Abstract
DNA polymerases are identified that copy a non-standard nucleotide pair joined by a hydrogen bonding pattern different from the patterns joining the dA:T and dG:dC pairs. 6-Amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone (dZ) implements the non-standard ‘small’ donor–donor–acceptor (pyDDA) hydrogen bonding pattern. 2-Amino-8-(1′-β-D-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (dP) implements the ‘large’ acceptor–acceptor–donor (puAAD) pattern. These nucleobases were designed to present electron density to the minor groove, density hypothesized to help determine specificity for polymerases. Consistent with this hypothesis, both dZTP and dPTP are accepted by many polymerases from both Families A and B. Further, the dZ:dP pair participates in PCR reactions catalyzed by Taq, Vent (exo−) and Deep Vent (exo−) polymerases, with 94.4%, 97.5% and 97.5%, respectively, retention per round. The dZ:dP pair appears to be lost principally via transition to a dC:dG pair. This is consistent with a mechanistic hypothesis that deprotonated dZ (presenting a pyDAA pattern) complements dG (presenting a puADD pattern), while protonated dC (presenting a pyDDA pattern) complements dP (presenting a puAAD pattern). This hypothesis, grounded in the Watson–Crick model for nucleobase pairing, was confirmed by studies of the pH-dependence of mismatching. The dZ:dP pair and these polymerases, should be useful in dynamic architectures for sequencing, molecular-, systems- and synthetic-biology.
INTRODUCTION
According to rules for double helix formation proposed by Watson and Crick in 1953, antiparallel DNA strands are held together by nucleobase pairs that obey two rules of complementarity: size complementarity (large purines pair with small pyrimidines) and hydrogen bonding complementarity (hydrogen bond donors from one nucleobase pair with hydrogen bond acceptors from the other) (1,2). The former permits the aperiodic crystal structure that underlies faithful replication. The latter helps achieve the specificity that gives rise to the simple rules for base pairing (‘A pairs with T, and G pairs with C’) that underlie genetics and molecular biology.
Some time ago, it was noticed that the DNA alphabet need not be limited to this architecture. For example, many groups, including those of Rappoport (3), Kool (4), Hirao et al. (5,6), Minakawa et al. (7), Romesberg (8) and Schultz (9), have shown that the Watson–Crick structural design can be drastically altered, removing hydrogen bonding, introducing steric determinants of specificity, or changing substantially the size of the nucleobases.
To date, however, the most useful ‘expanded genetic alphabets’ have come from more subtle modifications of the Watson–Crick architecture. One class of these involves simply rearranging hydrogen bond donor and acceptor groups within a pair while retaining the overall Watson–Crick geometry (Figure 1) (10,11). By doing this, 12 nucleobases forming six base pairs joined by mutually exclusive hydrogen bonding patterns are readily available within that geometry. Figure 1 shows the standard and non-standard hydrogen bonding patterns obtained by this rearrangement, together with a nomenclature to designate them (12).
Figure 1.
One example of an ‘artificially expanded genetic information system’ (AEGIS). Nucleobase pairs in this system have a Watson–Crick geometry, with large purines or purine analogs (indicated by ‘pu’) pairing with small pyrimidines or pyrimidine analogs (indicated by ‘py’) joined by hydrogen bonds. The hydrogen-bonding acceptor (A) and donor (D) groups are listed from the major to the minor groove as indicated. The heterocycles shown are the currently preferred implementations of the indicated hydrogen bonding patterns; others are conceivable. The electron densities presented to the minor groove that are believed to be potential recognition sites for polymerases is shown by the shaded lobes. Note that some non-standard pyrimidines do not present this density. The nucleotides implementing the pyDDA:puAAD hydrogen bonding pattern, the topic of this article, are at the bottom right.
These non-standard nucleotides and the pairs that they form have had particular value as ‘orthogonal binders’, recognition elements that bind with DNA-like specificity, but without interference by natural DNA. This orthogonality substantially lowers noise in a range of nucleic acid-targeted assays. For example, non-standard nucleotides that implement the pyAAD:puDDA hydrogen bonding pattern (Figure 1) are used in the ‘branched DNA’ diagnostic assay developed at Chiron and Bayer. Having now FDA approval, this diagnostic helps manage the care of some 400 000 patients annually infected with the HIV, hepatitis B and hepatitis C viruses (13–15).
The binding properties of these artificially expanded genetic information systems (AEGIS) could have still greater value, however, if their components could be incorporated as dynamic parts of architectures to detect, quantitate and sequence nucleic acids. For example, expanded genetic alphabets could support architectures for highly multiplexed amplification of DNA and RNA, the movement of PCR-amplified nucleic acids to specific spots on microarrays (‘binning’) and low-cost, high capacity re-sequencing of the genomes of individual patients. For these architectures to be practical, however, the components of an expanded genetic alphabet must interact with standard DNA polymerases with sufficient efficiency that they can be copied, and their copies copied.
To expand the potential application of expanded genetic alphabets in dynamic assays in molecular-, systems- and synthetic-biology, we returned to the structure of DNA polymerases. Studies in many laboratories suggested that polymerases might ‘scan’ the minor groove of a growing DNA duplex searching for the electron density that is presented by nitrogen-3 of adenine and guanine, and by the 2-position exocyclic carbonyl group of cytosine and thymine (16–20). This is a structural feature shared by the four standard nucleobases, making it potentially a convenient ‘handle’ for a polymerase, even though it does not seem to be a mandatory specificity determinant.
As can be seen by inspecting Figure 1, many of the non-standard nucleotides that are pyrimidine analogs do not have a 2-position exocyclic carbonyl group. Therefore, they do not present electron density to the minor groove at this position, as do thymine and cytosine. However, one non-standard pair ‘does’ present electron density at this position by ‘both’ components; this is the pair implementing the pyDDA:puAAD hydrogen bonding pattern (Figure 1).
For this reason, we focused on the pyDDA:puAAD hydrogen bonding pattern in our most recent work developing dynamic DNA sequencing, detection and quantitation architectures. A heterocycle to implement the pyDDA hydrogen-bonding pattern proved to be difficult to find, however. For example, implementation of the pyDDA hydrogen-bonding pattern was first attempted on a simple pyridine heterocycle; this failed to give a species that was stable to oxidation (21). The same pattern implemented on a pyrazine gave a nucleoside analog that was prone to specific acid-catalyzed epimerization (22). The same pattern implemented on a pyrimidine heterocycle gave rise to tautomeric ambiguity (Figure 2).
Figure 2.
Four alternative implementations of the pyDDA hydrogen bonding pattern. The implementation on a pyrimidine heterocycle suffers from tautomeric ambiguity (left). The implementation on a pyrazine suffers from facile epimerization (right). The implementation on a simple pyridine is too basic and prone to oxidation (top center). The preferred implementation is the nitropyridine heterocycle (discussed here, bottom center), which is stable to oxidation, is not basic, and does not epimerize near neutral pH.
Implementing the pyDDA hydrogen-bonding pattern on a nitropyridine heterocycle solved these problems, however. We recently reported that 6-amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone (trivially designated dZ) could implement the pyDDA hydrogen bonding pattern (23). The nitro group rendered the otherwise electron-rich heterocycle stable against both oxidation and epimerization under standard conditions. When paired with the corresponding puAAD nucleotide, duplexes were formed with stabilities that, in many cases, were higher than those observed in comparable strands incorporating the dG:dC nucleobase pair (24).
