Abstract
Despite a growing number of splicing mutations found in hereditary diseases, utilization of aberrant splice sites and their effects on gene expression remain challenging to predict. We compiled sequences of 346 aberrant 5′splice sites (5′ss) that were activated by mutations in 166 human disease genes. Mutations within the 5′ss consensus accounted for 254 cryptic 5′ss and mutations elsewhere activated 92 de novo 5′ss. Point mutations leading to cryptic 5′ss activation were most common in the first intron nucleotide, followed by the fifth nucleotide. Substitutions at position +5 were exclusively G>A transitions, which was largely attributable to high mutability rates of C/G>T/A. However, the frequency of point mutations at position +5 was significantly higher than that observed in the Human Gene Mutation Database, suggesting that alterations of this position are particularly prone to aberrant splicing, possibly due to a requirement for sequential interactions with U1 and U6 snRNAs. Cryptic 5′ss were best predicted by computational algorithms that accommodate nucleotide dependencies and not by weight-matrix models. Discrimination of intronic 5′ss from their authentic counterparts was less effective than for exonic sites, as the former were intrinsically stronger than the latter. Computational prediction of exonic de novo 5′ss was poor, suggesting that their activation critically depends on exonic splicing enhancers or silencers. The authentic counterparts of aberrant 5′ss were significantly weaker than the average human 5′ss. The development of an online database of aberrant 5′ss will be useful for studying basic mechanisms of splice-site selection, identifying splicing mutations and optimizing splice-site prediction algorithms.
INTRODUCTION
Mutations that influence pre-mRNA splicing represent a substantial proportion of gene alterations leading to Mendelian disorders (1). cDNA-based mutation studies of disease genes that have a large number of introns showed that splicing mutations accounted for about half of mutated alleles (2,3). In contrast, estimates derived from DNA-based mutation screening designed to scan coding regions and flanking intronic sequences have generally been lower (1,4). As a significant fraction of mutated alleles in both recessive and dominant conditions has not been identified, and the availability of RNA samples from affected individuals and their families is often problematic, the overall contribution of intronic alterations acting at the level of pre-mRNA splicing could be substantial. In addition to single-gene disorders, DNA variants that influence splicing may modify the risk of developing complex diseases and their phenotypic manifestations, but the overall role of this variability in the pathogenesis of such conditions is only beginning to be explored (5–8).
The most common consequence of splicing mutations is skipping of one or more exons, followed by the activation of aberrant 5′ (donor) splice sites (5′ss), 3′ (acceptor) splice sites (3′ss) and full intron retention (1,9,10). Mutation-induced aberrant splice sites found in disease genes often involve disruption of the consensus sequence of the authentic sites, while activating a cryptic splice site nearby. However, aberrant splice sites can also be generated by mutations that create splice-site consensus sequences. As described earlier (11), we refer to these aberrant splice sites as cryptic and de novo, respectively, even though the distinction between cryptic and de novo sites may occasionally be vague, because disruption of the authentic site can also create a new splice site consensus.
Cryptic 3′ss are preferentially located in exons whereas de novo 3′ss usually reside in introns, which has been attributed to splicing signal sequences upstream of the 3′ss that are required for selection of acceptor sites, including the polypyrimidine tract (PPT) and the branch point sequence (BPS) (12). In contrast to cryptic 3′ss, cryptic 5′ss have a similar frequency distribution in exons and introns and their number decreases with increasing distance from the authentic 5′ss (11). The human 5′ss consensus sequence is MAG|GURAGU (M is A or C; R is purine), spanning from position −3 to position +6 relative to the exon–intron junction. This sequence is critical but often insufficient for accurate 5′ss recognition, and may require auxiliary sequences in both introns and exons. These sequences can repress or activate splicing and are referred to as splicing silencers or enhancers, respectively (13–17). The complementarity of the 5′ss consensus to the 5′ end of U1 small nuclear RNA (snRNA) exerts a dominant effect on 5′ss selection, but auxiliary sequences may exhibit a more prominent role in selection of competing 5′ss with lower base-pairing complementarity (18,19). In addition, the intrinsic structural properties of the RNA molecule may hinder 5′ss availability for basal splicing factors, thus controlling splicing efficiency (20–22). Moreover, 5′ss selection can also be influenced by the presence of sequence motifs specific for RNA-binding proteins (23) and by the rate at which the pre-mRNA is transcribed (21).
A variety of methods have been used to computationally predict the 5′ss strength and recognition, including nucleotide frequency matrices (24,25), machine-learning approaches and neural networks (NNs) (26,27) and methods employing putative base-pairing interactions of 5′ss with U1 snRNP (28–30) and interdependence between adjacent or more distant positions of the splicing consensus sequences (31). Exon-prediction algorithms that take into account protein-coding information may perform better than those that rely only on signals present in the splice sites (32). However, it is unknown which models best predict the localization of cryptic or de novo 5′ss that were activated in vivo.
In the present study, we compiled nucleotide sequences of cryptic and de novo 5′ss that have been reported previously in human disease genes since the first description of disease-causing aberrant splice sites (33–35). This resource is being made available to the public through an online retrieval and submission tool termed DBASS5 (database of aberrant 5′ splice sites). In addition, we provide a detailed characterization of the underlying mutation pattern, a comparison of the nucleotide composition of aberrant and corresponding authentic 5′ss, and we evaluate the performance of computational tools that predict their utilization.
MATERIAL AND METHODS
Compilation of mutation-induced aberrant 5′ss in human disease genes
Aberrant 5′ss were identified by searching home pages of peer-reviewed journals and PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). They were included in the database and selected for further analysis if (i) they resulted from disease-causing or -predisposing mutations or variants in human genes; (ii) aberrant RNA products spliced to new 5′ss were verified by nucleotide sequencing; and (iii) their sequences or reliable identifiers were published in peer-reviewed journals between 1981 and January 2007. We also included 22 cases of aberrant 5′ss that were confirmed by minigene assays with wild-type and mutated reporter constructs transfected into mammalian cells, but from which patients’ RNA samples were not available. These criteria were similar to those used for a recently published analysis of aberrant 3′ss (36).
Aberrant 5′ss were manually validated by mapping the information in the literature to sequences in the Human Genome Project databases. Nucleotide sequences of authentic, mutated and aberrant 5′ and 3′ss are available online in the Database of Aberrant Splice Sites http://www.dbass.org.uk/, which consists of the recently described DBASS3 (36) and the newly developed DBASS5.
