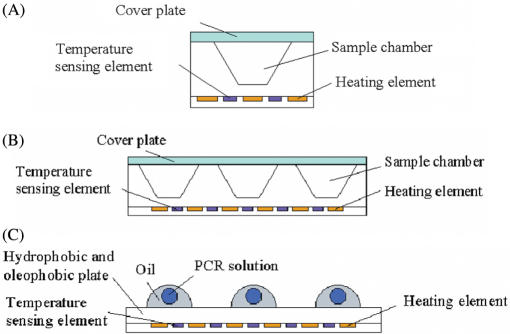
In this format of PCR chip, the PCR solution is kept stationary and the temperature of the reaction chamber is cycled between different temperatures. The first PCR chip developed by Northrup et al. in 1993 was based on such a stationary chamber (84). Since then, this format has been widely replicated and improved (2–4). During the past two years, the miniaturization of such PCR devices has been of growing interest. In general, they can be classified as single chamber stationary PCR chip (Figure 1A) (10,12–16,18,21,22,24,26,27,29,32,37,39,40,43,45,46,48,51–53,57,62,63,68–70) and multi chamber stationary PCR chip (Figure 1B) (11,17,19,20,25,28,31,33–36,38,47,49,54,78). The former can perform very well in terms of fluidic and thermal controls and offer beneficial properties such as reduced thermal and fluidic crosstalk between chambers. However, they cannot realize high-throughput and cannot readily be used for special purposes such as single cell gene expression analysis. To circumvent these issues, great efforts have been made to develop multi-chamber stationary PCR. This type of PCR chip allows generation of standard curves, use of multiple primer sets and optimization of microfluidic PCR. However, special care must be taken to achieve the thermal optimization of chamber array in order to obtain homogeneous temperature fields between chambers. In addition, precise handling and processing of sample microfluid on such PCR chips still face challenges (17,28).
Figure 1.
Stationary chamber-based PCR chip. (A) Single chamber PCR chip. (B) Multi chamber PCR chip. (C) VRC PCR. The PCR sample is introduced into the single/multiple/virtual chamber(s). The chip is then heated and cooled to provide thermocycling conditions.
Chamber-based PCR chips are constructed as closed systems where the reaction chamber(s) are actually microfabricated on the chip. Their design does not differ significantly from the model presented by Northrup et al. (84). Recently, Guttenberg et al. proposed a completely different approach to handle small-scale sample using a hydrophobic/oleophobic surface to provide virtual fluid confinement (77). The concept of virtual reaction chamber (VRC) (Figure 1C) was applied for the first time on the PCR chip in 2005, in which each PCR sample droplet was covered with a drop of mineral oil. Neuzil et al. further optimized this approach by placing a disposable microscope glass cover slip on top of a silicon chip to eliminate cross-contamination between samples (79,80).
Dynamic continuous-flow-based PCR amplification
The chamber-based stationary PCR chips lack the flexibility to change the PCR speed. The dynamic continuous-flow-based PCR amplification can overcome this issue by utilizing the ‘time-space conversion’ concept. The nucleic acid amplification occurs as the sample is continuously pumped through a microfluidic channel during each temperature cycle. The attractive features of this approach include: (i) The analysis processes of nucleic acids can be performed in a dynamic format on an integrated PCR chip. (ii) The temperature transition times depend only on the sample flow rate and the time needed for the sample to reach a thermal equilibrium. (iii) The heat inertia of PCR system is decreased to a minimum because only the sample's thermal mass need to be taken into consideration. (iv) The reaction volume can range from several microliters to several tens of microliters.
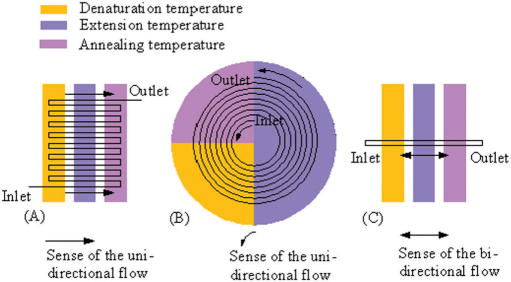
The continuous-flow PCR concept of using a capillary passing through different temperature baths was introduced by Nakano et al. in 1994 (85). In 1998, Kopp et al. reported a continuous-flow PCR chip using a serpentine channel passing through three thermostable copper blocks (Figure 2A) (86). Since then, especially during the last two years, such PCR chips have undergone substantial improvements (4,30,44,50,55,58,60,66,67,87,88). However, continuous-flow PCR poses several problems during thermal cycling: (i) Gas bubbles are easily generated in the microchannels, which adversely affect PCR amplification. (ii) Pressure-driven flow easily produces a hyperbolic flow profile that may lead to progressive sample dispersion (56), and often requires an external bulky syringe pump (23,30,41,42,44,50,58,66,67,71,72,74–76,88), which adversely affect the development of compact, portable and integrated continuous-flow PCR chips. (iii) The rate at which the PCR solution travels between different temperature zones is difficult to regulate, and thus most of the continuous-flow PCR systems lack the flexibility to regionally control fluid flow velocity so as to meet different PCR requirements. (iv) Highly integrated continuous-flow PCR chips are rarely reported due to high fabrication cost and difficulties in controlling the continuous liquid flow. In order to overcome these problems, some alternative approaches have been utilized. For example, by introducing a highly viscous fluorinated oil cap immediately before the introduction of the PCR sample, Nakayama et al. have overcome the generation of air bubbles in the continuous-flow microchannel (50). Gui and Ren have proposed the possibility of performing a novel EOF-driven continuous-flow PCR on a serpentine channel chip on the basis of numeric simulation (87). Similar work was also reported by Chen et al. (56). Li et al. have reported a continuous-flow PCR chip with a serpentine channel of varying width for ‘regional velocity control’ (30). Most recently, a hybrid chip incorporating the continuous-flow PCR, LDR and hybridization assays has been proposed by Hashimoto et al. to perform analysis of single base mutations in genomic DNA (60). This is a significant work because it opens the door to the integration of the continuous-flow PCR with pre-PCR or post-PCR analytical step.
Figure 2.
Continuous-flow PCR. (A) The serpentine channel continuous-flow PCR. (B) The spiral channel-based continuous-flow PCR. The sample is introduced at the inlet and pumped unidirectionally towards the outlet. (C) The straight channel oscillatory-flow PCR. The sample is introduced in the inlet and pumped back and forth in a straight channel. Temperature zones are provided by three heaters.
For serpentine channel continuous-flow PCR chips, parallelization is not easily realized as it complicates the chip architecture and most likely increases the chip footprint. In addition, the number of thermal cycles is usually not adjustable. Importantly, in these PCR chips the temperature zones are linearly arranged and are easy to establish a smooth temperature gradient. In this case, however, a melted single-stranded DNA sample is very likely to form double strands with the template strands or their complementary fragments when passing the extension zone, which compromises the PCR efficiency.
Several new approaches have been developed to partially or fully circumvent these problems. The roles of spiral-channel-based continuous-flow PCR devices (Figure 2B) have been recently emphasized by several groups since this approach can effectively avoid the possible formation of the DNA double strands (41,42,56,59,61,71,72,74–76). But it is still hard to perform parallel PCR amplifications using such devices. An oscillatory-flow-based approach (Figure 2C) (23,64,65) not only combines the cycling flexibility of the stationary chamber PCR with quick temperature transitions associated with the continuous-flow PCR, but also provides the possibility of performing high-throughput PCR amplifications in a parallel format (64). Microfluidic digital PCR (23,71,72,89–93) represents another example of the power of microfluidic PCR chips. This technology can provide precise control of sample volume and high-throughput analysis of serial (23,71,72,89–91) or parallel (93,92) format. In the serial format, the PCR solutions can flow continuously through a reaction channel path as a ‘droplet train’. However, such systems easily suffer from cross-contamination between samples and sample dispersion, and consequently appropriate two-phase flow systems are often required (71,72). In spite of this shortcoming, it is still possible to implement the totally automated, contamination-free, reusable and robust microfluidic digital systems for high-throughput PCR (71). Recently, Quake and colleagues reported another type of microfluidic digital PCR, where digital PCR was performed in parallel microarray format (92,93). The array chips used are commercially available, thus allowing single usage to effectively prevent carryover contamination. Importantly, the micropumps and microvalves not only can be conveniently used to distribute PCR fluid into a number of isolated reaction chambers for high-throughput PCR, but can also effectively seal the individual reaction chambers so as to realize a cross-contamination free microfluidic digital PCR system. In addition, for these microfluidic digital PCR systems, quantification relies only on binary, positive/negative calls for each subreaction within the partitioned analyte. This affords an absolute readout of DNA copy number with single-molecule resolution (93), allowing for transcription factor profiling in individual hematopoietic progenitors (93) and multigene analysis of individual environmental bacteria (92). Of course, such microfluidic digital PCR can also be used for other potential applications such as detection of base substitution mutations, chromosomal translocations, alternatively spliced products. It is also useful for allelic discrimination and detection of allelic imbalance (94).