We therefore developed chemistry to efficiently prepare dZ, together with its nucleoside complement, 2-amino-8-(1′-β- d-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (implementing puAAD, trivially designated as dP) (24). These syntheses made dZ and dP efficiently available as both their triphosphates and their protected phosphoramidites suitable for solid phase DNA synthesis. It also yielded their alpha-thiotriphosphates.
We report here studies of the interaction between DNA polymerases and the dZ:dP pair. Following a survey of polymerases, we found that both dZTP and dPTP are accepted by DNA polymerases representative of both Families A and B. We also showed that the dZ:dP pair can participate in PCR amplification using Taq DNA polymerase with >94.4% retention per round; using Vent (exo-) and Deep Vent (exo-) polymerases, 97.5% retention per round is measured. A study of the pH-dependence of mismatches suggests that the principal route for the loss of the dZ:dP pair is via a transition to a dC:dG pair through a mismatch between dP and protonated dC (at low pH), or a mismatch between dG and deprotonated dZ (at high pH). Here, the canonical Watson–Crick model for the nucleobase pair, which includes both size and hydrogen bonding complementarity, is adequate to explain these behaviors. Further, this level of fidelity is sufficient to allow the dZ:dP pair to participate as a dynamic component of many architectures for multiplexed detection and sequencing of DNA.
MATERIALS AND METHODS
Oligonucleotides (Table 1), except those containing dZ and dP (Z-Temp and P-Temp), were synthesized by Integrated DNA Technologies (Coralville, IA). All oligodeoxynucleotides were purified by PAGE (10–20%). Z-Temp (containing dZ at position 26) and P-Temp (containing dP at position 26) were synthesized in-house on an Expedite-8900 DNA synthesizer employing standard β–cyanoethylphosphoramidite chemistry using the dZ and dP protected phosphoramidites reported recently (24). Other reagents were purchased from Glen Research (1 μmol scale, CPG 1000 column).
Table 1.
Oligonucleotides used in primer extensions and PCRs
| Z-RS-S16: 5′-GCGTAATACGACTCAC*TATAG |
| Z-RS: 5′-GCGTAATACGACTCACTATAG |
| Z-SS: 5′-GCGTAATACGACTCACTATAGACGA |
| Z-Temp: 3′-CGCATTATGCTGAGTGATATCTGCTZGCATGAAATCACTCCCAATTAAGCG-5′ |
| P-RS-S16: 5′-GCGAATTAACCCTCAC*TAAAG |
| P-RS: 5′-GCGAATTAACCCTCACTAAAG |
| P-SS: 5′-GCGAATTAACCCTCACTAAAGTACG |
| P-Temp: 3′-CGCTTAATTGGGAGTGATTTCATGCPAGCAGATATCACTCAGCATAATGCG-5′ |
| G-Temp: 3′-CGCTTAATTGGGAGTGATTTCATGCGAGCAGATATCACTCAGCATAATGCG-5′ |
The * indicates the position of the phosphorothioate linker.
The triphosphates and α-thiotriphosphates of dZ and dP were prepared as described by Eckstein et al. (25). The SP- and RP-diastereoisomers of dPTPαS were separated by preparative rp-HPLC (Nova-Pak® HR C18 Column (7.8 × 300 mm). Natural deoxynucleoside triphosphates were purchased from Promega (Madison, WI).
Klenow Fragment (exo−), Bst, Taq, VentR®, Deep VentR®, 9°N, Phusion (high-fidelity DNA polymerase) and DyNAzyme™ EXT DNA polymerases were purchased from New England Biolabs (Beverly, MA). Tfl, Tth and Tli DNA polymerases were purchased from Promega (Madison, WI). Pfu (exo−), native Pfu and cloned Pfu DNA polymerases were purchased from Stratagene (La Jolla, CA). Exonuclease III was purchased from Promega (Madison, WI). They were generally used in the buffers provided by the supplier of the polymerase.
Recognizing that the pH of Tris buffers, which are routinely recommended by manufacturers (for example, Thermopol buffer used for Taq, VentR®, Deep VentR® and 9°N DNA polymerases is 20 mM Tris–HCl, pH 8.5 measured at 25°C, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100) is known to strongly vary with temperature (see ‘Results’), the pH of buffers was measured at the elevated temperatures (using a temperature calibrated Accumet® AB15 pH Meter, Fisher Scientific) used for the extension reactions and PCRs. As expected, these pHs were ca. 1.4 units below those measured in the same buffer at room temperature.
Primer extension experiments
In standing start primer extension experiments, 5′-32P-labeled primer, Z-SS (25-mer), or P-SS (25-mer) (4 pmol, final concentration 400 nM) was annealed to the complementary template, Z-Temp (dZ in position 26), or P-Temp (dP in position 26) (5 pmol, final concentration 500 nM) in polymerase reaction buffer by heating the mixture at 95°C for 5 min and allowing the solution to cool over 1 h to room temperature. Non-standard nucleoside triphosphate, dPTP or dZTP (2 nmol, final concentration 200 μM) was then added, followed by the polymerase (1 U), to give a final reaction volume of 10 μl. The mixture was immediately incubated at 72°C for 2 min and 10 min (except for Klenow Fragment at 37°C and Bst at 65°C). The reaction was quenched with 10 mM EDTA in formamide loading buffer (10 μl). Samples were resolved using a 20% PAGE (7 M urea). Gels were quantitated using MolecularImager software.
Running start primer extension experiments were similar, except that shorter primers were used. Here, 5′-32P-labeled primer, Z-RS (21-mer), or P-RS (21-mer) (2 pmol, final assay concentration 100 nM) was annealed to the corresponding template, Z-Temp (dZ in position 26), or P-Temp (dP in position 26) (3 pmol, final concentration 150 nM) in polymerase reaction buffer by heating the mixture at 95°C for 5 min, followed by cooling over 1 h to room temperature. Four natural dNTPs (2 nmol, final concentration 100 μM), and dPTP or dZTP (2 nmol, final concentration 100 μM) were added to the solution at room temperature. The mixture was pre-incubated at 72°C for 30 s; polymerase (0.2 to 0.25 U) was then added (final reaction volume of 20 μl). Aliquots (10 μl), withdrawn at 30 s and 60 s, were quenched with 10 mM EDTA in formamide loading buffer (12 μl). The products were resolved by 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software. The negative control reactions were performed using water instead of dZTP and dPTP.
Extension of primers with alpha-thiotriphosphates
5′-32P-labeled primer, Z-RS-S16, or P-RS-S16 (both containing phosphorothioate linker joining nucleotides 16 and 17) (2 pmol, final concentration 200 nM) was annealed to the corresponding template, Z-Temp (dZ in position 26), or P-Temp (dP in position 26) (3 pmol, final concentration 300 nM) in polymerase reaction buffer by heating the mixture at 95°C for 3 min and then allowing the solution to cool over 1 h hour to room temperature. Four natural dNTPs (each 100 μM final conc.), plus dPTPαS (resolved Sp diastereoisomer, 200 μM final concentration), or dZTPαS (a mixture of diastereoisomers, final 200 μM), were added at room temperature, followed by polymerase (2 or 2.5 U), to give a final reaction volume of 10 μl. The reaction was immediately incubated at 72°C for 3 min and then cooled to 4°C on a Peltier thermal cycler (DNAEngine®, Bio-Rad, CA). The reaction was diluted with PN buffer (100 μl) and purified by QIAquick Nucleotide Remove Kit (Qiagen, Valencia, CA). The DNA was eluted from the spin column using 60 μl of EB buffer (10 mM Tris pH 8.5). Samples (10 μl) were mixed with formamide loading buffer (10 μl) and resolved using a 20% PAGE (7 M urea). The gel was analyzed using the MolecularImager software (Figure 3, left).