Computational methods to predict aberrant 5′ss
Validated sequences of aberrant and corresponding authentic 5′ss were used as input files for seven publicly available splice-site prediction algorithms. The Shapiro and Senapathy (S&S) matrix is based on nucleotide frequencies of 5′ss and assumes independence between individual positions of the 9-nt consensus (24,25). The S&S matrix scores were computed using an online tool available at http://ast.bioinfo.tau.ac.il/SpliceSiteFrame.htm. To take into account known dependencies between adjacent and non-adjacent positions of the 5′ss consensus, the compiled sequences were analysed using the first-order Markov model (MM) and the maximum entropy (ME) model (31). The former method considers dependencies between adjacent positions, whereas the latter model approximates short-sequence motif distributions with the ME distribution and may include dependencies between non-adjacent as well as adjacent positions. The maximum dependence decomposition model (MDD) is a decision-tree approach that accentuates the strongest dependencies in the early branches of the tree (37). The MM, ME, MDD and weight-matrix (WMM) scores, which extract single nucleotide probabilities for each position from a training set (38), were computed using online tools at http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html. The HBond algorithm, which analyses individual hydrogen-bonding patterns to the U1 snRNA 5′ end irrespective of nucleotide frequencies and assumes that the threshold values for U1 snRNP binding are influenced by specific SR proteins (29) was computed using a web application available at http://www.uni-duesseldorf.de/rna/html/hbond_score.php. The NN algorithm is a machine-learning approach that recognizes sequence patterns once it is trained with DNA sequences encompassing authentic splice sites (27). We used the NN splice site predictor NNSPLICE (v. 0.9) at http://www.fruitfly.org/seq_tools/splice.html. The free energy (ΔG) of predicted 5′ss/U1 base-pairing was computed using OligoArrayAux (39), which is available at http://www.bioinfo.rpi.edu/applications/hybrid/Oligo-ArrayAux.php. Finally, the number of H bonds (#H) between 5′ss and U1 was computed using a web tool at http://ast.bioinfo.tau.ac.il/SpliceSiteFrame.htm.
To compare the strength of aberrant or authentic 5′ss with a large number of human 5′ss, we used the sequences of 8415 5′ss reported previously (31). The non-parametric Wilcoxon–Mann–Whitney rank test (Stat-200, v. 2.01, Biosoft Ltd., UK) was employed to test the significance of score differences between authentic and aberrant 5′ss in each category.
DBASS5 construction
DBASS5 (database of aberrant 5′ splice sites) is an online retrieval and submission tool for mutation-induced aberrant 5′ss available at http://www.dbass.org.uk/5/, complementing a recently described sister database of aberrant 3′ss, termed DBASS3 (36). The web application was created using the Microsoft ASP and ASP.Net server technology (http://www.asp.net), and Microsoft SQL Server database software (http://www.microsoft.com/sql/). In addition to aberrant 5′ss induced by disease-associated germline and somatic mutations, DBASS5 contains naturally occurring DNA variants that were shown to modify both the relative expression of RNA products spliced to alternative 5′ss and the disease predisposition. Polymorphisms that control exon skipping levels or full intron retention events have not been included in DBASS5.
RESULTS
Mutations that activate aberrant 5′ss
An exhaustive search for previously published aberrant 5′ss identified 305 unique aberrant 5′ss in 166 genes (Table 1). They were generated by a total of 26 deletions/duplications, 3 insertions, 2 complex alterations and 315 point mutations (Table 2). These alterations were described in a total of 264 publications.
Table 1.
Summary of aberrant 5′ splice sites
| Location of cryptic or de novo 5′ splice sites | Exon | Intron | Both | ||
|---|---|---|---|---|---|
| Mutation | In the 9-nt 5′ss consensus (cryptic) | Elsewhere (‘de novo’) | In the 9-nt 5′ss consensus (cryptic) | Elsewhere (‘de novo’) | All mutations |
| Number of genes | 80 | 33 | 71 | 36 | 166 |
| Number of phenotypes | 80 | 37 | 79 | 38 | 192 |
| Number of cryptic and de novo 5′ ss (%) | 121 (35.0) | 46 (13.3) | 133 (38.4) | 46 (13.3)a | 346 (100.0) |
| Number of unique 5′ss (%) | 114 (37.4) | 44 (14.4) | 101 (33.1) | 46 (15.1) | 305 (100.0) |
| Number of aberrant 5′ss affecting terminal exons (%) | 6 (50) | 1 (8) | 5 (42) | 0 (0) | 12 (4) |
| Median distance (nt) between authentic and aberrant 5′ splice sites | −46 | −56 | 49 | 16 | −7 |
| Change in the reading frame for unique aberrant 5′ splice sitesb | |||||
| 0 | 45 | 16 | 41 | 7 | 109 |
| +1 | 41 | 14 | 31 | 6 | 92 |
| +2 | 28 | 13 | 27 | 2 | 70 |
aThe number of pseudoexons (cryptic exons) was 31 (9.0% of all reported aberrant 5′ss and 10.1% of unique aberrant 5′ss).
bThe distribution of the reading frame changes among unique aberrant 5′ss (excl. cryptic exons) was not significantly different from that expected (P = 0.75).
Table 2.
Summary of mutations leading to aberrant 5′ splice sites
| Location of cryptic or de novo 5′ splice sites | Exon | Intron | Both | ||
|---|---|---|---|---|---|
| Mutation | In the 9-nt 5′ss consensus (cryptic) | Elsewhere (‘de novo’) | In the 9-nt 5′ss consensus (cryptic) | Elsewhere (‘de novo’) | All mutations |
| Number of deletions/duplications | 7 | 4 | 10 | 5 | 26 |
| Number of insertions | 1 | 1 | 1 | 0 | 3 |
| Number of complex mutations | 0 | 0 | 2 | 0 | 2 |
| Number of single-nucleotide substitutions | 113 | 41 | 121 | 40 | 315 |
| Wild-type nucleotide | |||||
| A | 12 | 9 | 2 | 17 | 40 |
| C | 3 | 19 | 0 | 13 | 35 |
| G | 81 | 10 | 96 | 8 | 195 |
| T | 17 | 3 | 23 | 2 | 45 |
| Mutated nucleotide | |||||
| A | 50 | 7 | 59 | 5 | 121 |
| C | 21 | 0 | 34 | 0 | 55 |
| G | 17 | 15 | 8 | 22 | 62 |
| T | 25 | 19 | 20 | 13 | 77 |
| Number of GT-creating mutations (%) | 65 (58) | 21 (51) | 60 (50) | 28 (70) | 174 (55) |
The number of reported cryptic 5′ss was almost three times higher than the number of de novo 5′ss (Table 1). Cryptic 5′ss were usually activated by single-nucleotide substitutions of guanosine (G) residues, which were ∼3-times more common than mutations of the remaining nucleotides (177 versus 57, P < 10−15, Table 2). Conversely, substituting adenosines accounted for almost every other point mutation. Among single-nucleotide substitutions leading to de novo 5′ss, cytosine was the most frequently mutated nucleotide (32/81, 40%). In contrast, no de novo 5′ss have thus far been reported to be created by a point mutation introducing cytosine (Table 2).
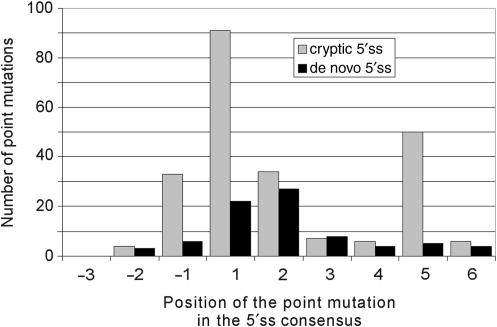
The overall distribution of unique point mutations within the 9-nt consensus sequence was highly non-random both for cryptic and de novo 5′ss (Figure 1). For cryptic 5′ss, point mutations were most common at the highly conserved position +1 relative to the natural intron/exon junctions (39.4%). Interestingly, the second most frequently mutated position was the fifth intron nucleotide (21.6%), followed by positions +2 (14.7%) and –1 (14.3%). Point mutations at positions +3, +4, +6 and −2 each accounted for <3% of all the single-nucleotide substitutions. In contrast to cryptic 5′ss, the most frequent point mutations resulting in de novo 5′ss were at the highly conserved first (28%) and second (34%) intron nucleotides (Figure 1). Single-nucleotide substitutions at position +5 were found only in 5/81 (6%) unique de novo 5′ss as opposed to 50/234 unique cryptic 5′ss (χ2 = 8.6, P = 0.003). The ratio of point mutations in cryptic over de novo 5′ss in the authentic and new 9-nt consensus, respectively, was highest for position +5 (10.0), followed by position −1 (5.5) and +1 (4.1), with an average ratio for all positions of 2.9.