ON-CHIP PCR REACTION VOLUME
One of the motivations for the development of on-chip PCR is to process a sample of small volume. Based on the reaction volumes of the conventional PCR, we define small volume as ≤3 μl. Within the last two years many small-volume PCRs have been performed (see Table 1). Quake's group performed 72 parallel reverse transcription (RT)-PCRs with a volume as low as 0.45 nl (49). An array of 3072 real-time, 33-nl RT-PCRs was recently reported by Morrison et al. (78). Matsubara et al. performed PCR in 40-nl reaction chambers (17). Neuzil et al. reported a VRC-based PCR system that could amplify a PCR sample of 100 nl covered by 1.0 μl of mineral oil (80). Similarly, Guttenberg et al. amplified a PCR sample of 200 nl covered by 5 μl of mineral oil to form a VRC (77). Landers's group developed a glass/PDMS hybrid PCR-CE chip with on-chip pressure injection using elastomeric valves and the product of 278 bp could be amplified within a PCR chamber of 280 nl (26). Recently, this group also reported a solid-phase extraction (SPE)-PCR-CE integrated glass/PDMS hybrid chip with a 550-nl reaction chamber (27). However, large-volume (>3 μl) on-chip PCRs are still in use (see Table 1). For example, Lee's group developed several PCR chip devices with different functions and volumes of 10 μl or more (38,40,45,47,48,89). A PCR chip device proposed by Shen et al. had a reaction volume of 25 μl (70). When working with samples containing a very low concentration of the target (e.g. in diagnostics), the large-volume PCR has several obvious advantages over small-volume PCR: (i) Evaporative loss of sample during thermal cycling on PCR amplification may be negligible. (ii) The large-volume PCR allows the routine detection techniques such as gel electrophoresis to be used for the analysis of PCR products. (iii) The large-volume PCR sampling is accomplished using a conventional manual sampling gun, making large-volume sample handling during reaction set up relatively easy. However, the large PCR reaction volume can be disadvantageous for low-molecule PCR amplification, especially for single-molecule or single-cell analysis. On the contrary, the submicroliter, nanoliter or picoliter PCR systems are robust in performing analysis with single molecule or cell sensitivity (17,31,35,43,49,78,92,93). However, as the sample volume is decreased, evaporation of sample solution and introduction of a small amount of solution into the reaction chamber/channel can be the major drawbacks for PCR analysis. In addition, as PCR volumes are decreased, amplification is increasingly prone to biochemical surface absorption problems at the chamber/channel walls due to the increasing surface-to-volume ratio.
Table 1.
Reaction volume and heating/cooling rates of PCR chips
| Reaction volume (nl) | Heating rate (°C/s) | Cooling rate (°C/s) | Amplification time (s)/Cycle number | Amplicon length (bp) | References |
|---|---|---|---|---|---|
| Chamber PCR | |||||
| Small volume | |||||
| 0.45 | 240 | (49) | |||
| 33 | (78) | ||||
| 40 | ∼6000 or ∼3720/40 | 74 | (17) | ||
| 100 | 175 | 125 | 340/40 | 82 | (80) |
| 200 | 10–50 | 5 | 600/30 | 150/1382 | (77) |
| 240 | >20 | >20 | 1320/35 | 690 | (32) |
| 280 | 4.8/7.8 | 5.0 | 708/30 | 278 | (26) |
| 380 | >15 | >10 | <1620/30 | (31,35) | |
| 420 | 3 | 2 | (39) | ||
| 550 | 660/30 | 211 | (27) | ||
| 900–7000 | 6.5 | 1.95 | (54) | ||
| 1000 | 44 | 20 | 208 | (79) | |
| 1170 | 522/30 | 300 | (13) | ||
| 1750 | 4.5 | 4.5 | 322 | (53) | |
| 2000 | 11 | 3.2 | 1350/30 | 330 | (11) |
| Large volume | |||||
| ∼3500 | 11 | 2.7 | ∼1680/30 | 209 | (14) |
| 3600 | ∼10 | ∼4.6 | ∼2400/30 | 183 | (51) |
| 8000 | 5 | 2.67 | 1740/25 | 305 | (62,63) |
| 10 000 | 305 | (57) | |||
| 10 000 | ∼20 | ∼10 | 900/20 | 240 | (40) |
| 10 000 | ∼20 | ∼10 | 840/20 | 273 | (47) |
| 10 000 | ∼20 | ∼10 | 1584/30 | 273 | (45) |
| 15 000 | 38 | 7.9 | 3300/25 | 511 | (89) |
| ∼20 000 | 11.5 | 5.5 | <900/20 | 273 | (38) |
| 25 000 | 8 | 8 | 1680/30 | 244 | (70) |
| 37 500 | 3.1 | 3.1 | 2100/40 | 544 | (37) |
| 50 000 | ∼20 | ∼10 | 511 | (48) | |
| Continuous-flow PCR | |||||
| (480 ∼ 1800)/30 | 430 | (44) | |||
| 500 | 1086/27 | 500 | (56) | ||
| 1000 | ∼900/35 | (23) | |||
| ∼6000 | 2100/25 | 145 | (41) | ||
| 4500 | ∼12 | ∼12 | 2280/23 | 240 | (64) |
| 8000 | 300/>40 | 372 | (65) |
Small-volume PCR should not only reduce the costs but also allow rapid thermal cycling. However, the use of small-volume PCR does not necessarily reduce amplification time if the heating method does not fully use the advantages of small-volume reaction system (17,43,49,78). On the contrary, the large-volume PCR can result in the reduction of amplification time when a better mode of heating is utilized (38,40,45,47,48,89). As yet, there is no definitive correlation between PCR time and PCR volume. PCR speed ultimately depends on the thermal mass of the entire PCR chip system. Although changeable in a wider range, the sample volume of continuous-flow PCR is usually <10 μl (see Table 1).
ON-CHIP PCR REACTION SPEED
Pursuing high-speed PCR is one of the major motivations in the development of on-chip PCR. The acquisition of high-speeds for PCR depends on reduction of the thermal mass of the entire PCR system. The basic methods used to decrease the system's thermal mass are to choose a better heating method and to adopt a desirable chip architecture.
For chamber-based stationary PCR chips, thermal cycling can be performed either with contact heating methods or noncontact heating methods. The former is defined as having heaters fabricated within the chip or in thermal contact with the outside of the chip, where the thermal mass of the microchip is in contact with the heating element. The latter uses a heating method that is not in physical contact with the PCR chamber (3). Among the contact heating methods, the MEMS-based film heating elements have smaller thermal mass, faster thermal response and higher heating rates (e.g. >10°C/s) (11,14,31,32,35,38,40,45,47,48,51,77,79,80,89) (see Table 1). To date, the fastest heating rate (175°C/s) and cooling rate (125°C/s) have been obtained using this method (80). However, since these heating elements are usually fabricated in a complicated process, the costs are considerably higher than other designs. To reduce costs, the temperature control chip can be physically separated from the PCR reaction chip and reused after initial temperature calibration (20,46,48,79,80). To develop a cost-effective heating element, interest was focused on the commercially available flexible thin film heaters with a heating rate of 6.5°C/s or higher (54,59,60,67,70). Alternatively, contact heating is also often realized by a Peltier device (17,37,43,53,62,63). Although its high thermal mass weakens the thermal response of the entire PCR chip, ∼5°C/s heating rates can be acquired and thus the system performances are better than most of Peltier effect or metal block-based PCR machines. A disadvantage associated with the contact heating is that a certain amount of thermal mass is added in the PCR chip assembly, which inevitably hinders fast thermal transitions. Moreover, when PCR and analytical function (e.g. CE) are integrated on a single chip, it is very difficult to confine the contact heating to the PCR chip itself and not analysis part of the chip. In order to overcome these issues, interest in noncontact heating continues to grow (4). Recently, Landers's group successfully realized the integration of noncontact infrared (IR)-mediated PCR with CE separation (26) or with SPE and CE separation (27) on a single glass chip. Hu et al. proposed a new method to control PCR thermal cycling using an alternating-electric-current induced buffer Joule heating effect without an external heater component (39). Although this approach obtained a low heating rate of 3°C/s and cooling rate of 2°C/s, there is still much room to improve the thermal response of Joule heating if the total thermal mass of the chip can be reduced.
As described above, the heat inertia of a continuous-flow PCR system is considered as only the sample thermal mass, and the temperature transition time depends only on the sample flow rate and its time to reach thermal equilibrium. Consequently, the speed of continuous-flow PCR is limited only by the synthesis rate of the DNA polymerase. For example, in the 30-cycle continuous-flow PCR chip proposed by Kim et al., only 8–30 min were required to produce a detectable amount of 430-bp PCR products (44). Münchow et al. reported that 372-bp PCR products could be achieved within 5 min by over 40 thermal cycles in the oscillatory-flow PCR chip (65). The fastest PCR reported to date was obtained on the continuous-flow PCR chip developed by Soper's group — amplification of a 500 bp λ-DNA fragment in 1.7 min and a 997-bp fragment in 3.2 min, respectively (59).
APPROACHES TO ELIMINATING CROSS CONTAMINATION
Due to the extremely sensitive nature of the PCR, minute amounts of nucleic acid contamination in PCR chips can result in a false positive PCR outcome. In order to eliminate sample-to-sample or run-to-run cross contamination in PCR microdevices, the most straightforward approach is the use of a disposable PCR chip. At minimum, the part of the chip device that comes into contact with the sample solution is discarded after a single use. The low cost and easy fabrication often allows disposal of the polymer-based PCR chips after a single use (38,46,54,57,62,63,70). Recently, Neuzil et al. used the disposable glass slip to perform the on-chip PCR amplification (79,80). In the case of the high costs of fabrication or substrate materials, the following economical and applicable approaches to eliminating cross contamination have been developed.