Figure 3.
Incorporation of dPTPαS (presented as its pure Sp diastereoisomer) or dZTPαS (presented as a 1:1 diastereomeric mixture) opposite template dZ or dP (respectively) to generate oligonucleotide products that have phosphorothioate linkages resistant to exonuclease digestion. Left: Running start extensions (by Taq and 9°N DNA polymerases, as indicated) of 5′-32P labeled 21-mer primers having a phosphorothioate linkage between nucleotides 16 and 17 and Z-Temp and P-Temp. Right: Exo III (100 U, final concentration 2.5 U/µl) digestion of full-length products (FLP) containing phosphorothioate linkages (digestion times indicated). A 26-mer band indicates pausing of Exo III digestion at phosphorothioate linkage arising via incorporation of dPTPαS or dZTPαS opposite dZ or dP (both at positions 26 in their respective templates). This shows that both dPTPαS and dZTPαS were incorporated opposite their partners. A 17-mer band appears only if Exo III digests through the 25–26 linkage, either because misincorporation at an earlier step prevents the respective thiotriphosphates from incorporating a phosphorothioate linkage to join nucleotides 25 and 26, or because Exo III has digested through a 25-26 phosphorothioate linkage having an Exo III-sensitive SP stereochemistry. Digestion will pause at the phophorothioate joining nucleotides 16 and 17 arising from the chemically synthesized primer (and therefore having both RP and SP stereochemistries). Extra pausing bands (between 51- and 26-mers) suggest that 9°N incorporates small amounts of dZTPαS opposite template dG. Low-weight molecular products arise by digestion through the SP phosphorothioates joining nucleotides 16 and 17. Lanes P-Taq and P-9°N indicate that extension was done with Taq or 9°N (respectively), and dPTPαS (in both). Lanes Z-Taq and Z-9°N indicate that extension was done with Taq or 9°N (respectively), and dZTPαS (in both).
Digestion by Exo III of oligonucleotides containing alpha-phosphorothioate linkages
To determine the position of the phosphorothioate linkages arising in an oligonucleotide product via incorporation of alpha-thiotriphosphates, samples (35 μl) were mixed with 10 × Exo III buffer (4 µl, final 66 mM Tris–HCl, pH 8.0 at 25°C, 0.66 mM MgCl2). Exonuclease III (100 U, final 2.5 U/µl) was then added at room temperature. Aliquots (6 µl) were withdrawn at intervals (2, 5, 15, 30 and 60 min), quenched with EDTA (2 µl, 0.5 M) and mixed with PAGE loading buffer (6 µl, formamide). Samples were resolved by electrophoresis using 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software (Figure 3, right).
PCR and Exo III digestion
Following the strategy in Figure 4, four parallel PCR mixtures containing four standard dNTPs (final 100 μM each), non-standard nucleoside triphosphates (dZTP and dPTP, each 200 μM), and DNA polymerase (2 U) were cycled (25 rounds, 45 s at 94°C, 45 s at 55°C and 2 min at 72°C) with identical amounts of primers (P-RS and Z-RS, each 10 pmol, 200 nM) and various concentrations of the template (P-Temp), obtained by 10-fold serial dilutions (1 pmol, 0.1 pmol, 0.01 pmol, 0.001 pmol). As each 10-fold dilution in template was equivalent to ∼3.32 rounds of amplification, the amount of retention of the dZ:dP pair could be determined as a function of the number of theoretical rounds of PCR. After PCR amplification, samples (7 μl) were taken from each reaction, mixed with agarose loading dye solution (2 μl), and separated on an agarose gel (3.2%). The gel was analyzed using the GeneSnap software (SynGene).
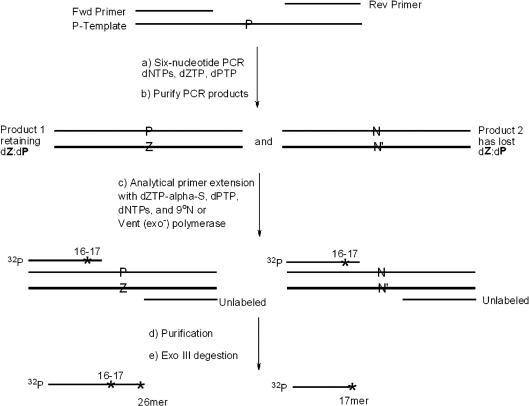
Figure 4.
Schematic showing the use of dZTPαS and Exo III digestion to estimate the amount of dP remaining in PCR products. (a) PCR amplification of template (P-Temp) for different numbers of PCR cycles using four standard dNTPs, dZTP and dPTP. (b) Purification of two PCR amplicon duplexes, one retaining the dZ:dP pair, the other having lost the dZ:dP pair. (c) Seven cycles of ‘analytical primer extension’ by 9°N or Vent (exo−) with dZTPαS, dPTP, and dNTPs converts 5′-32P labeled primer, P-RS-S16, (with a phosphorothioate linkage joining nucleotides 16 and 17 to standardize the Exo III digestion products) to FLP containing a phosphorothioate joining nucleotides 25 and 26 (if amplicon retained dP) (d) Purification of phosphorothioate-containing labeled oligonucleotide products. (e) Exo III digestion of the products; ratio of bands at positions 26 and 17 reflects the ratio of PCR products that retained/lost dP. N and N′ indicate standard nucleobases arising via misincorporation of dNTPs opposite dZ or dP in the amplicon. An * indicates the position of the phosphorothioate linker.
The remaining reaction mixture (43 μl) was purified by QIAquick Nucleotide Remove Kit (Qiagen, Valencia, CA) after diluting with PN buffer (400 μl). The products from the PCR were eluted from the spin column using EB buffer (40 μl, 10 mM Tris pH 8.5).
The products from four parallel PCR amplifications (10 μl, about 1.7 to 2.1 mol) were then used as the templates for seven rounds of ‘analytical primer extension’ with non-standard alpha-thiotriphosphates. The PCR products from above were mixed with 5′-32P labeled forward primer, P-RS-S16, and reverse primer, Z-RS (each, 1 pmol, final concentration 50 nM). Four natural dNTPs (each 100 μM), dPTP (200 μM), dZTPαS (200 μM), 10 × Thermopol buffer (2 μl), 9°N or Vent (exo−) DNA polymerase (2 U) were added at room temperature, and the mixtures were cycled according to the following profile: 2 min at 94°C, 7 cycles of 45 s at 94°C, 45 s at 55°C and 2 min at 72°C.
Control experiments were performed in parallel. Double-stranded DNA synthesized on a solid phase (Z-Temp and P-Temp, each 2 pmol) having dZ or dP at defined positions (position 26) served as templates for both control reactions. For the positive control reaction, dZTPαS and dPTP (each 200 μM final concentration) were used; for the negative control reaction, dZTP and dPTP (each, 200 μM final concentration) were used. The remaining components (primers, dNTPs and polymerase) were the same as above.