Figure 1.
Distribution of point mutations activating cryptic and/or creating de novo 5′ss. The x-axis shows the position of point mutations in the 9-nucleotide 5′ss consensus relative to the natural exon–intron junctions (cryptic 5′ss) or to the newly created exon–intron boundary (de novo 5′ss). The total number of point mutations at each position of the 5′ss consensus is shown on the y-axis. Grey bars denote the number of cryptic 5′ss; black bars denote the number of de novo 5′ss. The corresponding number of splicing mutations logged in the HGMD was 11, 51, 320, 1445, 456, 163, 63, 347 and 46 for positions –3 through +6, respectively (4).
The overall proportion of point mutations in patients with aberrant 5′ss that created the 5′GT consensus was ∼55% (Table 2). Newly created 5′GT dinucleotides were utilized by the spliceosome in 100% of the observed cases. In contrast, although mutations generating 3′AG dinucleotides found in individuals with aberrant 3′ss are also present in about half of the cases, only ∼95% are used in vivo, owing to the presence of ‘AG exclusion zones’ downstream of the BPS (36).
Tables 3 and 4 show the breakdown of point mutations by nucleotide and by highly conserved positions of the 5′ss consensus. Transitions (R-to-R or Y-to-Y, Y is pyrimidine), which account for 62.5% of point mutations in human disease genes (40), were found in 58.7% of cases (Table 3). Comparison of mutations in highly conserved positions of the 5′ss consensus with those expected based on previously published mononucleotide mutation rates corrected for a number of confounding effects (40) suggested that the biased distribution is unlikely to be fully explained by differential mutability (Table 4; P = 0.002 and 4.3 × 10−7 for position +1 and +2, respectively). However, comparison with the published dinucleotides rates that take into account nearest-neighbour effects no longer showed a significant P-value for position +1, consistent with a severe block of splicing following mutations to any nucleotide (41). Nevertheless, the distribution of point mutations at position +2 was still unlikely to be fully explained by differential mutabilities (Table 4, P = 0.035), raising the possibility that the observed under-representation of +2C/A among cryptic 5′ss may be attributed to higher residual levels of accurately spliced pre-mRNAs with 5′GC or 5′GA dinucleotides. This would be consistent with a previously observed +2T>+2C>+2A>+2G hierarchy in splicing efficiency (42,43) and with efficient recognition of the 0.56% of mammalian introns that have 5′GC-3′AG splice sites (44). Finally, the distribution of point mutations at positions +1 and +2 and that for all splicing mutations in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php) were not significantly different (P = 0.77 for position +1 and P = 0.15 for position +2, Table 4). This suggests that the mutation spectrum of the 5′GT dinucleotide is similar for aberrant 5′ss and exon skipping events, which represent the bulk of HGMD entries.
Table 3.
Breakdown by type of point mutations leading to aberrant 5′ss
| Mutated nucleotide | |||||
|---|---|---|---|---|---|
| A | C | G | T | ||
| Wild-type nucleotide | A | – | 1 (0.3) | 35 (11.1) | 4 (1.3) |
| C | 2 (0.6) | – | 10 (3.2) | 23 (7.3) | |
| G | 109 (34.6) | 36 (11.4) | – | 50 (15.9) | |
| T | 10 (3.2) | 18 (5.7) | 17 (5.4) | – | |
The number of point mutations for each nucleotide substitution is followed by a percentage in parentheses.
Table 4.
Number of single-nucleotide substitutions in highly conserved positions of the 5′ss consensus that resulted in cryptic 5′ss activation
| Observed numbers for cryptic 5′ss | Expectedb | |||
|---|---|---|---|---|
| Location of cryptic 5′ splice site | Exon | Intron | Both | |
| Point mutations at position IVS-1 | 13 | 20 | 33 | |
| −1G>A | 6 | 9 | 15 | |
| −1G>C | 4 | 9 | 13 | |
| −1G>T | 3 | 2 | 5 | |
| Point mutations at position IVS+1 | 50 | 41 | 91 | |
| +1G>A | 30 | 27 | 57 (1024)a | 65.3 (60.0) |
| +1G>C | 5 | 7 | 12 (273)a | 14.9 (12.2) |
| +1G>T | 15 | 7 | 22 (411)a | 10.9 (18.8) |
| Point mutations at position IVS+2 | 15 | 19 | 34 | |
| +2T>C | 4 | 9 | 13 (270)a | 22.5 (16.2) |
| +2T>G | 8 | 6 | 14 (144)a | 6.0 (9.2) |
| +2T>A | 2 | 4 | 6 (121)a | 5.5 (8.6) |
| Point mutations at position IVS+5 | 17 | 33 | 50 | |
| +5G>C | 5 | 6 | 11 | 8.2 |
| +5G>T | 3 | 8 | 11 | 6.0 |
| +z5G>A | 9 | 19 | 28 | 35.9 |
aThe numbers in parentheses refer to all splicing mutations logged in the Human Gene Mutation Database (4). (http://www.hgmd.cf.ac.uk/ac/hoho2/php).
bThe expected numbers were calculated from the relative mononucleotide mutability rates corrected for codon frequencies and other confounding effects (40). The expected numbers calculated from the previously published estimates of the relative substitution rates at the dinucleotide level (94) are in parentheses in the same table column.
Frequent occurrence of IVS+5G mutations leading to cryptic 5′ss activation
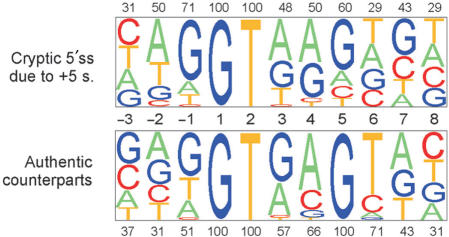
Interestingly, all point mutations at position +5 of authentic 5′ss that activated cryptic 5′ss were substitutions of G, and not any other nucleotide (Figure 2), raising the possibility that 5′ss with +5G are more susceptible to aberrant splice-site activation than 5′ss with +5H (non-G). However, assuming ∼78% occupancy of this nucleotide in human 5′ss (30) and a G/C substitution rate of ∼70% derived from the HGMD data (40), the expected number of +5H substitutions among authentic sites whose mutation induces cryptic 5′ss activation would only be ∼4 in our dataset and not significantly different from zero (P = 0.1, Fisher's exact test). A prominent influence of differential mutability rates on the mutation spectrum was also supported by the observed predominance of +5G>A transitions over transversions (Table 4). In addition, the distribution of point mutations activating cryptic 5′ss was significantly different from that resulting in de novo sites (P < 0.0001, Figure 1), with the latter showing peaks in the most conserved positions +1 and +2 and exclusive +5A>G transitions relative to new 5′ss that were located both in exons (45) or introns (46–48). However, unique point mutations in the 9-nt consensus logged in the HGMD (4) and in our sample (Figure 1) had significantly different distributions (χ2 = 27.7, P = 0.0005), with position +5 clearly overrepresented in our dataset (∼22% versus ∼12%). This suggests that the mutation spectra underlying cryptic 5′ss activation and exon skipping events are distinct.