Creation of liquid–liquid two-phase reaction systems. Viovy's group performed continuous-flow, high-throughput PCR amplification using droplets in an immiscible, fluorinated oil/fluorosurfactant solvent, without any detectable contamination between droplets (72,71). In such systems, the stability of droplet trains is essential for eliminating cross contamination between droplets.
Use of differential hydrophilic/hydrophobic coating. In this approach, the interior surface of each reaction chamber remains hydrophilic, while the exterior surface is rendered hydrophobic by coating a hydrophobic layer onto the substrate or using a substrate with hydrophobic nature (e.g. silicon) (17,78). This differential hydrophilic–hydrophobic coating approach facilitates precise loading and isolated retention of PCR solution in individual chambers, thus avoiding cross contamination between chambers. However, this approach maybe is not robust enough to be of practical usefulness. Recently, Prakash and Kaler developed a novel ‘stripping and re-silanizing’ (SRS) approach for chip reuse (33). The SRS process can be repeated many times between PCR experiments and can avoid PCR carryover cross-contamination since it imparts a fresh coating prior to each PCR experiment. It should be noted that the deep hydrophilic reaction chambers can also avoid the contamination between chambers during PCR (83).
Use of microvalves on chip. The microvalves can be used to isolate each reaction chamber or to separate the PCR reaction chamber with other functional units such as CE and SPE, thus effectively avoiding possible cross contamination (10,19,27,20,26,31–33,35,43,52,92,93). Valving systems that contact the PCR mixture could increase the risk of contamination arising from small protrusions and crevices that may trap DNA and reagents from a previous run. Therefore, the actuating elements of microvalves should not be in direct contact with the PCR mixture (43,52,53), or the actuating elements of microvalves should be replaced for a new run (31,35).
Cleaning of PCR chip. If the chips are cleaned with organic solvents or strong detergents without causing damages, the carryover cross contamination from run-to-run can be effectively eliminated, and as a result the PCR chip can be reused (31,35,43,77).
Pretreatment of reaction system with biological reagents. For example, Cho et al. incubated the PCR chip with uracil-N-glycosylase (UNG) to destroy potential carryover contamination (10). Finally, it needs to be noted that if each PCR chip is sealed with a cover (for example a tape) immediately after loading the sample, contamination is less likely.
FLUIDIC AND TEMPERATURE CONTROLS AND MEASUREMENTS OF ON-CHIP PCR SAMPLE
Thermal Insulation
Thermal interaction or ‘crosstalk’ has emerged as an important issue as chip device size decreases. Most DNA-based assays [e.g. PCR, restriction endonuclease reaction, temperature gradient gel electrophoresis (TGGE) and PCR-sequence-specific oligonucleotide polymorphism (SSOP)] are highly temperature sensitive and require precise temperature control. When integrating these analytical components on a single chip, thermal crosstalk will deteriorate chip device's performance and the thermal insulation is often required. The thermal isolation should also be used to eliminate or weaken the thermal crosstalk between chambers within the PCR array chip because PCR conditions vary slightly from one gene target to another although DNA hybridization occurs at the same temperature for many different gene targets. In addition, the thermal isolation between the temperature zones and the substrate is also usually considered to prevent heat loss to the surroundings. For these reasons, there have been increasing attempts to develop thermal isolation solutions on PCR chips. Shih et al. recently proposed a novel technology using parylene-cross-linking structure to achieve air gap thermal isolation for on-chip continuous-flow PCR (81). This technology provides excellent thermal isolation efficiency. Its simplicity of integration with other analytical components also makes applications of micro total analysis systems (μTAS) feasible. Burn's group reported in detail two cost-effective thermal isolation techniques: the thermal conduit technique based on a selective conduction mechanism (32,36), and the silicon back-dicing technique based on a selective insulation mechanism (36). They are inexpensive alternatives to the silicon back etching technique. In addition, most existing thermal isolation techniques also adopt thin substrate structures (e.g. cantilever beam (79,80) or deep trenches (14,23,26,63)) to thermally insulate the PCR region. These structures can provide excellent thermal isolation due to their high thermal resistance, but they usually have the low mechanical stability and require complicated microfabrication processes. Recently, Zou et al. proposed a simple conductive polymer flip-chip bonding technique to accomplish the thermal isolation of multi-chip array to allow the PCR chambers to be thermally and independently controlled (25).
Evaporation and its Inhibition Measures of PCR Sample
Sample evaporation is often problematic because PCR volumes are usually very small. Especially as denaturation temperatures approach 100°C the sample evaporation is so rapid that the sample would dry up quickly under standard atmospheric pressure. To circumvent such evaporation, a number of measures can be taken, but all have their respective advantages and disadvantages. A mineral oil cover layer is frequently used as a vapor barrier to prevent evaporation (17,26,27,29,54,77,79,80). The mineral oil is a suitable liquid cover because it has a boiling point far above 100°C and a density slightly below 1.0 g/cm3. However, its applicability is questionable for highly integrated PCR systems. Another approach is using a solid cover or valve to resist the internal pressure generated during PCR (10,19,20,31,32,35,43,51,52). It is known that the evaporation rate decreases with the increase of the gas pressure around a liquid. Recently, Cheng et al. extended this concept to the oscillating-flow PCR chip (64). In their approach, a single opening serves for both sample loading and syringe pump port. When the sample plug is pumped to high-temperature zones, the internal pressure increases by six times and thus the sample evaporation is greatly reduced. Noteworthy is that within the continuous-flow PCR chip, the relative sample evaporation loss can be decreased because of the decrease in the free surface area of the liquid. In addition, sample evaporation is also affected by the PCR chip substrates. For example, the water diffusion/vapor loss property of PDMS could lead to the sample evaporation loss and thus special approaches such as vapor barrier (52,53) should be considered to reduce the sample loss at elevated temperatures.
Formation of Gas Bubbles and Inhibition Measures
An important drawback of the PCR chip microsystems is the generation of air bubbles. which not only cause large temperature difference in the sample but also expel the sample from the PCR chamber. The formation of air bubbles has two prerequisites: the liquid must be superheated and there must be nucleation site(s). Several methods have been used to avoid the prerequisites and inhibit the bubble generation: (i) The structural design of PCR chamber. A diamond-shaped or rhomboidal chamber is superior to a circular chamber in preventing bubble formation (10,51). Recently, Gong et al. reported that the deeper the PCR chamber, the more difficult it is for the PCR solution to flow into the chamber without trapping bubbles. However, the size of the chamber or the shape and size of the inlet and outlet have little or no influence on the bubble formation (28). (ii) The surface treatment of the PCR chamber. In general, the wetting properties of the PCR chamber and its inlet/outlet have an obvious effect on the bubble formation. When the chamber surface is highly hydrophilic, the PCR sample can flow into the chamber smoothly and rapidly without bubble formation (28,31,35,51). (iii) The sealing pressurization of the PCR chamber. Under pressurization and high-temperature, the gas solubility will increase and the dissolved gases and microbubbles in the PCR sample cannot grow up in volume, thus preventing the air bubble formation (37,50,57,62,63). (iv) Degasification of the PCR sample. This process can eliminate non-condensable gases in the PCR sample before loading and consequently decrease the risk of bubble formation (56). (v) The addition of high boiling-point biocompatible reagents to the PCR sample. When a solvent with a boiling point above 100°C (e.g. glycol, glycerol or poly(ethylene glycol)) is included in the PCR sample, the boiling point of the resulting sample is increased, and thus preventing bubble formation at high temperatures (56).
Semi-Invasive or Noninvasive Temperature Measurement for PCR Chips Based on Optical Techniques
Since the PCR is temperature sensitive, it is essential to choose a proper temperature measurement technique to determine or at least to estimate the temperature within the PCR chip. In general, the measurement techniques can be classified into three categories based on the nature of the contact mode between the temperature measuring element and the PCR chip (solid) or PCR solution (liquid): (i) Invasive. The measuring element is in direct contact with the PCR chip or solution. (ii) Semi-invasive. The PCR solution is treated in some manner to enable remote observation, e.g. inclusion of dye whose color changes with temperature. (iii) Noninvasive. The PCR solution is observed remotely, e.g. IR thermography. Due to their low cost and convenience, invasive temperature measurements are still most widely used (4). However, there are several obvious challenges associated with these measuring techniques: First, regardless of the nature of the temperature sensing element, it will add some thermal mass to the PCR chip, ultimately decreasing the chamber PCR thermal cycling rates. Second, they can obtain temperature data only at a few discrete points or lines and thus do not reflect the overall temperature field of the PCR chip or solution. Third, the temperature sensor within the PCR solution will inhibit the PCR and increase the risk of sample cross contamination. Fourth, the use of invasive temperature measurement involves a disturbance, which manifests itself as a difference between the temperature being measured and that which would exist in the absence of the temperature-measuring element. To address these issues, attention has been given to the semi-invasive or noninvasive temperature (and fluidic) measurement techniques, which are usually accomplished by the optical techniques.
Semi-invasive temperature measurement techniques for PCR chips
In these techniques, temperature-sensitive materials such as thermochromic liquid crystals (TLCs) or fluorescence dye indicators are included in the PCR solution, variations in optical properties can be observed remotely. These techniques are categorized as semi-invasive since they involve a modification of the component in the solution and therefore could cause disturbances to the temperature field.