In all cases, reactions were quenched with EDTA (2 μl, 100 mM) and purified by QIAquick Nucleotide Remove Kit (Qiagen, Valencia, CA). The DNA was eluted from the spin columns using EB buffer (60 μl, 10 mM Tris pH 8.5). The products were resolved using a 20% PAGE (7 M urea). The gel was analyzed using the Molecular Imager software.
For Exo III digestion, sample (35 μl) was mixed with 10 × Exo III buffer (4 µl, 66 mM Tris–HCl, pH 8.0, 0.66 mM MgCl2). Exo III (20 U, final 0.5 U/µl) was added at room temperature. After 10 min, the reaction was quenched by adding aqueous EDTA (3 µl, 0.5 M) and then PAGE loading buffer (40 µl, formamide). Products were resolved by electrophoresis using a 20% PAGE (7 M urea). The gel was analyzed using MolecularImager software.
Six-nucleotide PCR as a function of pH with Taq DNA polymerase
Four parallel PCRs were performed in 1 x Thermopol buffer at four different pHs (7.5, 7.8, 8.0 and 8.5 at 25°C). The PCR mixtures containing identical amounts of primers, (P-RS and Z-RS, each 10 pmol, 200 nM final), template, (P-Temp, 0.0001 pmol), dNTPs (each 100 μM), non-standard nucleotide triphosphates, dZ/PTP (each 200 μM), and Taq DNA polymerase (2.5 U) were cycled (30 rounds, 45 s at 94°C, 45 s at 55°C and 2 min at 72°C). After PCR amplification, samples (7 μl) were taken from each mixture, placed in agarose loading dye (2 μl) and analyzed on a 3.2% agarose gel. The gel was analyzed using the GeneSnap software (SynGene). The left over reaction mixture (43 μl) was diluted with PN buffer (400 μl) and purified by QIAquick Nucleotide Remove Kit. The products of PCR amplification were eluted from the spin column using 40 μl of EB buffer (10 mM Tris pH 8.5).
The products from four parallel PCRs (10 μl, about 1.7–2.1 pmol) served as the templates for incorporation of alpha-thiotriphosphates as before. A mixture was prepared with 5′-32P-labeled forward primer, P-RS-S16, and reverse primer, Z-RS (each, 1 pmol, final concentration 50 nM). Four natural dNTPs (each 100 μM final), dPTP (200 μM), dZTPαS (200 μM final), 10 × Thermopol buffer (2 μl, pH 8.5), and Vent (exo−) DNA polymerase (2 U) were added at room temperature. The mixtures were cycled according to the following profile: 2 min at 94°C, 7 cycles of 45 s at 94°C, 45 s at 55°C and 2 min at 72°C. For the control experiment, double-stranded DNA, Z-Temp and P-Temp (each 2 pmol) served as template, and the reactions were performed in parallel. After the primer extension, reactions were quenched with EDTA (2 μl, 100 mM) and purified by QIAquick Nucleotide Remove Kit. The DNA was eluted from the spin columns using 60 μl of EB buffer (10 mM Tris pH 8.5). The products were resolved using a 20% PAGE (7 M urea). The gel was analyzed using the Molecular Imager software. Exo III digestion was performed as described above.
RESULTS
dZTP and dPTP are substrates for DNA polymerases
Four Family A polymerases (Bst, Taq, Tfl and Tth, all exo−) and 10 Family B polymerases [Vent and Deep Vent (both exo− and exo+)], Pfu (exo−, native and cloned), 9°N, Tli (exo+) and Phusion (exo+) were initially screened for their ability to incorporate dZTP opposite template dP, and dPTP opposite template dZ (data not shown). In general, the incorporation of dZTP opposite template dP appeared to be more facile than the incorporation of dPTP opposite template dZ. These experiments identified ‘polymerases of interest’ from both families, in particular, Taq, Bst, Vent (both exo− and exo+), Deep Vent (both exo− and exo+), and 9°N. Based on our experience with other screens that sought to incorporate nucleoside variants that do ‘not’ present electron density in the minor groove, (10,26) finding this number of polymerases able to incorporate the dZ:dP pair was surprising.
We then sought to determine how well a dZ:dP pair survives in duplex DNA after multiple rounds of PCR. To do this, we wished to apply a strategy that combines the incorporation of alpha-thiotriphosphates and Exo III digestion to estimate the amount of dZ and dP in an oligonucleotide (Yang et al., in press). This strategy is based on the fact that phosphorothioate linkages, incorporated into an oligonucleotide by a polymerase from the corresponding S-α-thiotriphosphate, can resist hydrolysis by Exo III. To the extent that dZ or dP is present in a template, therefore, primer extension on that template with dPTPαS or dZTPαS, respectively, will generate products containing phosphorothioate linkages at the positions where dPTPαS or dZTPαS is incorporated. Exo III digestion of these products, in turn, will give bands in a gel at positions where the dPTPαS or dZTPαS were incorporated. If that nucleotide is dZ, this band implies the presence of dP surviving in the PCR product. If that nucleotide is dP, this band implies the presence of dZ surviving in the PCR product.
Some stereochemical features of the analysis are relevant to the interpretation of these experiments. First, a phosphorothioate linkage having an RP configuration is believed to be highly resistant to Exo III cleavage. Conversely, the SP configuration is believed to be sensitive to Exo III digestion, with the extent of degradation depending on the amount of Exo III used and time of the digestion. Given the reasonable assumption that polymerases invert configuration at the alpha phosphorus of the triphosphate being incorporated, SP-α-thiotriphosphates should deliver the cleavage-resistant RP-phosphorothioate linkage to the product oligonucleotide, while RP-α-thiotriphosphates should deliver the cleavage-prone SP-phosphorothioate linkage. Therefore, the analysis is expected to work best if pure SP-α-thiotriphosphates are used.
Unfortunately for the application of this strategy here, while the diastereomers of dPTPαS could be resolved using HPLC resolution, the diastereomers of dZTPαS could not. Also unfortunately, both Taq and 9°N DNA polymerases were found to accept ‘both’ S and R isomers of dPTPαS (and therefore presumably dZTPαS) to some extent (Yang et al., in press). This means that the use of dZTPαS in a primer extension will give some Exo III sensitive product (a phosphorothioate linkage with SP configuration), which implies an underestimation of the amount of dP remaining in the PCR product.
The stereochemical details also are relevant to the design of an internal standard to control for these factors. To quantitate total product, a phosphorothioate linkage is incorporated into the primer by chemical synthesis. Chemical synthesis delivers phosphorothioate linkages as a ≈50:50 mixture of RP and SP diastereomers. Thus, only ca. 50% of the oligonucleotide will be highly resistant to degradation. This must be considered when interpreting a reference band arising from a chemically introduced phosphorothioate linkage.
To implement the phosphorothioate–exonuclease combination analysis, we first examined the ‘polymerases of interest’ for their ability to incorporate the alpha-thiotriphosphates of both dZ and dP. Both Taq and 9°N were shown to allow primers to be fully extended past templates containing dP and dZ with dZTPαS and dPTPαS, respectively, after incubation for 3 min by (Figure 3, left). The full-length product (FLP) was then treated with Exo III (100 U) and the digestion products were resolved by gel. As shown in Figure 3 (right), the 9°N polymerase generates more phosphorothioate-containing product than Taq, as determined by a greater intensity of the band at 26-mer (relative to the intensity of the 17-mer reference band). Indeed, the faintness of bands at position 26 when Taq was used suggested that Taq mismatched standard nucleotides opposite template dZ and template dP rather than accept dZTPαS or dPTPαS.