Figure 2.
Cryptic 5′ss generated by point mutations at position IVS+5. (A) Nucleotide composition of cryptic 5′ss activated by point mutations at position +5 (upper panel) and their authentic counterparts (lower panel). Sequence alignments of unique 5′ss were analysed by a pictogram utility available at: http://genes.mit.edu/pictogram.html. The size of a pictogram character is proportional to the frequency of each nucleotide, with the most frequent at the top. The percentage of the most frequent nucleotide is shown above or below each panel.
A search for literature reports of point mutations that produce either aberrant 5′ss activation or exon skipping in the same 5′ss consensus revealed several discordant cases. For example, the FBN1 substitution IVS46+5G>A resulted in cryptic 5′ss activation 33-nt downstream of the authentic exon–intron junction (49), whereas the IVS46+1G>A mutation caused exon 46 skipping (50). Similarly, the PTEN mutation IVS7+1G>A activated a cryptic 5′ss 75-nt downstream of the authentic exon–intron boundary (51), whereas mutation IVS7+2T>G in the same 5′ss led exclusively to exon 7 skipping (52). In the latter case, IVS7+2T>G creates a putative splicing silencer containing the AGGG motif, which may prevent activation of the downstream cryptic 5′ss, whereas IVS7+1G>A results in no consecutive Gs in the 5′ss consensus.
The presence of IVS+5H in authentic 5′ss, which is not predicted to base-pair with U1 or U6 snRNAs, was proposed to be compensated by having G at the last exon position (–1G) (53). The –1G can base-pair to U1 snRNA (30) and is almost completely conserved (97.5%) in IVS+5H 5′ss (53). The +5/−1 association was confirmed with a large sample of homologous human-mouse 5′ss (30). In our dataset, only 18/35 (51%) of unique authentic 5′ss that were repressed by mutations of IVS+5G in favour of a cryptic 5′ss had -1G. This percentage is significantly (χ2 = 10.9, P = 0.001) lower than for a large set of authentic 5′ss (5142/6716, ∼77%). In addition to position –1, adenosine –2 was less frequent in our sample (31%; 11/24) as compared with 57% in average 5′ss (3830/6716, P = 0.002), while the number of uracils at position +6 was higher (25/35; 71% versus 3415/6716; 51%; χ2 = 5.1, P = 0.02). These results are consistent with previously described +5 dependencies (30,53) and suggest that authentic 5′ss that are susceptible to IVS+5 mutations are less likely to make sufficient contacts between positions –1/–2 and their interacting factors, but may exhibit stronger putative base-pairing interactions between U1/U6 snRNAs and intron position +6.
Comparison of computational tools to predict mutation-induced aberrant 5′ss in vivo
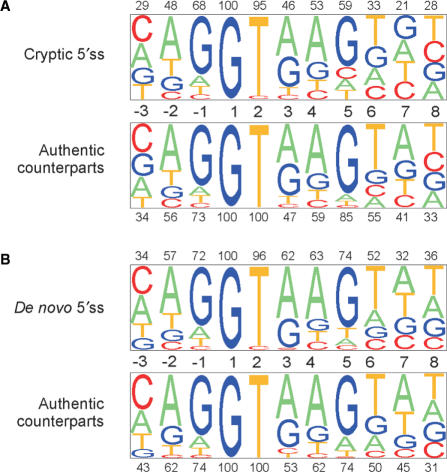
Figure 3 shows the relative representation of each nucleotide in the consensus sequence of aberrant 5′ss (upper panels) and the corresponding authentic sites (lower panels). The consensus sequence of cryptic 5′ss had lower proportions of conserved residues than for authentic 5′ss at each position, except for the invariant position +1 (Figure 3A). This difference was much reduced in de novo 5′ss, in which conserved nucleotides at positions +3 through +6 had even higher frequencies than those in their authentic counterparts (Figure 3B). Sequence alignments of cryptic and de novo sites generated in exons and introns are shown in Supplementary Figures 1–3 together with their authentic counterparts.
Figure 3.
Sequence alignment of authentic and aberrant 5′ss. Sequence alignment of cryptic (A, upper panel) and de novo (B, upper panel) 5′ss with their matching authentic counterparts (lower panels). The nucleotide alignments were analysed by a pictogram utility as in Figure 2. The size of a pictogram character is proportional to the frequency of each nucleotide, with the most common at the top. The percentage of the most frequent nucleotides in aberrant and authentic sites is shown above and below each panel, respectively.
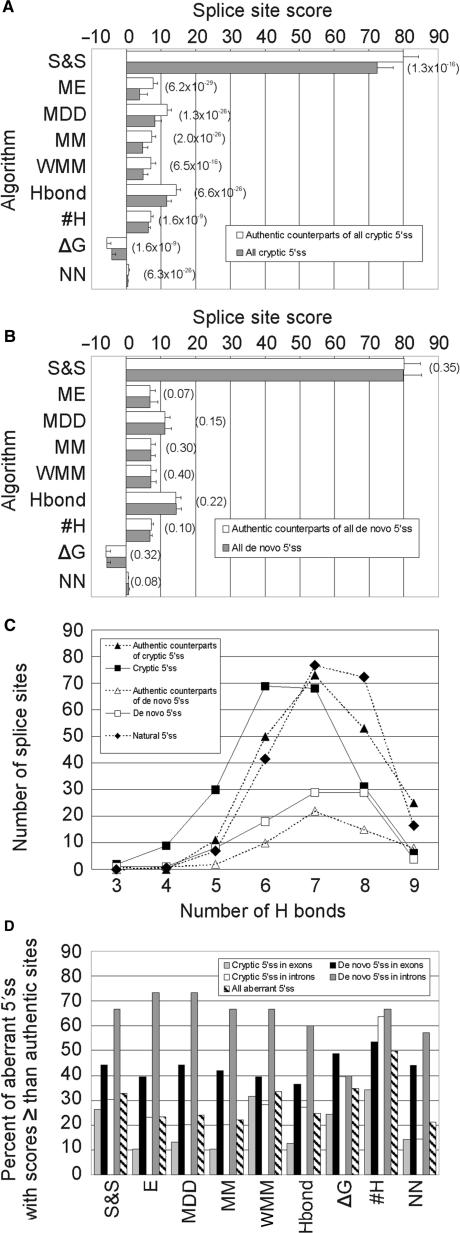
Apart from ΔG and #H between 5′ss and U1 snRNA, we used seven different algorithms that predict utilization of 5′ss in multiple sequences and are publicly available (Figure 4A,B). Cryptic 5′ss had significantly lower scores with each algorithm, lower #H and higher ΔG than their authentic, wild-type counterparts. Cryptic 5′ss were most effectively discriminated from the authentic sites by the ME model, followed by MDD and MM algorithms. P-values obtained for the HBond and NN scores were higher, even when we disregarded cryptic 5′ss with non-canonical 5′ss dinucleotides to obtain the scores and replaced them with group means. All these models clearly outperformed the matrix-based prediction scores—S&S and WMM. The #H and ΔG values gave the poorest, albeit still significant discrimination. The weakness of cryptic 5′ss was well illustrated by a shift of the #H peak frequency from seven in the authentic counterparts to six in the cryptic 5′ss (Figure 4C).