Semi-invasive temperature measurement based on TLCs
TLCs are substances with a molecular structure intermediate between that of a crystalline solid and an isotropic liquid. They possess both mechanical properties of liquid and optical properties of crystalline solids. Of particular relevance to heat transfer studies are the cholesteric liquid crystals (commonly called TLCs), which are optically active since they react to changes in temperature and shear stress by changing their color. Encapsulated TLCs are commercially available and have proved to be very useful for research on PCR chips (4). Recently, Noh et al. utilized TLC thermometry to investigate the spatial temperature uniformity inside the PCR chamber. The chip used in this work has the smallest volume (ca. 1 μl) that has been ever reported in TLC thermometry (18). Kim et al. used the TLC thermometry to measure the temperature at the Raman calibration point to calibrate the Raman intensity of O–H stretch band of water with temperature (15). Additionally, TLC solution has also been used to determine the rate of temperature change in the PCR microchannel (64). Although this technique can directly obtain the spatial and temporal temperature distribution with a high precision of 0.1°C, it usually has a small measurement range. The TLC's encapsulated spheres typically have a diameter of several ten micrometers and consequently it is hard to apply them in the PCR system with smaller characteristic dimensions. In addition, TLCs have to coexist with a PCR solution, which might affect the biological activity of the medium.
Semi-invasive temperature measurement based on a fluorescence dye indicator
The principle of this temperature measuring technique has been described by Sakakibara and Adrian (95). Although this technique has been well studied, it was not applied for the temperature measurement within the PCR chips until recently. Guttenberg et al. used a thermochrome dye chromazone-based temperature measuring technique to yield more temperature information inside the PCR sample droplet (77). Neuzil et al. investigated the temperature measurement of the on-chip PCR sample in SYBR Green-based system or 6-FAM-based system (80). SYBR Green allows melting curve analysis (MCA) to be routinely used for PCR sample temperature monitoring, but it can be used with high precision only around the DNA melting point. By contrast, the fluorescence intensity of 6-FAM is proportional to its temperature up to 90°C and thus it is more suitable to estimate the PCR sample temperature over a wide range. Most recently, rhodamine B, one of most widely used temperature sensitive dyes, has been utilized to evaluate the accuracy of the in-channel temperature measured by K-type thermocouple or by numerical simulation (39). This dye-based measuring technique has advantages of simplicity, a wide range of temperature measurement, fast response speed, good spatial resolution and great sensitivity. However, its measurement accuracy is low.
Noninvasive temperature measurement techniques for PCR chips
Most of the noninvasive techniques measure temperature using the electromagnetic spectrum. For example, IR devices are sensitive to the spectrum in the infrared region. Optical techniques such as absorption and emission spectroscopy are sensitive in the visible region.
Noninvasive temperature measurement technique based on IR thermography
Temperature measurements by monitoring thermal radiation in the IR spectrum have been widely applied in the PCR chips, especially within the recent two years (10,14,17,30,32,47,64,65,67,70,77,79). Such IR thermography-based technique usually consists of the PCR chip, the environment, the air transmission medium and the measurement device. Factors such as radiation attenuation, background radiation, uncertainty of the chip emissivity and errors from the thermometer itself can affect the accuracy of an IR thermometer. The IR thermometer must be calibrated to compensate for the environmental effects. Although this technique can only obtain the surface temperature distribution of the PCR chip, the actual temperature of the PCR sample can be reasonably deduced with the help of the heat transfer law. Attractive characteristics of this technique also include rapid response, continuous temperature readings, high spatial resolution, no interference with the object observed and the ability to determine the temperature of moving component.
Noninvasive temperature measurement technique based on Raman spectroscopy
If a molecule is excited by incident radiation from the ground state to a higher unstable vibrational state, it can either return to the original state (i.e. Rayleigh scattering), or to a different vibrational state (i.e. Raman scattering). There are two basic methods for determining the temperature by Raman scattering: the Stokes Raman method and the Stokes to anti-Stokes ratio method (96). The Raman spectroscopy-based temperature measurement has been successfully applied to the PCR chips as a new in situ and noninvasive temperature measuring technique (15). The diameter of the laser beam in the micro-Raman system can be adjusted to 1 μm and the perturbation of the fluid condition can be negligible. The laser beam can be focused anywhere inside the chamber/channel to allow the whole field temperature measurement.
Semi-Invasive Fluid Measurement for PCR Chips Based on Optical Techniques
It is essential to know the flow state of the micro fluids in the PCR chip since results depend on the fluid distribution and variation in the reaction system. Less attention has been devoted to the fluid mechanics of micro PCR devices than to other aspects. One notable advance has been to use semi-invasive fluid measurement techniques to measure flow fields inside a continuous-flow PCR chip. Curtin et al. used the micro particle image velocimetry (μ-PIV) and pressure measurements to study the effect of PCR on biofluid viscosity in continuous-flow PCR chips (58). It is reported that for low molecular weight substances, the biofluid viscosity will not increase after PCR. Li et al. used μ-PIV to measure the velocity fields inside the continuous-flow PCR microchannel at various downstream locations (30). Using μ-PIV in the PCR chips represents a novel application of existing measurement techniques. For this technique, the size choice of the fluorescence seed particle is essential, as a particle that is too large or too small will cause undesirable effects on flow field.
Numerical Simulation of Temperature and Fluid Fields in PCR Chips
Due to the small dimensions of the PCR chips, the direct temperature/fluid measurement remains a challenge. Moreover, direct measurement cannot provide any static and/or dynamic information on temperature/fluid before micromachining of PCR chips. In order to address this issue, numerical simulation is thought to be a very effective approach and can provide the opportunity to evaluate flow, thermal and even chemical processes during PCR (4). Within the past two years, this approach has been widely used on the chamber-based stationary PCR chips (10,12–14,20,21,24,25,36,39,51,70). Meanwhile, a great attention has been focused on the numerical simulation of the continuous-flow PCR chips (23,30,41,42,66,87,88). For example, Gui and Ren developed a 3D model to simulate the electrical potential field, flow field and temperature field in an electro-osmosis-based continuous-flow PCR chip (87). Tsai and Sue numerically simulated the flow field at the continuous-flow channel turning corners, as well as the performance of designed heaters (66). Wang et al. described a 2D numerical model to study the effects of chip geometries, materials, heater temperatures, flow rates and boundary conditions on the thermal performance of the continuous-flow PCR chip (88). Commercial software often used in the numerical simulation include ANSYS (30,36,41,42,66,70,79), CFD-RC (66), CFD-ACE (+) (10,20,45), FLUENT (41,42), FLOTHERM (12), CoventorWare (13,14,21,25) and COSMOS (51). ConventorWare (IMAG Inc.) comprises a simulation module devoted to the microfluidics, and thus can simplify the development of the microfluidic mathematical model and accelerate the study of the microfluidics theory. FLOTHERM (Flometrics Inc.) is a thermal analysis software tool developed for the electronics industry and it has been widely used for the thermal modeling of microfluidic systems.
DNA DETECTION METHODS FOR PCR CHIPS
Off-line detection methods
All existing DNA detection methods can be used for off-line detection of on-chip PCR products. The use of intercalators in combination with gel electrophoresis is the most widely used method for the detection of post-PCR products (14,23,25,28,30,38,40,44–48,50,51,54,56–60,62–64,66,70,72,77,89). In this combination, the DNA molecules are effectively labeled with an intercalating dye and subsequently separated according to their sizes. Upon binding to double-stranded DNA, the intercalator molecules exhibit significant enhancement in their fluorescence quantum efficiencies. Ethidium bromide (EtBr) and SYBR Green I are the most popular intercalator dyes, but other intercalating dyes, such as GoldView™, have also been investigated (74–76). The use of intercalator to detect DNA molecules has two main advantages: real-time detection (10,20,77,79,80) and versatility. However, indiscriminate binding is also a major disadvantage: both specific and nonspecific PCR products can produce the same type of signal and it is difficult to differentiate between them. In addition, this technique is time-consuming and labor intensive.
CE is another widely used off-line detection technique (97–100). The use of CE for DNA separation detection has several obvious advantages including low operating costs, high separation efficiency, small sample volume, short analysis time, versatility and simplicity. Incorporation of CE on a chip fully utilizes these advantages. CE chips were first demonstrated in 1990 by Manz et al. (101) and have now become commercially available. One notable example is the Agilent 2100 Bioanalyzer (Agilent Technologies). Recently, this commercial CE chip has been used for off-line detection of PCR products (10,11,13,16,69,73). Other microchip electrophoresis systems include the Hitachi SV 12-channel electrophoresis microchip (Hitachi Electronics Co.) (73), LabChip® 90 Automated Electrophoresis System (Caliper Lifesciences) (20) and CE chips with laser-induced fluorescence (LIF) detection (41,52,53,56). The off-line CE chip platform can offer walkaway and unattended analysis of DNA molecules, eliminate the time-consuming and messy slab gel process, generate much more reproducible and high quality data, and allow high-throughput laboratory analysis; however, the manual sample loading may increase the risk of cross contamination and the total analysis time.