Less prominent features in Figure 3 are low molecular weight products that presumably arise following digestion past the 16–17 phosphorothioate linker that has the Exo III-sensitive stereochemistry. This is expected, as this phosphorothioate linker, introduced by chemical synthesis, is expected to be a mixture of the SP and RP diastereomers. These were noticeable in these experiments, where 100 U of Exo III were used; they were greatly reduced when the amount of Exo III was reduced to 20 U (Figures 5 and 8, below).
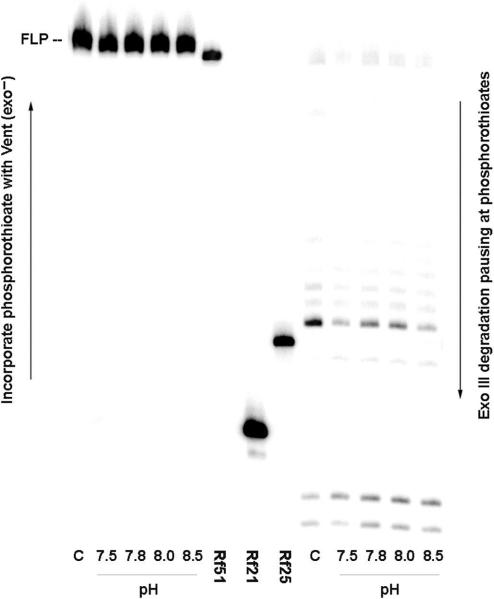
Figure 5.
Retention of the dZ:dP pair in products generated by 16.6 theoretical PCR cycles (using Taq) as a function of pH. Left: Seven cycles of ‘analytical primer extension’ by Vent (exo−) with dZTPαS, dPTP and dNTPs converts 5′-32P labeled primer (P-RS-S16) to FLP with a phosphorothioate linkage joining nucleotides 25 and 26 (arising via incorporation of dZTPαS opposite retained dP in the amplicon) at pH 8.5. Right: Digestion of FLP using Exo III (20 U, final concentration 0.5 U/µl), giving pause bands where an phosphorothioate linker is present, either from dZTPαS incorporation (expected between nucleotides 25 and 26) or from the synthetic primer (between nucleotides 16 and 17). Control (C) used synthetic templates (containing 100% dZ:dP pair at position 26). Lanes Rf51, Rf21 and Rf25 hold reference 51-, 21- and 25-mers. Noticeably stronger are Exo III digestion bands indicating greater retention of dZ:dP pair at pH 7.8 and 8.0 than at 7.5 or 8.5.
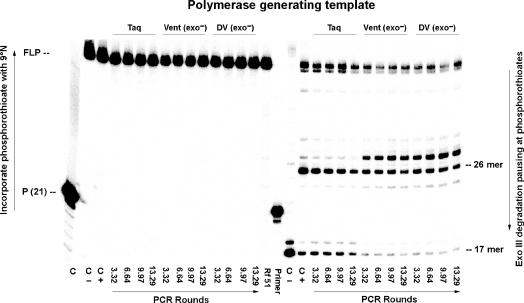
Figure 8.
Retention of the dZ:dP pair in products generated by the indicated number of theoretical rounds of PCR using Taq, Vent (exo−) and Deep Vent (exo−) at pH 8.0. Left: FLP are formed by extension by 9°N polymerase of 5′-32P labeled primer (P-RS-S16) with unlabeled primer (Z-RS) and dZTPαS, dPTP and dNTPs using PCR amplicons generated by Taq, Vent (exo−) and DV (exo−) as templates (shown in Figure 7). Right: PAGE (20%) resolution of products from Exo III (20 U, final concentration 0.5 U/µl) digestion of the FLP from Left. A band indicates presence of a phosphorothioate linkage, either arising through the incorporation of dZTPαS opposite dP at position 26, or from the synthetic primer joining nucleotidies 16 and 17, or through misincorporation of dZ:dP pair during PCR. Controls (lanes marked C) are: (i) C, incubation without 9°N; the absence of extended primer indicates successful removal of Taq polymerase (lane 1, Figure 7) by the QIAquick Nucleotide Remove Kit. (ii) C −, extension by 9°N using synthetic templates containing dZ and dP (at position 26) with standard (oxygen-containing) dZTP and dPTP; absent a phosphorothioate linkage in the extension, Exo III degradation gives a 17-mer due to presence of a phosphorothioate only from the synthetic primer. (iii) C +, extension using synthetic templates as before, but with dZTPαS, generating phosphorothioate linkage in the product, Exo III degradation gives a 26-mer due to the phosphorothioate linkage arising from the incorporation of dZTPαS opposite dP in the synthetic template. P (21) indicates the 21-mer, 5′-32P labeled primer (P-RS-S16).
Last, extra pausing bands (larger than the 26-mers) are evident in the 9°N lanes using dZTPαS. These are positioned in a way that suggests that 9°N incorporates small amounts of dZTPαS opposite dG in the template.
Inspection of Figure 3 also suggested that dZTPαS was incorporated more efficiently than dPTPαS by both polymerases, but in particular by 9°N. This is indicated by the greater intensity of the band at position 26 relative to the intensity of the band at position 17 in the appropriately compared lanes (Figure 3, right). Therefore, we decided that the incorporation of dZTPαS by 9°N is a more reliable analytical tool to detect dP in a template coming from multiple rounds of PCR, than the incorporation of dPTPαS is to detect dZ in the PCR product.
Various unknowns make the reference (the band at position 17) especially important. First, the rate at which Exo III digests DNA need not be independent of local sequence or secondary structure. For example, Linxweiler and He et al. (27,28) reported that Exo III digests through nucleotides in the order C > A,T > G. Second, dZTPαS, as presented to this assay, is a mixture of unresolvable diastereoisomers. If the polymerase accept both isomers (as indicated by a study of Taq and 9°N for dPTPαS), both the RP (Exo III resistant) and SP (Exo III sensitive) phosphorothioate linkages will result. While we suspect that polymerases prefer the S isomer of dZTPαS over the R isomer, we do not know by how much. Hence, the reference band at position 17 is needed to normalize for this unknown as well.
Other Family A (Bst, Tth, Tfl, all exo−) and Family B polymerases (Vent, Deep Vent, and Pfu, all exo−) were also examined using this assay. We found also Vent (exo−) and Deep Vent (exo−) were comparable to 9°N in their ability to incorporate dZTPαS opposite dP in the template (data not shown).
Incorporation of dZ and dP in multiple rounds of PCR as a function of pH
Figure 4 shows the strategy used to apply the phosphorothioate–exonuclease combination to determine the amount of dP remaining in oligonucleotides after a specified number of PCR cycles at different pHs. The starting material was a synthetic template (P-Temp) that contained a single dP at position 26 (the length of the extended primer after dZ is incorporated opposite dP). Forward and reverse primers (P-RS and Z-RS, respectively) were designed as usual (Table 1). PCR cycles were then done with P-Template at concentrations that generated a theoretical number of cycles ranging from 3.3 to 16.6, using dZTP, dPTP and the four natural dNTPs.