Figure 4.
Comparison of computational tools to predict aberrant 5′ss. (A) cryptic 5′ss, (B) de novo 5′ss. S&S, Shapiro-Senapathy matrix score; ME, Maximum Entropy model; MDD, maximal dependence decomposition model; MM, First-order Markov model; WMM, weight matrix model; Hbond, HBond score; #H, number of H bonds; ΔG, predicted free energy; NN, neural network model. P-values of the Wilcoxon–Mann–Whitney rank tests comparing authentic and aberrant 5′ss are shown in parentheses for each algorithm. Pseudoexons were not included in this comparison. (C), #H distribution in cryptic/de novo 5′ss and their authentic counterparts. The expected #H distribution of natural 5′ss (closed diamonds) was calculated from the #H frequencies observed for a large collection of human 5′ss (31). (D), The proportion of aberrant 5′ss with equal or higher splice-site scores than their authentic counterparts.
In contrast to cryptic 5′ss, de novo 5′ss were not distinguished from their authentic counterparts by any of the tested algorithms (Figure 4B). Although the number of de novo 5′ss was smaller than cryptic 5′ss (Table 1), random selections of the same number of cryptic 5′ss and their comparison with authentic sites gave consistently significant discrimination with several algorithms (data not shown), indicating that computational prediction of de novo 5′ss is poor. However, newly created 5′ss activating pseudoexons had higher ME scores than the remaining de novo 5′ss (8.66 ± 3.00 versus 6.07 ± 4.83, P = 0.0002) or the remaining intronic de novo 5′ss (Table 5, P = 0.002). The corresponding 3′ss of these pseudoexons were slightly stronger than intronic de novo 3′ss (ME scores 6.79 ± 3.39 versus 5.24 ± 4.50, P = 0.04) ascertained previously (36), but were not significantly different from exonic de novo 3′ss or their authentic counterparts. Thus, activation of cryptic exons through de novo 5′ss use requires their high strength and may be facilitated by intrinsically stronger decoy 3′ss across the newly formed exon.
Table 5.
Comparison of the strength of wild-type authentic, mutated and aberrant 5′ss
| Location of aberrant 5′ splice sites a | Exon | Intron | Both | |||
|---|---|---|---|---|---|---|
| Mutation | In the 9-nt 5′ss consensus (cryptic) | Elsewhere (‘de novo’) | In the 9-nt 5′ss consensus (cryptic) | Elsewhere ('de novo') | All mutations | |
| Shapiro and Senapathy matrix score (S&S) | AU (SD) | 80.3 (8.3) | 81.6 (9.4) | 79.6 (9.1) | 75.7 (7.4) | 80.0 (8.8) |
| AB (SD) | 70.8 (9.7) | 77.7 (10.6) | 74.3 (8.7) | 81.7 (10.3) | 74.6 (10.3) | |
| P-value | 4.8 × 10−13 | 0.09 | 6.1 × 10−6 | 0.008 | 7.6 × 10−11 | |
| MU (SD) | 65.4 (11.6) | 81.6 (9.4) | 63.1 (11.7) | 75.7 (7.4) | 67.7 (12.9) | |
| Maximum entropy model (ME) | AU (SD) | 7.9 (2.2) | 7.3 (2.9) | 7.5 (2.9) | 5.8 (2.9) | 7.6 (2.6) |
| AB (SD) | 3.2 (5.1) | 6.1 (4.6) | 4.6 (3.9) | 7.8 (4.2) | 4.8 (4.8) | |
| P-value | 3.8 × 10−21 | 0.26 | 4.8 × 10−10 | 0.003 | 3.6 × 10−16 | |
| MU (SD) | 0.6 (6.3) | 7.3 (2.9) | –0.8 (6.0) | 5.8 (2.9) | 1.4 (6.3) | |
| Maximum-dependence decomposition model (MDD) | AU (SD) | 11.7 (2.5) | 11.6 (2.9) | 11.7 (2.7) | 9.9 (2.6) | 11.6 (2.7) |
| AB (SD) | 7.9 (3.9) | 10.4 (3.7) | 8.9 (3.5) | 11.8 (4.3) | 9.2 (4.0) | |
| P-value | 1.0 × 10−18 | 0.11 | 1.1 × 10−9 | 0.001 | 1.0 × 10−15 | |
| MU (SD) | 4.6 (5.5) | 11.6 (2.9) | 3.7 (5.3) | 9.9 (2.6) | 5.7 (5.7) | |
| First-order Markov model (MM) | AU (SD) | 7.5 (2.0) | 7.5 (2.8) | 7.2 (2.4) | 6.1 (2.5) | 7.3 (2.3) |
| AB (SD) | 4.2 (3.6) | 6.5 (3.0) | 5.1 (2.5) | 7.6 (2.7) | 5.4 (3.3) | |
| P-value | 2.6 × 10−19 | 0.13 | 1.4 × 10−9 | 0.016 | 2.9 × 10−16 | |
| MU (SD) | 1.3 (4.7) | 7.5 (2.8) | 0.6 (4.4) | 6.1 (2.5) | 2.3 (4.9) | |
| Weight matrix model (WMM) | AU (SD) | 7.2 (2.5) | 7.6 (2.9) | 6.9 (2.9) | 6.0 (2.5) | 7.1 (2.7) |
| AB (SD) | 4.3 (3.2) | 6.5 (3.1) | 5.3 (2.7) | 7.6 (2.9) | 5.4 (3.2) | |
| P-value | 5.4 × 10−12 | 0.09 | 6.2 × 10−6 | 0.020 | 1.8 × 10−10 | |
| MU (SD) | 1.3 (4.5) | 7.6 (2.9) | 0.7 (4.5) | 6.0 (2.5) | 2.4 (4.9) | |
| HBond score | AU (SD) | 14.7 (2.7) | 14.6 (3.0) | 14.2 (2.3) | 13.3 (2.5) | 14.4 (2.6) |
| AB (SD) | 11.2 (2.8) | 13.8 (3.2) | 12.2 (2.5) | 15.0 (3.1) | 12.5 (3.1) | |
| P-value | 1.4 × 10−18 | 0.17 | 2.0 × 10−8 | 0.015 | 1.8 × 10−15 | |
| Free energy (ΔG) | AU (SD) | −5.8 (2.5) | −5.8 (2.9) | −5.5 (2.2) | −5.7 (2.1) | −5.7 (2.4) |
| AB (SD) | −3.9 (2.3) | −5.5 (2.2) | −4.7 (2.4) | −5.6 (2.2) | −4.6 (2.4) | |
| P-value | 3.5 × 10−9 | 0.23 | 9.3 × 10−3 | 0.46 | 4.2 × 10−7 | |
| MU (SD) | −2.7 (2.2) | −5.8 (2.9) | −1.9 (1.9) | −5.7 (2.1) | −3.1 (2.7) | |
| Number of H bonds (#H) | AU (SD) | 7.3 (1.2) | 7.3 (1.1) | 6.9 (1.0) | 6.9 (1.0) | 7.2 (1.1) |
| AB (SD) | 6.1 (1.2) | 7.0 (1.2) | 6.8 (1.0) | 7.0 (1.1) | 6.6 (1.2) | |
| P-value | 5.7 × 10−12 | 0.07 | 0.19 | 0.34 | 2.1 × 10−8 | |
| MU (SD) | 6.3 (1.3) | 7.3 (1.1) | 5.7 (1.3) | 6.9 (1.0) | 6.3 (1.4) | |
| Neural network (NN) | AU (SD) | 0.78 (0.25) | 0.74 (0.30) | 0.82 (0.25) | 0.55 (0.38) | 0.78 (0.27) |
| AB (SD) | 0.41 (0.35) | 0.66 (0.39) | 0.45 (0.38) | 0.83 (0.31) | 0.52 (0.39) | |
| P-value | 7.2 × 10−16 | 0.15 | 2.4 × 10−12 | 0.003 | 4.1 × 10−15 | |
Means and standard deviations (SD) of splice-site prediction scores for wild-type authentic (AU) and aberrant (AB) 5′ss are followed by the P-values (corrected for ties) of the Wilcoxon–Mann–Whitney rank tests comparing AB and AU. A small number of missing values for the HBond and NN scores due to non-canonical nucleotides at positions +1 and +2 were treated as a group mean. P-values below 0.05 were considered significant and are shown in bold. The splice-site strength for authentic 5′ss with mutations (MU) is shown only for algorithms that score non-canonical 5′ss.