On-line detection methods
In order to circumvent the drawbacks of off-line DNA detection methods, great efforts have been made to develop on-line DNA detection methods. Some of the on-line detection methods described below may soon be routinely implemented for on-chip quantitative and/or qualitative PCR detection.
Fluorescence-based DNA detection method
Fluorescence-based real-time and end-point DNA detection is a powerful and important detection technique. The real-time detection kinetically observes the fluorescence signals from the interaction between the dye or probe and the increasing amount of double-stranded DNA molecules during PCR (10,16,20,37,39,50,54,77–80); the end-point detection measures the fluorescence only before and after PCR amplification, by which it can be established whether the DNA template is successfully amplified (17,49,71). Although the end-point method is simple and can be used for single cell or molecule analysis (17,49), it is difficult to obtain reliable results if the DNA template concentrations are calculated only by the fluorescence signals. The real-time method has become popular. It uses an analysis procedure called threshold method. The threshold value (also called CT value) represents the number of cycles necessary to produce an exact amount of DNA. By using a series of different CT values, one can calculate the DNA template concentrations and obtain the PCR efficiency. If the CT value is further used as a criterion for the PCR efficiency, the dynamic parameters of the on-chip PCR can be optimized (80). However, since the sequence-nonspecific fluorescence dyes are often utilized for the on-chip real-time PCR systems, both specific and nonspecific products will produce the same fluorescence signals that are difficult to differentiate. In order to overcome this challenge, after on-chip PCR a MCV can be performed to determine the purity of the PCR and to identify the specific target products (10,16,20,78,79). Other on-chip fluorescence-based detection technologies are also being developed. One such example was the DNA—‘up-converting’ phosphor particles (UPT)—lateral flow (LF) assay on a single chip proposed by Wang et al. (63), in which the PCR products were labeled with the UPT reporter particles in an incubation chamber, applied to the LF strip, bound to pre-immobilized ligands and detected with an IR laser.
On-chip CE separation integrated with PCR
One of the most common components integrated with PCR on a single chip is CE. A hybrid DNA analytical chip that performs PCR in a silicon microheater and CE in a glass chip was first reported by Woolley et al. in 1996 (102). Since then, a variety of chips with different degrees of functional integration have been reported (4). Nowadays, the integrated PCR-CE chip is still a burgeoning research area (26,27,31–33,35,40,43). For example, Easley et al. proposed on-chip pressure injection for integration of IR-mediated PCR amplification with CE separation (26). It was reported that the total analysis time was ∼12 min, representing the fastest PCR-CE chip integration achieved to date. Within another integrated chip recently reported by Easley et al. (27), nucleic acid purification through SPE, followed by PCR amplification and microchip CE amplicon separation and detection, was completed in less than 30 min. Huang et al. presented an integrated chip for DNA/RNA amplification, CE separation and on-line optical detection (40). This work has two important improvements: (i) The sample template can be DNA or RNA and (ii) using a buried optic fiber instead of the LIF to optically detect the PCR products can avoid the bulky optical detection apparatus. Another notable work is the development of four-lane PCR-CE chip that is capable of performing multiple (RT-) PCR-CE analyses in parallel (31,35). Recently, Prakash and Kaler reported an integrated PCR-CE chip that performed nine PCR reactions simultaneously in separate chambers and then sequentially analyzed the PCR product from each chamber by CE (33). Those works may represent the development trend of the future PCR-CE chips: high analytical speed, high functional integration without the need for the external bulky equipments and improved parallelism.
On-chip DNA hybridization integrated with PCR
DNA hybridization is a highly selective and sensitive reaction which takes place between target DNA/RNA and a DNA probe. With the advent of microfabrication techniques, the miniaturization of the DNA probe detection has proven to be feasible, and both DNA hybridization and PCR amplification can be incorporated on a single chip to directly detect the PCR products. For example, Guttenberg et al. used the on-chip SAW pump to combine the PCR amplification with the PCR product hybridization detection (77). Hashimoto et al. applied the PCR and/or LDR chips combined to hybridization microarrays to detect low-abundance DNA point mutations in a continuous-flow format (59,60). The coupling of functional biochip and informational biochip not only allows functional operations such as sample preparation and PCR amplification by controlling the biological microfluids, but also provides information on the amount, sequence and even DNA/RNA target source through DNA hybridization microarrays.
HIGH INTEGRATION OF FUNCTIONAL COMPONENTS TOWARDS PORTABLE PCR DEVICES
One of the main challenges in miniaturization of PCR is the integration of functional components to perform several operations without the need of external macro apparatus or manual operation. With the advance of the PCR chips, more and more on-chip PCR systems have been developed. Lien et al. reported a device which could accommodate the following operations: virus loading, virus capture, virus purification, cell lysis, plus RT and stationary PCR (48). In addition, micropumps, microvalves, micromixers, microcoils, film-resistive heaters and film temperature sensors were integrated on the chip. Another system developed by Hashimoto et al. possessed following capabilities: continuous-flow PCR, LDR and hybridization (60). Other integrated PCR chips have also been investigated (see Table 2). The stationary chamber PCR seems to be preferred for integrated systems when compared to the continuous-flow PCR system. This might be due to an increased simplicity in sample handling and structure designing for the former system. The most commonly integrated physical elements are film-resistive heaters, film-temperature sensors, micropumps and microvalves (see Table 2).
Table 2.
Integrated operation and components of miniaturized PCR chips
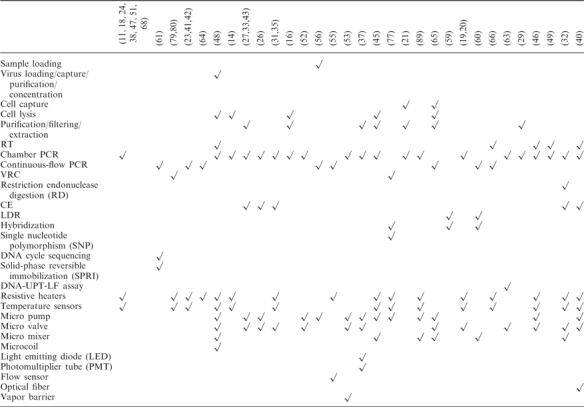 |
Although the partially integrated PCR chips have been successfully developed, the PCR-based ‘complete’ lab-on-a-chip still requires further development. The bottlenecks blocking the realization of a truly and highly integrated PCR chip include on-chip pre-PCR sample preparation and on-chip PCR product detection. Since the source of raw template samples is varied and the sample preparation methods are diverse, the miniaturization of conventional sample preparation and functionalities on a chip remains a challenge (103). As for the on-chip on-line DNA detection, the bulky optical detection systems such as charge-coupled device (CCD) and LIF are difficult to miniaturize onto a single chip. However, along with the development of MEMS technology, the optical-MEMS apparatus such as optical fiber, LED and PMT can be applied onto the PCR chip to realize a portable DNA analysis device (37,40).
BIOLOGICAL SAMPLES USED IN PCR CHIPS AND POTENTIAL APPLICATIONS OF PCR CHIPS
Starting DNA/cDNA samples used in PCR chips include bacterial genomic DNA (for example, Escherichia coli (E. coli) (17,39,42,51,54,58,68), Salmonella typhimurium (S. typhimurium) (26), Bacillus anthracis (B. anthracis) (30), cyanobacterial genomic (11), Streptococcus pneumoniae (S. pneumoniae) (38,47), Staphylococcus aureus (S. aureus) (47), Streptococcus pyogenes (S. pyogenes) (47), Neisseria meningitides (N. meningitides) (47), Bacillus cereus (B. cereus) genomic (57,62,63) DNA), viral genomic DNA (for example, human papilloma virus (HPV) (23), hepatitis C virus (HCV) (42,70), hepatitis B virus (20), human immunodeficiency virus (HIV)-1 (33), Dengue II virus (89), Haemophlius influenzae (H. influenzae) (47) and severe acute respiratory syndrome (SARS) (28) DNA), λ phage DNA (13), yeast genomic DNA (53), human genomic DNA (11,17,22,32,60) and others (31,32,50,53,69,77).
For RNA analysis, cDNA are reversed transcription from the corresponding RNAs. They may include cDNA from sorted CD19+ malignant B cells (52), cDNA from brain tissue from transgenic GFAP–GFP mice (80), cDNA from human breast cancer cell lines T47D (71), cDNA from human heart and human liver samples (78) and influenza viral cDNA (32).
In order for a PCR microfluidic chip to have value in clinical diagnostics or genetic profiling analysis, it must be capable of accepting the crude biological samples (other than purified DNAs) as an analytical target, such as cells, total RNA, virus, whole blood, urine, sperm, or nasal aspirate (see Table 3). As seen from a wide range of biological samples being on-chip amplified and analyzed, the PCR chips could be used for broad applications including molecular diagnostics of diseases (10,32,43,47,60), gene expression analysis (35,71,78,92,93), forensics, environmental testing, food safety testing and biothreat sensing. For example, the PCR/LDR/hybridization chip reported by Hashimoto et al. has been used to detect low-abundant DNA mutations in gene fragments (K-ras) that carry point mutations with high diagnostic value for colorectal cancers (60). The lowest mutant:wide-type ratio that could be detected by this chip was up to 1:80, and the total assay time was ∼50 min, including 18.7 min for PCR, 8.1 min for LDR, 5 min for hybridization, 10 min for washing and 2.6 min for fluorescence scanning (60). Since the sample preparation processes have been integrated on a single chip, along with the robust product detection techniques, the total analysis process can be completed within less than 25 min (less than 10 min for DNA extraction, 11 min for PCR and less than 3 min for injection, CE separation and detection) (27). In addition, due to high integration and miniaturization of PCR chips, the biological sample and costly reagent consumption, as well as the possible contamination resulting from manual processes can be decreased. In principle, the PCR chips can be applied in any field where minute amounts of nucleic acid sample needs to be rapidly amplified and subsequently analyzed.