At this point, with all primer consumed, the duplex PCR products were separated from the polymerase and excess triphosphates. Then, dZTPαS was added to incorporate phosphorothioate linkages opposite any dP remaining in the PCR products using 9°N or Vent (exo−), dPTP, dNTPs, radiolabeled forward primer containing a synthetic phosphorothioate joining nucleotides 16 and 17 (P-RS-S16, 50% of the amount of PCR product) and unlabelled reverse primer. Seven additional cycles of ‘analytical primer extension’ were used to convert all of the radiolabeled primer to FLP suitable for Exo III digestion. The presence of reverse primer ensured that sufficient dP-containing template was present to lead to full conversion of radiolabeled primer (P-RS-S16). The 2-fold excess of PCR product over P-RS-S16 primer also served this purpose. These products were then isolated and subjected to Exo III digestion.
To determine whether the pH influenced the amount of dP surviving in the PCR products after multiple rounds of PCR, the strategy shown in Figure 4 was applied at pH 7.5, 7.8, 8.0 and 8.5 for 16.6 theoretical rounds of PCR. The buffers were Tris–HCl (20 mM), as provided by the manufacturer for these polymerases. These must be regarded as ‘nominal pH's’, as they were measured at room temperature, and the pH of Tris-HCl buffers is well known to change as a function of temperature (29,30).
Figure 5 shows the result of these experiments where Taq polymerase was used to generate the PCR products, and Vent (exo−) was used to incorporate the phosphorothioate linkage. The five lanes on the left show that the fully extended products (obtained by primer extension of the PCR amplicons obtained at various pHs) co-migrated with the product from the positive control. These products were then subjected to Exo III digestion, and the intensity of the bands indicative of a phosphorothioate linkage joining nucleotides 25 and 26 (and therefore indicative of the incorporation of alpha-thiotriphosphate of dZTP, and therefore indicative of the survival of dP in the PCR products) relative to the bands arising from the phosphorothioate joining nucleotides 16 and 17 (introduced into the primer by chemical synthesis) showed a clear pH dependence, with an optimum at nominal pH between 7.8 and 8.0 (actual pH during elongation is ∼1.4 pH units lower). This is graphed in Figure 6.
Figure 6.
Percent retention of dZ:dP pair after 16.6 theoretical PCR cycles with Taq as a function of pH. Data from Figure 5 were standardized by the positive control (which, by synthesis, contained 100% dZ:dP pair at position 26 before the ‘analytical primer extension’).
All other polymerases of interest were then examined using a similar strategy. Figure 7 shows that Taq and Deep Vent (exo−) convert all of the PCR primers into the expected PCR product at pH 8.0. 9°N and Vent (exo−) also effected this conversion (data not shown). The amounts of PCR products were, for the most part, the same when dZ and dP were used as in the positive control, which incorporated only standard nucleotides in the templates and standard triphosphates; Taq appeared to perform better than Deep Vent in this regard.
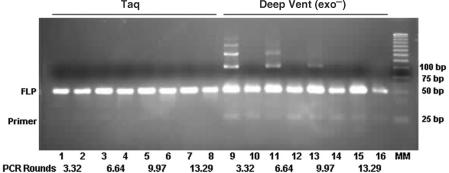
Figure 7.
Ethidium bromide-stained agarose (3.2%) gel showing amplicon formation by six-nucleotide PCR (including four standard dNTPS, dZTP and dPTP) using Taq and Deep Vent (exo−) polymerases (Figure 4a) at pH 8.0. Lanes 2, 4, 6 and 8 hold products from four different concentrations of template (P-Temp) amplified by Taq with identical amounts of primers, giving the indicated number of theoretical rounds of PCR. Lanes 1, 3, 5 and 7 hold products from amplification of G-Temp (positive control), identical except that the amplicon had dC:dG instead of dZ:dP. Lanes 10, 12, 14 and 16 hold products from amplification of P-Temp with dZTP, dPTP and dNTPs by Deep Vent (exo−); Lanes 9, 11, 13 and 15 hold products from the positive control using Deep Vent (exo−). MM: molecular weight marker.
These PCR products were then used as templates for the ‘analytical primer extension’ step (Figure 4) to introduce a phosphorothioate linkage from dZTPαS using, in this case, 9°N instead of Vent (exo−) (Figure 8 left). These products were then digested with Exo III (Figure 8, right).
Figure 8 (right) has three prominent features. First, with the amplicon derived from PCR using Taq, the ratio of intensities of the bands at positions 26 and 17 (including band at position 18) was quite high, but decreased with the number of PCR cycles used to generate the amplicons. This provides direct evidence for the gradual loss of the dZ:dP pair over multiple rounds of PCR (additional loss during the ‘analytical primer extension’ steps was normalized through the reference).
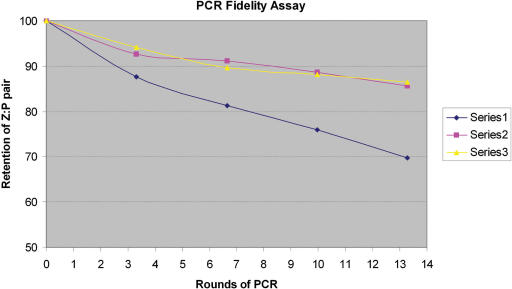
A similar loss was not obvious with Vent (exo−) and Deep Vent (exo−). Here, the relative intensity of the bands at positions 26 and 17 (including the band at position 24) does not decrease with increasing PCR cycles as much as seen with Taq (Figure 9). This implies that both of these polymerases retain the dZ:dP pair better than does Taq. We then fit the data to the equation y = (0.5 + fidelity/2)r (31) where y is the fraction of the original dP remaining in the PCR product, and r is the number of theoretical rounds of PCR. This formula correctly reflects the fact that after r rounds of PCR only half of the PCR product has survived r rounds of PCR; a half of this remainder has survived (r−1) rounds, a half of the next remainder has survived (r−2) rounds, and so on. The estimated values for fidelity per round for Taq were 94.4%, 97.5% for Vent (exo−) and 97.5% for Deep Vent (exo−).
Figure 9.
Retention of dZ:dP pair at pH 8.0 as a function of PCR rounds with three different polymerases. Series 1 (Taq), Series 2 [Vent (exo−)] and Series 3 [Deep Vent (exo−)]. Data from Figure 8 were standardized by the positive control (which, by synthesis, contained 100% dZ:dP pair at position 26 before the ‘analytical primer extension’).
Interestingly, both Vent (exo−) and Deep Vent (exo−) generated PCR products that gave an Exo III degradation pause band at position 28 as well as at position 26. This suggests that the amplicons contain a small amount of a phosphorothioate linkage joining nucleotides 27 and 28. This, in turn, implies the incorporation of dZ at position 28. This position should complement a dG, however, not dP. Thus, the presence of this band suggests that during the PCR, dG must have been replaced by dP at this position. This infidelity is not general, however, as other dG:dC pairs are not replaced; this phenomenon was not further explored.
Further, for Vent (exo−) and Deep Vent (exo−), it appears as if the amount of dP inferred at position 28 in the PCR product ‘increases’ as the number of PCR cycles increases. This implies that during PCR with these polymerases, a dC:dG nucleotide pair is gradually converted to a dZ:dP nucleotide pair. Thus, the higher level of retention of the dZ:dP pair by Vent (exo−) and Deep Vent (exo−) at the site where retention is desired is paralleled by a higher level of misincorporation of dZ and/or dP opposite dG and/or dC, respectively.