aPseudoexons were not included in this comparison.
We then tested each computational method separately for aberrant 5′ss in exons and introns (Table 5). Although cryptic 5′ss in exons were best discriminated by the ME scores, the lowest P-values for cryptic 5′ss in introns were achieved by the NN model. To test whether this could be explained by having to disregard 5′GC splice sites for the NN method in both datasets and replace them by group means, we recalculated the NN and ME scores after removing 5′GC splice sites, but we obtained a similar result (P = 1.2 × 10−12 versus 1.0 × 10−10, respectively). Authentic counterparts of intronic de novo 5′ss were intrinsically weak and therefore less likely to challenge newly created competitors. However, this was not evident for exonic de novo sites, strongly suggesting that their activation is more reliant on splicing regulatory sequences in exons rather than on the intrinsic strength of the 5′ss consensus (Table 5). The overall performance of ME, MDD, MM, HBond and NN models for the whole set of aberrant 5′ss was very similar, with minimal differences in P-values. Finally, mutated authentic 5′ss were on average weaker than cryptic 5′ss, confirming an earlier observation (11). Again, the lowest P-values of the non-parametric test were observed for the ME model (Table 5 and data not shown). Thus, as shown for 3′ss (36), the ME algorithm discriminated best both wild-type and mutated authentic 5′ss from cryptic 5′ss (Table 5), thus providing a method of choice for computational prediction of aberrant splice sites.
Next, we carried out pair-wise comparisons of cryptic and de novo 5′ss with their authentic counterparts. For each computational algorithm, we determined the proportion of aberrant 5′ss that showed equal or higher scores than their respective wild-type authentic sites (Figure 4D). This proportion was on average significantly higher for de novo 5′ss than for cryptic 5′ss and roughly reflected the ability of each method to discriminate between aberrant 5′ss and their authentic counterparts. The percentage of exonic cryptic 5′ss with equal or higher scores than their authentic counterparts was lowest for the ME algorithm (10.5%). For intronic cryptic 5′ss, the same proportion was lowest for the NN method (14.4%, Figure 4D). Using the best-performing algorithms, ∼12.3% of cryptic 5′ss were computationally stronger than their wild-type authentic counterparts, yet they were used in vivo only if the wild-type 5′ss consensus was inactivated or weakened by mutation. This underscores the importance of factors that repress utilization of decoy splice sites that are present in excess over natural sites in the genome.
Importantly, the authentic counterparts of cryptic 5′ss were significantly weaker than a large collection of 8415 natural 5′ss (31), with the ME scores of 7.75 ± 2.50 and 8.37 ± 2.08, respectively (P = 2 × 10−6; Wilcoxon–Mann–Whitney rank test). The distribution of #H in the authentic counterparts of cryptic 5′ss and natural 5′ss was also significantly different (P = 0.02, χ2 = 13.5, 5 df for 4–9 #H), with a maximum difference at 8 #H (Figure 4C). The relative weakness of the authentic counterparts of cryptic 5′ss is consistent with the notion that mutations in less conserved positions of stronger 5′ss produce, on average, higher amounts of natural transcripts and less severe phenotypes than identical alterations in intrinsically weaker 5′ss.
The predicted strengths of authentic sites that were mutated at position +5 were significantly lower than the average authentic 5′ss (ME scores 7.55 ± 1.81 versus 8.37 ± 2.08, P = 0.0002) and also somewhat lower than the authentic counterparts of all unique cryptic 5′ss in our dataset (7.55 ± 1.81 versus 7.75 ± 2.52), despite all having +5G and a higher than average relative frequency of +6T (Figure 2). Guanine at position +5 was proposed not to be obligatory for 5′ss selection if the two preceding positions are purines (28); nevertheless 24/35 (69%) of unique authentic 5′ss with point mutations of +5G had only purines at positions +3 and +4 and only a single authentic counterpart had pyrimidines at both positions.
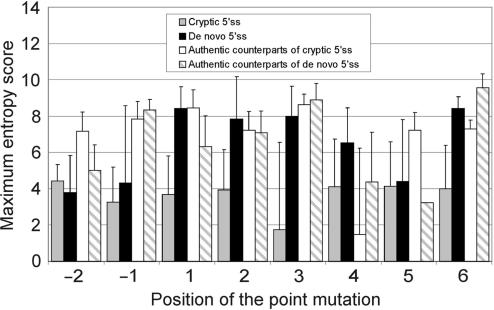
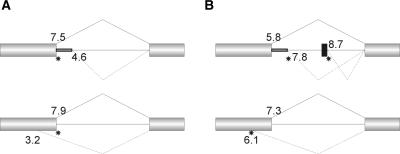
Figure 5 shows a comparison of the ME scores of cryptic and de novo 5′ss by mutated position in the authentic and new 5′ss consensus, respectively. Cryptic 5′ss had similar ME values irrespective of the location of the point mutation (P > 0.05, F-test). In contrast, de novo sites created by mutations in a subset of intronic positions of the new 5′ss consensus tended to be stronger, with statistically significant differences between de novo and cryptic 5′ss for the highly conserved positions +1 and +2. In addition, we compared the ME scores of aberrant 5′ss with their respective counterparts (Figure 5). The authentic counterparts of cryptic 5′ss induced by point mutations at positions +2 and +5 of natural sites were weaker than the authentic counterparts of cryptic 5′ss activated by substitutions at position +1 (7.24 ± 1.93 for position +2 and 7.22 ± 2.06 for position +5 versus 8.34 ± 2.21 for position +1, P < 0.01 for both comparisons). This is consistent with the notion that mutations at less conserved positions of authentic 5′ss are less likely to completely inactivate the 5′ss and result in recognizable phenotypes than mutations at position +1. The authentic counterparts of de novo sites induced by mutations at position +1 were significantly weaker than the authentic counterparts of cryptic 5′ss induced by mutations at the same position (6.33 ± 3.39 versus 8.34 ± 2.21). The number of mutations for the remaining positions of the 5′ss consensus was too small for meaningful comparisons. The average intrinsic strength of aberrant and authentic 5′ss in each category is schematically summarized as the mean ME score in Figure 6.
Figure 5.