Table 3.
Examples of integrated PCR chips that can accept the crude biological samples
| Crude biological samples accepted on-chip | Approaches to processing crude samples on-chip | References |
|---|---|---|
| E. coli K12 MG1655 cells | (31) | |
| S. pneumoniae cells | Thermal lysis | (45) |
| Whole blood or sperm cells | μ-SPE | (29) |
| E. coli cells | Thermal lysis | (14) |
| Bacterial cells (Streptococcus mutans, Staphylococcus epidermidis and E. coli BL21) and HBV | Laser-irradiated magnetic bead system (LIMBS) | (16) |
| Listeria monocytogenes cell lysate | μ-SPE | (37) |
| E. coli cells or total RNA, and human breast total RNA | Reverse transcription | (35) |
| Dengue virus type-2 and enterovirus 71 (EV 71) RNA | Reverse transcription | (46) |
| Dengue virus serotype 2 and enterovirus (EV) 71 viruses | Antibodies-conjugated magnetic bead capture and reverse transcription | (48) |
| Human whole blood | Micropillar array filter | (21) |
| White blood cells or whole blood from a patient's blood | μ-filter | (65) |
| Mice whole blood, and Nasal aspirate from a patient symptomatic of whooping cough | μ-SPE | (27) |
| Urine samples from renal transplant patients | (43) |
PRACTICAL ISSUES RELATED TO THE DEVELOPMENT OF PCR CHIPS
Despite great progresses in many aspects of PCR chips that have made them a central part of μTAS, PCR chips also face some practical issues. First, due to the complicated MEMS process, many PCR chips are relatively expensive and thus not disposable. Therefore, cross contamination between samples is difficult to avoid. Second, fully integrated PCR chips are difficult to make. Many PCR chips can only perform the DNA/RNA amplification. The development of the highly integrated PCR chips is limited by complicated design and fabricating process. Third, the product detection methods have not advanced as rapidly as other aspects of chip development. Most of the PCR chips still utilize the conventional gel electrophoresis techniques to detect the products. With the decrease in PCR volume, detecting the products by this technique becomes a challenge. Fourth, intellectual property issues, which may limit the ability to combine different technologies in a single system have yet to be fully explored. Finally, when PCR chips become widely used clinically, ethical issues such as genetically-ally-based employment discrimination will become more urgent and protection legislation will need to be considered.
SUMMARY
Our survey of recent literatures on miniaturized PCR chips concludes that such PCR chips have been well developed and have found important applications in miniaturization techniques. The autonomous or quasi-autonomous high-speed implementation of nucleic acid amplification and analysis in the case of a small-volume biological sample will provide continued impetus for the development and improvement of miniaturized PCR chips. Although the PCR chip provides many advantages over the conventional PCR device, the miniaturization also raises some challenging issues. Thus, many problem-solving strategies are required to tackle challenges such as adsorption of the reagents to the chip surface, being prone to evaporation of the sample solution and formation of gas bubbles, the requirement of precise temperature control and the patent dispute. Despite these obstacles, the potential of miniaturized PCR, as a future nucleic acid amplification and analysis tool, is still attractive. As the development of PCR chips continues, newly designed microstructures should make a full use of the better MEMS procedures to meet the requirements of each bioassay procedure to enable them to be widely customized to accomplish specific bioassays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support by the National Natural Science Foundation of China (30670507; 30600128; 30470494) and the Natural Science Foundation of Guangdong Province (015012). Funding to pay the Open Access publication charges for this article was provided by the National Natural Science Foundation of China (30470494).
Conflict of interest statement. None declared.
REFERENCES
- 1.Saiki RK, Scharf F, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 2.Auroux P.-A, Koc Y, deMello A, Manz A, Day PJR. Miniaturized nucleic acid analysis. Lab Chip. 2004;4:534–546. doi: 10.1039/b408850f. [DOI] [PubMed] [Google Scholar]
- 3.Roper MG, Easley CJ, Landers JP. Advances in polymerase chain reaction on microfluidic chips. Anal. Chem. 2005;77:3887–3894. doi: 10.1021/ac050756m. [DOI] [PubMed] [Google Scholar]
- 4.Zhang CS, Xu JL, Ma WL, Zheng WL. PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster … better. Curr. Opin. Struct. Biol. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Hong JW, Quake SR. Integrated nanoliter systems. Nat. Biotechnol. 2003;21:1179–1183. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- 7.Kartalov EP, Anderson WF, Scherer A. The analytical approach to polydimethylsiloxane microfluidic technology and its biological applications. J. Nanosci. Nanotechnol. 2006;6:2265–2277. doi: 10.1166/jnn.2006.504. [DOI] [PubMed] [Google Scholar]
- 8.Quake SR, Scherer A. From micro- to nanofabrication with soft materials. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- 9.Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77:977–1026. [Google Scholar]
- 10.Cho YK, Kim J, Lee Y, Kim YA, Namkoong K, Lim H, Oh KW, Kim S, Han J, et al. Clinical evaluation of micro-scale chip-based PCR system for rapid detection of hepatitis B virus. Biosens. Bioelectron. 2006;21:2161–2169. doi: 10.1016/j.bios.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Consolandi C, Severgnini M, Frosini A, Caramenti G, De Fazio M, Ferrara F, Zocco A, Fischetti A, Palmieri M, et al. Polymerase chain reaction of 2-kb cyanobacterial gene and human anti-α1-chymotrypsin gene from genomic DNA on In-Check single-use microfabricated silicon chip. Anal. Biochem. 2006;353:191–197. doi: 10.1016/j.ab.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Gnanappa AK, Cathy K, Slattery O, Sheehan M. Thermal performance analysis of a silicon microreactor for rapid DNA analysis.. The Tenth Intersociety Conference on Thermal and Thermomechanical Phenomena in Electronics Systems (ITHERM’06); San Diego, CA, USA. 2006. pp. 1330–1335. [Google Scholar]
- 13.Hataoka Y, Zhang LH, Yukimasa T, Baba Y. Rapid microvolume PCR of DNA confirmed by microchip electrophoresis. Anal. Sci. 2005;21:53–56. doi: 10.2116/analsci.21.53. [DOI] [PubMed] [Google Scholar]
- 14.Ke C, Kelleher AM, Berney H, Sheehan M, Mathewson A. Single step cell lysis/PCR detection of Escherichia Coli in an independently controllable silicon microreactor. Sens. Actuators B Chem. 2007;120:538–544. [Google Scholar]
- 15.Kim SH, Noh J, Jeon MK, Kim KW, Lee LP, Woo SI. Micro-Raman thermometry for measuring the temperature distribution inside the microchannel of a polymerase chain reaction chip. J. Micromech. Microeng. 2006;16:526–530. [Google Scholar]
- 16.Lee JG, Cheong KH, Huh N, Kim S, Choi JW, Ko C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara Y, Kerman K, Kobayashi M, Yamanura S, Morita Y, Tamiya E. Microchamber array based DNA quantification and specific sequence detection from a single copy via PCR in nanoliter volumes. Biosens. Bioelectron. 2005;20:1482–1490. doi: 10.1016/j.bios.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Noh J, Sung SW, Jeon MK, Kim SH, Lee LP, Woo SI. In situ thermal diagnosis of the micro-PCR system using liquid crystals. Sens. Actuators A Phys. 2005;122:196–202. [Google Scholar]
- 19.Oh KW, Park C, Namkoong K. A world-to-chip microfluidic interconnection technology with dual functions of sample injection and sealing for a multichamber micro PCR chip.. IEEE 18 International Conference on Micro Electro Mechanical Systems (MEMS 2005); Miami. 2005. pp. 714–717. [Google Scholar]
- 20.Oh KW, Park C, Namkoong K, Kim J, Ock KS, Kim S, Kim YA, Cho YK, Ko C. World-to-chip microfluidic interface with built-in valves for multichamber chip-based PCR assays. Lab Chip. 2005;5:845–850. doi: 10.1039/b503437j. [DOI] [PubMed] [Google Scholar]
- 21.Panaro NJ, Lou XJ, Fortina P, Kricka LJ, Wilding P. Micropillar array chip for integrated white blood cell isolation and PCR. Biomol. Eng. 2005;21:157–162. doi: 10.1016/j.bioeng.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Park SH, Jung MY, Yoon TH, Pyo HB. Cylindrical pillars in silicon PCR chip enhance the performance of DNA amplification.. The 13th International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers’05); June 5–9; Seoul, Korea. 2005. pp. 1604–1607. [Google Scholar]