The third feature of the gel in Figure 8 (right) is the apparent resistance of all products to initial digestion by Exo III, giving pause bands at the length expected for a 51-mer (approximately; the bands are also doubled). These bands may arise because 9°N added a terminal N + 1 thiotriphosphate in an untemplated extension reaction, a process seen with certain polymerases (32,33). The fact that these bands disappear with Vent (exo−), which is believed to be less prone to non-templated addition (Figure 5), is consistent with this analysis.
The resistance of the product to initial Exo III degradation also suggests, however, the misincorporation of dZTPαS opposite a dG at position 49 or 51, possibly in the ‘analytical primer extension’ by 9°N (Figure 3, right). Such misincorporation might be expected to occur most easily at the end of a template, where the nucleobase pairing is perhaps the least specific.
DISCUSSION
To date, just three examples have reported where six different nucleotides have been introduced into a PCR. The extra base pair in the first example was joined by the pyDAD:puADA hydrogen bonding pattern implemented on 2,4-diaminopyrimidine and xanthine heterocycles (26). This required a double mutant of HIV reverse transcriptase to achieve. As HIV reverse transcriptase is unstable to heating, it was necessary to add new enzyme after each cycle of thermal denaturation. Therefore, only five rounds of PCR were reported. The reported fidelity per round (99%) was quite high.
The pyAAD:puDDA hydrogen bonding pattern implemented on 5-methylisocytidine and isoguanosine was used in the next two examples of six-nucleotide PCR (31,34). The success of one of these PCRs was mitigated slightly by fact that isoguanine exists in two tautomeric forms (35), a keto tautomer (presenting the puDDA hydrogen bonding pattern) that is complementary to 5-methylisocytosine (as desired), and the enol tautomer (presenting the puDAD hydrogen bonding pattern) that is complementary to thymidine (creating the possibility of an isoG:T mismatch) (36). Consistent with the hydrogen bonding ambiguity of isoguanine, significant loss of the nonstandard base pair was observed after multiple PCR cycles (34). Johnson et al. (34) reported a 96% retention per round, but obtained this number by assuming that the slope of a plot of the amount of non-standard nucleotide remaining directly indicates fidelity, an assumption that overlooks the fact that much of the PCR product present in a mixture after r theoretical rounds of cycling is derived from fewer than r cycles. Applying a correct formula to the same data gives a fidelity of ca. 93%, a fidelity per round that is comparable to the amount of minor tautomer of isoguanosine present at equilibrium (ca. 10%).
Sismour et al. (31) managed the tautomerism of isoguanine by exploiting the fact that adenine forms only two of the three canonical hydrogen bonds. Sismour et al. (31) replaced thymidine by 2-thiothymidine in a six letter PCR that included 5-methylisocytidine and isoguanosine. This strategy assumed that the thiol group makes an unfavorable sulfur-proton interaction with the 2-hydroxyl group of the undesired (enol) tautomer of isoguanosine (37), thereby destabilizing the isoguanosine:thymidine mismatch. Because adenine does not present a hydrogen bonding opportunity to the C = S unit, the 2-thioT:A match is not destabilized similarly, creating enhanced specificity. Sismour et al. reported substantially higher fidelity (98% per round) with 2-thiothymidine than with thymidine (a per round fidelity of 93%), permitting over 30 cycles of PCR. The principal feature (and possible disadvantage) of this strategy is that it produces PCR products that are rich in sulfur.
The 6-amino-5-nitro-2(1H)-pyridone heterocycle (dZ, implementing the pyDDA hydrogen bonding pattern), paired with the 2-amino-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one heterocycle (dP, implementing the puAAD hydrogen bonding pattern), appears to successfully support a six-nucleotide PCR in a way that shares certain advantages, and avoids certain disadvantages, of these other examples. Most noticeable among the advantages is the fact that many native polymerases accept this non-standard pair; this extends even to some exo+ polymerases (data not shown). While it is clear that presenting electron density to the minor groove of a non-standard pair is not an absolute requirement for a non-standard nucleobase to be accepted by all polymerases, experience with many over the past decade makes it noteworthy how easily the dZ:dP pair is accepted. This suggests that minor groove scanning, as discussed by Joyce, Steitz et al. (16–20), is a feature of many polymerases that contributes to nucleoside recognition, even if it is not an absolute requirement.
It is difficult to compare directly the reported fidelities of the different literature six-nucleotide PCR experiments, as different methods were used to estimate fidelity for different expanded genetic alphabets, and these methods were often specific to the non-standard nucleobase pair incorporated. In the first example (26), where a 99% fidelity per cycle was reported, a polymerase was available that stopped extension when it encountered the pyDAD non-standard nucleotide. This made estimation of the amount of non-standard nucleotide remaining direct.
Analysis of the loss of pyAAD after multiple rounds of PCR likewise relied on a specific chemical feature of this non-standard nucleotide, its sensitivity to acid (31,34). Using comparable formulas, both Johnson et al. (34) and Sismour et al. (31) arrived at comparable retention per rounds (93%) for the isocytidine-isoguanosine pair using standard dA, dG, dC and T. As acid sensitivity was also used to measure fidelity in a system that exploited thiothymidine to suppress infidelity arising from tautomerism, these 93% values can be directly compared with the 98% fidelity observed with thiothymidine replacing T.
While the phosphorothioate-exonuclease analysis tool is in principle general for all non-standard nucleotides, it has limitations that are apparent in this work. Polymerases must be found that incorporate the alpha-thiotriphosphate of the non-standard nucleotide, as well as the non-standard nucleotide itself. The results must be quantitated against a reference to manage a relatively large number of unknown parameters. It is best when the thiotriphosphate that is incorporated is also the thiotriphosphate that is easily resolved into its diastereomers.
For these reasons, the 94.4%, 97.5% and 97.5% fidelities reported here for Taq, Vent (exo−) and Deep Vent (exo−), respectively, are best used in comparison with each other. In particular, the analysis is adequate to support comparison across a series of closely related experiments, such as those used to determine the pH-dependence of fidelity.
These pH-dependency experiments suggest that the acid-base properties of the components of the dZ:dP pair contribute most to infidelity. The canonical Watson–Crick model (which assigns an important role to inter-strand hydrogen bonding) is able to explain the results of experiments to detect infidelity. Specifically, a route for the conversion of a dZ:dP pair to a dC:dG pair may involve deprotonated dZ (presenting a pyDAA hydrogen bonding pattern) complementing dG (presenting a puADD hydrogen bonding pattern) (Figure 10). Conversely, protonated dC (presenting a pyDDA hydrogen bonding pattern) can complement dP (presenting a puAAD hydrogen bonding pattern). Given this model, one expects fidelity to be lowest at the extremes of pH, and highest at a pH between the pKa of protonated cytidine (ca. 4.5) (38) and the pKa of dZ (ca. 7.8).
Figure 10.
Explanation for the conversion of the dZ:dP pair to a dC:dG pair (and vice versa) involving deprotonation of dZ to give a nucleobase presenting a pyDAA hydrogen bonding pattern complementary to dG, and protonation of dC to give a nucleobase presenting a pyDDA hydrogen bonding pattern complementary to dP.