Splice-site strength of authentic and aberrant 5′ss by position of the point mutation. The ME scores of cryptic and de novo 5′ss induced by point mutations at positions -2 through +6 relative to the authentic and the new intron–exon boundary, respectively, are followed by the ME scores of their authentic counterparts. Unlike the authentic counterparts, aberrant 5′ss included pseudoexon sequences. Error bars represent standard deviations (except for a single value entry for authentic counterparts of de novo 5′ss generated by a mutation at position +5).
Figure 6.
Summary of the splice-site strength by category of aberrant 5′ss. (A) cryptic 5′ss in introns (upper panel) and exons (lower panel); (B) de novo 5′ss in introns (upper panel) and exons (lower panel). The average ME scores are shown next to the corresponding category of 5′ss. Exons are represented by boxes, introns are represented by lines. A cryptic exon is denoted by a black box. Canonical and aberrant RNA products are shown above and below the primary transcript, respectively. Mutations in the 9-nt consensus of authentic 5′ss (A) or elsewhere (B) are schematically represented by stars.
Taken together, cryptic 5′ss generated in vivo were best predicted by models that accommodate nucleotide dependencies in the 5′ss, particularly by the ME algorithm, which takes into account non-adjacent positions (Figure 4A). Discrimination of exonic cryptic 5′ss from their authentic counterparts was more efficient than that for intronic cryptic 5′ss, because the former category of aberrant 5′ss was weaker than the latter (P = 0.02), for which the NN model gave the best performance (Table 5). Computational discrimination of de novo 5′ss and their authentic counterparts was poor (Figure 4B) as de novo 5′ss were, on average, stronger than cryptic 5′ss (Table 5), particularly when generated by point mutations in highly conserved intronic positions of the new 5′ss consensus (Figure 5). The intrinsic strength of exonic de novo 5′ss could not be distinguished from their authentic sites at all, pointing to the importance of exonic regulatory sequences in their selection. Finally, the authentic counterparts of aberrant (both cryptic and de novo) 5′ss were weaker than a large collection of human 5′ss, highlighting the practical importance of ranking splice sites in human disease genes using efficient computational tools. We propose that their systematic categorization may facilitate identification of intronic mutations or polymorphisms that affect pre-mRNA splicing, improve the interpretation of unknown alterations and, ultimately, increase the cost-effectiveness of mutation screening.
DBASS5
The DBASS5 (http://www.dbass.org.uk/5) provides access to the database of aberrant 5′ss through the search option (Supplemental Figure 4A). DBASS5 can be searched by phenotype, gene, mutation, location of aberrant 5′ss and their distance from authentic 5′ss. If more than one database entry is found, the user can manually choose the details page (Supplemental Figure 4B), which shows nucleotide sequences flanking the authentic and aberrant 5′ss, the estimated strength of both authentic and aberrant 5′ss and literature references with a PubMed hyperlink. DBASS5 visitors can register to obtain regular updates by email and can submit published data through a submission tool. Potential applications of DBASS5 include optimization of splice-site prediction algorithms, leading to improved prediction of aberrant 5′ss, identification of genes and gene segments frequently involved in aberrant splice-site activation, detection of splicing mutations in a gene or phenotype of interest and selection of in vitro models for studying basic mechanisms of 5′ss utilization.
DISCUSSION
The database of mutation-induced aberrant splice sites
This report presents the first comprehensive and publicly available database of aberrant splice sites in human disease genes. Together with a recently described database of aberrant 3′ss (36), this combined resource now contains over 600 unique mutations that create or activate a total of 562 aberrant splice sites.
The overall number of reported aberrant 5′ss was higher than aberrant 3′ss, consistent with sequence limitations imposed by additional signal sequences upstream of 3′ss (BPS and PPT) that are important for recognition of splice acceptor sites. The relative ratio of non-repetitive aberrant 5′ss (n = 305) and 3′ss (n = 257) [(36) and I.V., (unpublished data)], was smaller than that reported for unique splicing mutations in the HGMD that were arbitrarily selected to reside in 5 exonic and 15 intronic nucleotides adjacent to natural splice sites (4), i.e, 1.2 versus 1.5, respectively. The lower ratio might reflect a reporting bias towards mutations closer to authentic splice sites for exon skipping events. Mutations located upstream of intronic splicing signals that are required for 3′ss selection could not be detected in many published mutation reports, because these regions were amplified only for a subset of introns or were not scanned at all. In addition, the lower ratio could be due to an under-representation of mutations leading to de novo splice sites in the HGMD as compared to our dataset. Also, the availability of suitable decoy splice sites near mutated sites is likely to determine if the outcome of a splicing mutation is exon skipping or aberrant splice-site activation (4).
The higher number of cryptic than de novo 5′ss (Table 1) can, probably to a large extent, be explained by a detection bias of DNA-based mutation screening, a method used to identify most aberrant 5′ss in this dataset, towards coding regions and flanking intronic sequences. As explained above, classification of aberrant 5′ss as cryptic and de novo 5′ss may occasionally be vague, but DBASS5 contains only two ambiguous examples (54,55). Both cases were induced by G+1-to-T+1 substitutions in 5′ss that had G at position −1, creating a new 5′GT 1-nt upstream of the authentic 5′'ss. Both cases were classified as cryptic 5′ss in our analysis. The rarity of such cases confirms the validity of the previously proposed (11) categorization of aberrant 5′ss.
Mutation pattern of aberrant 5′ss
The most frequent point mutations that activated aberrant 5′ss were purine transitions, accounting for 45.7% cases (11.1% A>G and 34.6% G>A mutations; Table 3). This figure seems to be somewhat lower (P = 0.08) than the ∼54% (113/211) observed for aberrant 3′ss (36), probably due to a higher prevalence of transitions in the 3′YAG than those in the 5′ss consensus. Cryptic 5′ss resulted from point mutations in each nucleotide of the 9-nt consensus except for position –3, consistent with this position being the least conserved. However, position –3 has previously been implicated in pathological exon skipping in well-documented cases (56–59), suggesting that –3 substitutions in weak 5′ss are also likely to result in aberrant 5′ss, although these cases must be rare and aberrant splicing and putative phenotypic manifestations could be subtle.
As for positions adjacent to the 9-nt 5′ss consensus, each of the reported single-base substitutions at intron positions +7 and +8 created new 5′GT dinucleotides in situ that were used in vivo (60,61). Despite position +7 exhibiting a predominance of purines after several rounds of functional 5′ss selection experiments (28), point mutations downstream of the 5′ss consensus resulting in activation of cryptic 5′ss have thus far not been reported. Among upstream substitutions in DBASS5, we found only a single case of an exonic de novo 5′ss generated by a C>T transition 11-nt upstream of an authentic 5′ss (62), consistent with a disruption of exonic splicing regulatory sequences.
Are point mutations at any position of the 5′ss consensus sequence particularly prone to aberrant 5′ss activation? As observed for mutations in the HGMD (4), position +1 led the frequency table in our dataset, with 49.8% and 39.4% mutations observed in the two studies, respectively (Figure 1). However, the overall distribution of mutations within the 5′ss consensus was significantly different between the two. In particular, the proportion of mutations at position +5 was almost twice as high among cryptic 5′ss than in the HGMD [Figure 1 and ref. (4)]. For unique point mutations leading to cryptic 5′ss activation, this position was in the second place and position +2 in the third, whereas this order was opposite for unique mutations in the HGMD (50 and 34 mutations versus 347 and 456, respectively; P = 0.004).