- 23.Wang W, Li ZX, Luo R, Lü SH, Xu AD, Yang YJ. Droplet-based micro oscillating-flow PCR chip. J. Micromech. Microeng. 2005;15:1369–1377. [Google Scholar]
- 24.Yan WP, Du LQ, Wang J, Ma LZ, Zhu JB. Simulation and experimental study of PCR chip based on silicon. Sens. Actuators B Chem. 2005;108:695–699. [Google Scholar]
- 25.Zou ZQ, Chen X, Jin QH, Yang MS, Zhao JL. A novel miniaturized PCR multi-reactor array fabricated using flip-chip bonding techniques. J. Micromech. Microeng. 2005;15:1476–1481. [Google Scholar]
- 26.Easley CJ, Karlinsey JM, Landers JP. On-chip pressure injection for integration of infrared-mediated DNA amplification with electrophoretic separation. Lab Chip. 2006;6:601–610. doi: 10.1039/b600039h. [DOI] [PubMed] [Google Scholar]
- 27.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, et al. A fully integrated microfluidic genetic analysis system with sample-in–answer-out capability. Proc. Natl Acad. Sci. USA. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong H, Ramalingam N, Chen L, Che J, Wang Q, Wang Y, Yang X, Yap PHE, Neo CH. Microfluidic handling of PCR solution and DNA amplification on a reaction chamber array biochip. Biomed. Microdevices. 2006;8:167–176. doi: 10.1007/s10544-006-7712-8. [DOI] [PubMed] [Google Scholar]
- 29.Legendre LA, Bienvenue JM, Roper MG, Ferrance JP, Landers JP. A simple, valveless microfluidic sample preparation device for extraction and amplification of DNA from nanoliter-volume samples. Anal. Chem. 2006;78:1444–1451. doi: 10.1021/ac0516988. [DOI] [PubMed] [Google Scholar]
- 30.Li SF, Fozdar DY, Ali MF, Li H, Shao DB, Vykoukal DM, Vykoukal J, Floriano PN, Olsen M, et al. A continuous-flow polymerase chain reaction microchip with regional velocity control. J. Microelectromech. Syst. 2006;15:223–236. doi: 10.1109/JMEMS.2005.859083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu CN, Toriello NM, Mathies RA. Multichannel PCR-CE microdevice for genetic analysis. Anal. Chem. 2006;78:5474–5479. doi: 10.1021/ac060335k. [DOI] [PubMed] [Google Scholar]
- 32.Pal R, Yang M, Lin R, Johnson BN, Srivastava N, Razzacki SZ, Chomistek KJ, Heldsinger DC, Haque RM, et al. An integrated microfluidic device for influenza and other genetic analyses. Lab Chip. 2005;5:1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- 33.Prakash R, Kaler KVIS. An integrated genetic analysis microfluidic platform with valves and a PCR chip reusability method to avoid contamination. Microfluid. Nanofluid. 2007;3:177–187. [Google Scholar]
- 34.Proff C, Rothschild MA, Schneider PM. Low volume PCR (LV-PCR) for STP typing of forensic casework samples. Int. Congr. Series. 2006;1288:645–647. [Google Scholar]
- 35.Toriello NM, Liu CN, Mathies RA. Multichannel reverse transcription-polymerase chain reaction microdevice for rapid gene expression and biomarker analysis. Anal. Chem. 2006;78:7997–8003. doi: 10.1021/ac061058k. [DOI] [PubMed] [Google Scholar]
- 36.Yang M, Pal R, Burns MA. Cost-effective thermal isolation techniques for use on microfabricated DNA amplification and analysis devices. J. Micromech. Microeng. 2005;15:221–230. [Google Scholar]
- 37.Cady NC, Stelick S, Kunnavakkam MV, Batt CA. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens. Actuators B Chem. 2005;107:332–341. [Google Scholar]
- 38.Hsieh TM, Luo CH, Lee GB, Liao CS, Huang FC. A micromachined low-power-consumption portable PCR system. J. Med. Biol. Eng. 2006;26:43–49. [Google Scholar]
- 39.Hu GQ, Xiang Q, Fu R, Xu B, Venditti R, Li DQ. Electrokinetically controlled real-time polymerase chain reaction in microchannel using Joule heating effect. Anal. Chim. Acta. 2006;557:146–151. [Google Scholar]
- 40.Huang FC, Liao CS, Lee GB. An integrated microfluidic chip for DNA/RNA amplification, electrophoresis separation and on-line optical detection. Electrophoresis. 2006;27:3297–3305. doi: 10.1002/elps.200600458. [DOI] [PubMed] [Google Scholar]
- 41.Jia XY, Niu ZQ, Chen WY. A rotary polydimethylsiloxane-based device for polymerase chain reaction. Anal. Lett. 2005;38:2143–2149. [Google Scholar]
- 42.Jia XY, Niu ZQ, Chen WY, Zhang WP. Polydimethylsiloxane (PDMS)-based spiral channel PCR chip. Electron. Lett. 2005;41:890–891. [Google Scholar]
- 43.Kaigala GV, Huskins RJ, Preiksaitis J, Pang X.-L, Pilarski LM, Backhouse CJ. Automated screening using microfluidic chip-based PCR and product detection to assess risk of BK virus-associated nephropathy in renal transplant recipients. Electrophoresis. 2006;27:3753–3763. doi: 10.1002/elps.200600061. [DOI] [PubMed] [Google Scholar]
- 44.Kim JA, Lee JY, Seong S, Cha SH, Lee SH, Kim JJ, Park TH. Fabrication and characterization of a PDMS-glass hybrid continuous-flow PCR chip. Biochem. Eng. J. 2006;29:91–97. [Google Scholar]
- 45.Lee CY, Lee GB, Lin JL, Huang FC, Liao CS. Integrated microfluidic systems for cell lysis, mixing/pumping and DNA amplification. J. Micromech. Microeng. 2005;15:1215–1223. [Google Scholar]
- 46.Liao CS, Lee GB, Liu HS, Hsieh TM, Luo CH. Miniature RT-PCR system for diagnosis of RNA-based viruses. Nucleic Acids Res. 2005;33:e156. doi: 10.1093/nar/gni157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao CS, Lee GB, Wu JJ, Chang CC, Hsieh TM, Huang FC, Luo CH. Micromachined polymerase chain reaction system for multiple DNA amplification of upper respiratory tract infectious diseases. Biosens. Bioelectron. 2005;20:1341–1348. doi: 10.1016/j.bios.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Lien KY, Lee WC, Lei HY, Lee GB. Integrated reverse transcription polymerase chain reaction systems for virus detection. Biosens. Bioelectron. 2007;22:1739–1748. doi: 10.1016/j.bios.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Marcus JS, Anderson WF, Quake SR. Parallel picoliter RT-PCR assays using microfluidics. Anal. Chem. 2006;78:956–958. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama T, Kurosawa Y, Furui S, Kerman K, Kobayashi M, Rao SR, Yonezawa Y, Nakano K, Hino A, et al. Circumventing air bubbles in microfluidic systems and quantitative continuous-flow PCR applications. Anal. Bioanal. Chem. 2006;386:1327–1333. doi: 10.1007/s00216-006-0688-7. [DOI] [PubMed] [Google Scholar]
- 51.Niu ZQ, Chen WY, Shao SY, Jia XY, Zhang WP. DNA amplification on a PDMS-glass hybrid microchip. J. Micromech. Microeng. 2006;16:425–433. [Google Scholar]
- 52.Pilarski PM, Adamia S, Backhouse CJ. An adaptable microvalving system for on-chip polymerase chain reactions. J. Immunol. Methods. 2005;305:48–58. doi: 10.1016/j.jim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Prakash AR, Adamia S, Sieben V, Pilarski P, Pilarski LM, Backhouse CJ. Small volume PCR in PDMS biochips with integrated fluid control and vapour barrier. Sens. Actuators B Chem. 2006;113:398–409. [Google Scholar]
- 54.Xiang Q, Xu B, Fu R, Li D. Real time PCR on disposable PDMS chip with a miniaturized thermal cycler. Biomed. Microdevices. 2005;7:273–279. doi: 10.1007/s10544-005-6069-8. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y, Zhang X. A novel pressure indicator for continuous flow PCR chip using micro molded PDMS pillar arrays. Mater. Res. Soc. Symp. Proc. 2005;845:AA5.10.1–AA5.10.6. [Google Scholar]
- 56.Chen JF, Wabuyele M, Chen HW, Patterson D, Hupert M, Shadpour H, Nikitopoulos D, Soper SA. Electrokinetically synchronized polymerase chain reaction microchip fabricated in polycarbonate. Anal. Chem. 2005;77:658–666. doi: 10.1021/ac048758e. [DOI] [PubMed] [Google Scholar]
- 57.Chen ZY, Wang J, Qian SZ, Bau HH. Thermally-actuated, phase change flow control for microfluidic systems. Lab Chip. 2005;5:1277–1285. doi: 10.1039/b508275g. [DOI] [PubMed] [Google Scholar]
- 58.Curtin DM, Newport DT, Davies MR. Utilising μ-PIV and pressure measurements to determine the viscosity of a DNA solution in a microchannel. Exp. Therm. Fluid Sci. 2006;30:843–852. [Google Scholar]