This model, together with the experiments reported here, suggest different conditions to achieve different goals. To achieve the highest retention of the dZ:dP, a nominal pH of 7.8–8.0 and Vent/Deep Vent (both exo−) are best. This retention comes, however, with the risk of converting dC:dG pairs to dZ:dP pairs. Conversely, for optimal fidelity overall, Taq appears to be better.
A PCR with a fidelity of 94.4% with Taq (which shows little evidence of any ability to convert dC:dG pairs to dZ:dP pairs) is certainly sufficient to allow non-standard nucleotides to participate as dynamic components of multiplexed DNA and RNA sequencing, detection, quantitation and characterization tools. These are widely sought in architectures for molecular biology, systems biology and synthetic biology. Several of these are presently being developed at the Foundation for Applied Molecular Evolution and the Westheimer Institute. Other polymerases, and Taq polymerase variants, are now being explored.
ACKNOWLEDGEMENTS
This project was supported in part through grants from the National Human Genome Research Institute (grants HG3581 and HG3579). Funding to pay the Open Access publication charges for this article was provided by the National Human Genome Research Institute (grants HG3579).
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson JD, Crick FHC. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FHC. General implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Rappaport HP. The 6-thioguanine/5-methyl-2-pyrimidinone base pair. Nucleic Acids Res. 1988;16:7253–7267. doi: 10.1093/nar/16.15.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kool ET. Replacing the nucleobases in DNA with designer molecules. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa M, Hirao I, Yokoyama S. Synthesis of 3-(2-deoxy-β-D-ribofuranosyl) pyridin-2-one and 2-amino-6-(N,N-dimethylamino)-9-(2-deoxy-β-D-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Lett. 2000;41:3931–3934. [Google Scholar]
- 6.Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. A two unnatural base pair system toward the expansion of the genetic code. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 7.Minakawa N, Kojima N, Hikishima S, Sasaki T, Kiyosue A, Atsumi N, Ueno Y, Matsuda A. New Base Pairing Motifs. The synthesis and thermal stability of oligodeoxynucleotides containing imidazopyridopyrimidine nucleosides with the ability to form four hydrogen bonds. J. Am. Chem. Soc. 2003;125:9970–9982. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 8.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting Nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Tae EL, Wu YQ, Xia G, Schultz PG, Romesberg FE. Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J. Am. Chem. Soc. 2001;123:7439–7440. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 10.Switzer CY, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989;111:8322–8323. [Google Scholar]
- 11.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Extending the genetic alphabet. Enzymatic incorporation of a new base pair into DNA and RNA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 12.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 13.Gleaves CA, Welle J, Campbell M, Elbeik T, Ng V, Taylor PE, Kuramoto K, Aceituno S, Lewalski E, et al. Multicenter evaluation of the Bayer VERSANT (TM) HIV-1 RNA 3.0 assay: Analytical and clinical performance. J. Clin. Virol. 2002;25:205–216. doi: 10.1016/s1386-6532(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 14.Elbeik T, Surtihadi J, Destree M, Gorlin J, Holodniy M, Jortani SA, Kuramoto K, Ng V, Valdes R, et al. Multicenter evaluation of the performance characteristics of the Bayer VERSANT HCV RNA 3.0 assay (bDNA) J. Clin. Microbiol. 2004;42:563–569. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson SC, Marshall DJ, Harms G, Miller CM, Sherrill CB, Beaty EL, Lederer SA, Roesch EB, Madsen G, et al. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 2004;50:2019–2027. doi: 10.1373/clinchem.2004.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 18.Morales JC, Kool ET. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson C, Devine K, Benner SA. Probing the necessity of minor groove interactions with three DNA polymerase families using 3-deaza-2′-deoxyadenosine 5′-triphosphate. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCain MD, Meyer AS, Schultz SS, Glekas A, Spratt TE. Fidelity of mispair formation and extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli. Biochemistry. 2005;44:5647–5659. doi: 10.1021/bi047460f. [DOI] [PubMed] [Google Scholar]
- 21.Piccirilli JA, Krauch T, MacPherson LJ, Benner SA. A direct route to 3-(ribofuranosyl)-pyridine nucleosides. Helv. Chim. Acta. 1991;74:397–406. [Google Scholar]
- 22.Voegel JJ, von Krosigk U, Benner SA. Synthesis and tautomeric equilibrium of 6-amino-5-benzyl-3-methylpyrazin-2-one. An acceptor-donor-donor nucleoside base analog. J. Org. Chem. 1993;58:7542–7547. [Google Scholar]
- 23.Hutter D, Benner SA. Expanding the genetic alphabet. Nonepimerizing nucleoside with the pyDDA hydrogen bonding pattern. J. Org. Chem. 2003;68:9839–9842. doi: 10.1021/jo034900k. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Hutter D, Sheng P, Sismour AM, Benner SA. Artificially expanded genetic information system. A new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34:6095–6101. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O (1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H- 1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 26.Sismour AM, Lutz S, Park J.-H, Lutz MJ, Boyer PL, Hughes SH, Benner SA. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linxweiler W, Horz W. Sequence specificity of exonuclease III from E. coli. Nucleic Aids Res. 1982;10:4845–4859. doi: 10.1093/nar/10.16.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He KZ, Porter KW, Hasan A, Briley DJ, Shaw BR. Synthesis of 5-substituted 2′-deoxycytidine 5′-(alpha-P-borano)triphosphates, their incorporationinto DNA and effects on exonuclease. Nucleic Aids Res. 1999;27:1788–1794. doi: 10.1093/nar/27.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Good NE, Winget GD, Winter W, Connolly TN, Izawa S, Singh RMM. Hydrogen ion buffers for biological research. Biochemistry. 1966;5:467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- 30.Day JP, Hammer RP, Bergstrom D, Barany F. Nucleotide analogs and new buffers improve a generalized method to enrich for low abundance mutations. Nucleic Acids Res. 1999;27:1819–1827. doi: 10.1093/nar/27.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sismour AM, Benner SA. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark JM, Joyce CM, Beardsley GP. Novel blunt-end addition-reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 33.Hu GX. DNA polymerase-catalyzed addition of nontemplated extra nucleotides to the 3′ end of a DNA fragment. DNA Cell Biol. 1993;12:763–770. doi: 10.1089/dna.1993.12.763. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: Inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sepiol J, Kazimierczuk Z, Shugar D. Tautomerism of iso-guanosine and solvent-induced keto-enol equilibrium. Z. Naturforsch. 1976;C 31:361–370. doi: 10.1515/znc-1976-7-803. [DOI] [PubMed] [Google Scholar]
- 36.Switzer CY, Moroney SE, Benner SA. Enzymatic recognition of the base-pair between isocytidine and isoguanosine. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 37.Lezius AG, Scheit KH. Enzymatic synthesis of DNA with 4-thio- thymidine triphosphate as substitute for dTTP. Eur. J. Biochem. 1967;3:85–94. doi: 10.1111/j.1432-1033.1967.tb19501.x. [DOI] [PubMed] [Google Scholar]
- 38.Young LY, Krawczyk SH, Matteucci MD, Toole JJ. Triple helix formation inhibits transcription elongation in vitro. Proc. Natl Acad. Sci. USA. 1991;88:10023–10026. doi: 10.1073/pnas.88.22.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]