- 59.Hashimoto M, Hupert ML, Murphy MC, Soper SA. Ligase detection reaction/hybridization assays using three-dimensional microfluidic networks for the detection of low-abundant DNA point mutations. Anal. Chem. 2005;77:3243–3245. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- 60.Hashimoto M, Barany F, Soper SA. Polymerase chain reaction/ligase detection reaction/hybridization assays using flow-through microfluidic devices for the detection of low-abundant DNA point mutations. Biosens. Bioelectron. 2006;21:1915–1923. doi: 10.1016/j.bios.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Chen JF, Zhu L, Shadpour H, Hupert ML, Soper SA. Continuous flow thermal cycler microchip for DNA cycle sequencing. Anal. Chem. 2006;78:6223–6231. doi: 10.1021/ac060568b. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Chen ZY, Mauk MG, Hong K.-S, Li MY, Yang S, Bau HH. Self-actuated, thermo-responsive hydrogel valves for lab on a chip. Biomed. Microdevices. 2005;7:313–322. doi: 10.1007/s10544-005-6073-z. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Chen ZY, Corstjens PLAM, Mauk MG, Bau HH. A disposable microfluidic cassette for DNA amplification and detection. Lab Chip. 2006;6:46–53. doi: 10.1039/b511494b. [DOI] [PubMed] [Google Scholar]
- 64.Cheng JY, Hsieh CJ, Chuang YC, Hsieh JR. Performing microchannel temperature cycling reactions using reciprocating reagent shuttling along a radial temperature gradient. Analyst. 2005;130:931–940. doi: 10.1039/b501061f. [DOI] [PubMed] [Google Scholar]
- 65.Münchow G, Dadic D, Doffing F, Hardt S, Drese KS. Automated chip-based device for simple and fast nucleic acid amplification. Expert Rev. Mol. Diagn. 2005;5:613–620. doi: 10.1586/14737159.5.4.613. [DOI] [PubMed] [Google Scholar]
- 66.Tsai NC, Sue CY. SU-8 based continuous-flow RT-PCR bio-chips under high-precision temperature control. Biosens. Bioelectron. 2006;22:313–317. doi: 10.1016/j.bios.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 67.Yao LY, Liu BA, Chen T, Liu SB, Zuo TC. Micro flow-through PCR in a PMMA chip fabricated by KrF excimer laser. Biomed. Microdevices. 2005;7:253–257. doi: 10.1007/s10544-005-3999-0. [DOI] [PubMed] [Google Scholar]
- 68.Yu HM, Chien CH. Miniaturized PCR-chip for DNA amplification with an external peristaltic pump. J. Mech. Med. Biol. 2005;5:81–87. [Google Scholar]
- 69.Poulsen CR, El-Ali J, Perch-Nielsen IR, Bang DD, Telleman P, Wolff A. Detection of a putative virulence cadF gene of campylobacter jejuni obtained from different sources using a microfabricated PCR chip. J. Rapid Meth. Aut. Mic. 2005;13:111–126. [Google Scholar]
- 70.Shen KY, Chen XF, Guo M, Cheng J. A microchip-based PCR device using flexible printed circuit technology. Sens. Actuators B Chem. 2005;105:251–258. [Google Scholar]
- 71.Chabert M, Dorfman KD, de Cremoux P, Roeraade J, Viovy J.-L. Automated microdroplet platform for sample manipulation and polymerase chain reaction. Anal. Chem. 2006;78:7722–7728. doi: 10.1021/ac061205e. [DOI] [PubMed] [Google Scholar]
- 72.Dorfman KD, Chabert M, Codarbox JH, Rousseau G, de Cremoux P, Viovy JL. Contamination-free continuous flow microfluidic polymerase chain reaction for quantitative and clinical applications. Anal. Chem. 2005;77:3700–3704. doi: 10.1021/ac050031i. [DOI] [PubMed] [Google Scholar]
- 73.Zhang LH, Dang FQ, Kaji N, Baba Y. Fast extraction, amplification and analysis of genes from human blood. J. Chromatogr. A. 2006;1106:175–180. doi: 10.1016/j.chroma.2005.10.071. [DOI] [PubMed] [Google Scholar]
- 74.Zhang CS, Xu JL, Wang JQ, Wang HP. Continuous-flow polymerase chain reaction microfluidics by using spiral capillary channel embedded on copper. Anal. Lett. 2007;40:497–511. [Google Scholar]
- 75.Zhang CS, Xing D, Xu JL. A continuous-flow PCR microfluidics for rapid DNA amplification based on thin film heater with low thermal mass. Anal. Lett. 2006 doi: 10.1080/00032710701298446. [Google Scholar]
- 76.Zhang CS, Xu JL, Wang JQ, Wang HP. Experimental study of continuous-flow polymerase chain reaction microfluidics based on polytetrafluoroethylene capillary. Chinese J. Anal. Chem. 2006;34:1197–1202. [Google Scholar]
- 77.Guttenberg Z, Müller H, Habermüller H, Geisbauer A, Pipper J, Felbel J, Kielpinski M, Scriba J, Wixforth A. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab on a Chip. 2005;5:308–317. doi: 10.1039/b412712a. [DOI] [PubMed] [Google Scholar]
- 78.Morrison T, Hurley J, Garcia J, Yoder K, Katz A, Roberts D, Cho J, Kanigan T, Ilyin SE, et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006;34:e123. doi: 10.1093/nar/gkl639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neuzil P, Pipper J, Hsieh TM. Disposable real-time microPCR device: lab-on-a-chip at a low cost. Mol. BioSyst. 2006;2:292–298. doi: 10.1039/b605957k. [DOI] [PubMed] [Google Scholar]
- 80.Neuzil P, Zhang CY, Pipper J, Oh S, Zhuo L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006;34:e77. doi: 10.1093/nar/gkl416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shih CY, Chen Y, Tai YC. Parylene-strengthened thermal isolation technology for microfluidic system-on-chip applications. Sens. Actuators A Phys. 2006;126:270–276. [Google Scholar]
- 82.Kartalov EP, Quake SR. Microfluidic device reads up to four consecutive base pairs in DNA sequencing-by-synthesis. Nucleic Acids Res. 2004;32:2873–2879. doi: 10.1093/nar/gkh613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu XM, Zhang DC, Li T, Hao L, Li XH. 3-D microarrays biochip for DNA amplification in polydimethylsiloxane (PDMS) elastomer. Sens. Actuators A Phys. 2003;108:103–107. [Google Scholar]
- 84.Northrup MA, Ching MT, White RM, Watson RT. DNA amplification in a microfabricated reaction chamber.. Proceedings of the 7th International Conference on Solid-State Sensors and Actuators (Transducers ‘93); 7–10 June; Yokohama, Japan. 1993. pp. 924–926. [Google Scholar]
- 85.Nakano H, Matsuda K, Yohda M, Nagamune T, Endo I, Yamane T. High-speed polymerase chain reaction in constant flow. Biosci. Biotechnol. Biochem. 1994;58:349–352. doi: 10.1271/bbb.58.349. [DOI] [PubMed] [Google Scholar]
- 86.Kopp MU, de Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 87.Gui L, Ren CL. Numerical simulation of heat transfer and electrokinetic flow in an electroosmosis-based continuous-flow PCR chip. Anal. Chem. 2006;78:6215–6222. doi: 10.1021/ac060553d. [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Li ZX, Guo ZY. Numerical simulation of micro flow-through PCR chip. Microscale Therm. Eng. 2005;9:281–293. [Google Scholar]
- 89.Chang YH, Lee GB, Huang FC, Chen YY, Lin JL. Integrated polymerase chain reaction chips utilizing digital microfluidics. Biomed. Microdevices. 2006;8:215–225. doi: 10.1007/s10544-006-8171-y. [DOI] [PubMed] [Google Scholar]
- 90.Griffith EJ, Akella S. Coordinating multiple droplets in planar array digital microfluidic systems. Int. J. Robot. Res. 2005;24:933–949. [Google Scholar]
- 91.Griffith EJ, Akella S, Goldberg MK. Performance characterization of a reconfigurable planar-array digital microfluidic system. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2006;25:340–352. [Google Scholar]
- 92.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 93.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl Acad. Sci. USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl Acad. Sci. USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakakibara J, Adrian RJ. Whole field measurement of temperature in water using two-color laser induced fluorescence. Exp. Fluids. 1999;26:7–15. [Google Scholar]
- 96.Childs PRN, Greenwood JR, Long CA. Review of temperature measurement. Rev. Sci. Instrum. 2000;71:2959–2978. [Google Scholar]
- 97.Hejerten S. Free zone electrophoresis. Chromatogr. Rev. 1967;9:122–219. doi: 10.1016/0009-5907(67)80003-6. [DOI] [PubMed] [Google Scholar]
- 98.Jorgenson JW, Lukacs KD. Zone electrophoresis in open-tubular glass capillaries. Anal. Chem. 1981;53:1298–1302. [PubMed] [Google Scholar]
- 99.Jorgenson JW, Lukacs KD. Capillary zone electrophoresis. Science. 1983;222:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- 100.Terabe S, Otsuka K, Ichikawa K, Ando T. Electrokinetic separations with micellar solutions and open-tubular capillaries. Anal. Chem. 1984;56:111–113. [Google Scholar]
- 101.Manz A, Graber N, Widmer HM. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990;1:244–248. [Google Scholar]
- 102.Woolley AT, Hadley D, Landre P, deMello AJ, Mathies RA, Northrup MA. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 1996;68:4081–4086. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 103.Lee TMH, Hsing IM. DNA-based bioanalytical microsystems for handheld device applications. Anal. Chim. Acta. 2006;556:26–37. doi: 10.1016/j.aca.2005.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]