G at position +5 is nearly invariant in Saccharomyces cerevisiae (63). In contrast, +5G is present in only ∼88% of S. pombae introns (64) and ∼78% of human introns (30), indicating that relief from the absolute requirement for G was an ancient evolutionary event. Comparison of exonized and non-exonized intronic Alu repeats revealed a higher number of +5Gs in exonized sequences (65). Mutations at position +5 have resulted in frequent activation of cryptic 5′ss both in yeasts (66–69) and humans (Figure 1, all references available at: http://www.dbass.org.uk). Our study is the first to provide statistical evidence that this position is important for distinct aberrant splicing outcomes. DBASS5 gives many examples of natural 5′ss in which different point mutations resulted in the same cryptic 5′ss. Similarly, there are numerous cases in the literature of identical exon skipping events caused by different point mutations in the same 5′ss. The identification of several exceptions in humans using the DBASS5 and HGMD data (49–52) is consistent with an earlier observation in S. cerevisiae, namely that cryptic 5′ss activation by +5G>A mutation was not replicated for another 5′ss point mutation in the same intron (67). These rare examples may provide important insights into the requirements for activation of aberrant 5′ss, as opposed to exon skipping events.
In addition to the local sequence context, the frequent occurrence of +5G>A substitutions underlying aberrant 5′ss activation (Figure 1 and Table 3) can be explained by a more severe splicing outcome of these transitions. More dramatic splicing defects for +5G>A transitions than +5G transversions were found in S. cerevisiae (69). In contrast, each IVS1 + 5G>H mutation in the human proinsulin gene promoted activation of a competing decoy 5′ss 26 nt downstream of the authentic 5′ss to the same extent, irrespective of the substituting nucleotide [(7); J.K. and I.V., unpublished data], consistent with a position effect.
What interaction(s) at position +5 is crucial for aberrant 5′ss activation? Authentic 5′ss in which mutation at position +5 generated cryptic 5′ss had a high proportion of +5G+6T (Figure 2). Interestingly, the +5G+6T dinucleotide signifies the most frequent location for alternative 5′ss across several species, and this preference was suggested to result from U1 binding rather than U6 binding (70). However, compensatory mutations in U1 snRNA that restore base-pairing with the mutated intron frequently fail to suppress aberrant splicing, suggesting that position +5 is engaged in additional interactions (28,71–74). Interaction of U6 snRNA with the 5′ss (75) at intron position +5 (76) was partially suppressed by U6 mutations predicted to increase base-pairing (77,78). Although the 5′ ss has very limited complementarity to the U6 ACAGAG motif, this interaction seems to be important for accurate 5′ss selection also in mammals (78–80), albeit not in all systems (81). In addition, cryptic splice sites have been induced by co-expression of splicing reporters with mutated snRNAs, including U1 (82), U5 (83,84) and U6 (77–79,85), both in yeast (77,82,83,85) and mammals (78,79,84).
As U1 snRNP binds sequences that are not used as 5′ss and is present in excess over U6, sequential occupancy by both snRNPs may be absolutely essential for accurate 5′ss utilization (86,87). This would be consistent with rate-limiting U6 snRNA interactions with the pre-mRNA observed for U1-independent splicing (78) and loose requirements for U1 binding in numerous introns (28,71,72,74). Mutations of IVS+5G that resulted in cryptic 5′ss occurred in relatively weak authentic 5′ss and they are likely to further reduce U1 binding so that it may no longer be sufficient for accurate 5′ss recognition. U6/U4.U5 snRNP would then bind to nearby pseudo-sites that are, on average, intrinsically stronger than the mutated authentic 5′ss [Table 5 and ref. (11)]. The strength of predicted base-pairing interactions at positions +5 and +6 in the authentic counterparts (Figure 2) may hamper the transfer of these 5′ss from U1 to U6. In fact, strong 5′ss were reported to be inhibitory in S. cerevisiae, potentially delaying the release of U1 and productive interactions with U6 (28), although extended complementarity between U1 snRNA and a human immunodeficiency virus 1 donor site did not inhibit splicing (81). Mutations that destabilized a yeast 5′ss/U6 duplex improved the second step of splicing and hyperstabilization of the 5′ss/U6 interaction had the opposite effect, suggesting that changing the stability of these interactions alters the equilibrium between the first and second step conformations (88). Suppression of 5′ss mutations by U6 in a hybrid reporter was more efficient when U1 could pair nearby than when pairing was restored further away (79). In addition, position +5 may directly interact with U6 residues that base pair to the BPS recognition region of U2 (89) as well as with other splicing factors, such as PRP8 (90,91). Finally, sequential recognition of position +5 is likely to require contacts with exon-bound factors that may substitute for U1 interactions (92,93), and that may be essential for spliceosome assembly at authentic 5′ss and contribute to the observed high number of cryptic sites as compared to de novo 5′ss (Figure 1 and Table 1).
In summary, we have shown that cryptic 5′ss in human disease genes are best predicted by computational methods that accommodate nucleotide dependencies and not by methods employing only nucleotide frequency matrices. Discrimination of intronic cryptic 5′ss from their authentic counterparts was less effective than for exonic cryptic 5′ss, as the former were intrinsically stronger than the latter. Computational prediction of exonic de novo 5′ss was poor, suggesting that their activation in vivo critically depends on exonic splicing enhancers or silencers, rather than on the strength of the 5′ss consensus, and that improved algorithms for their prediction will need to accommodate auxiliary splicing sequences. The authentic counterparts of both de novo and cryptic 5′ss were weaker than the average human 5′ss, highlighting the practical importance of ranking splice sites in disease genes to improve detection of splicing mutations. The mutation spectra of cryptic and de novo 5′ss were distinct and differed also from that underlying exon skipping events, implicating point mutations at position +5 in frequent activation of cryptic 5′ss. Finally, the development of an online database of aberrant 5′ss will facilitate detection of introns and exons frequently involved in aberrant splicing, identification of auxiliary sequences that control selection of aberrant splice sites, fine-tuning of splice-site prediction algorithms, identification of splicing mutations, as well as studies of the basic mechanisms of splice-site selection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Juvenile Diabetes Research Foundation International (1-2006-263), Telethon Onlus Foundation (GGP02453 and GGP06147), FIRB (RBNE01W9PM), and by the EC grant EURASNET-LSHG-CT-2005-518238. A.R.K. acknowledges support from NIH grant GM42699. We thank S. Mills, R. Sood, P. Gibbs and T. Bryant for technical help. Funding to pay the Open Access publication charges for this article was shared equally by the above funding sources.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cooper DN, Krawczak M. Human Gene Mutation. Oxford: BIOS Scientific Publishers; 1993. [Google Scholar]
- 2.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal Ö., Bernatowska E, et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 4.Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 5.Cooper TA, Mattox W. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nissim-Rafinia M, Kerem B. Splicing regulation as a potential genetic modifier. Trends Genet. 2002;18:123–127. doi: 10.1016/s0168-9525(01)02619-1. [DOI] [PubMed] [Google Scholar]
- 7.Královičová J, Gaunt TR, Rodriguez S, Wood PJ, Day INM, Vořechovský I. Variants in the human insulin gene that affect pre-mRNA splicing: is -23HphI a functional single nucleotide polymorphism at IDDM2? Diabetes. 2006;55:260–264. doi: 10.2337/diabetes.55.01.06.db05-0773. [DOI] [PubMed] [Google Scholar]
- 8.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 9.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 10.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roca X, Sachidanandam R, Krainer AR. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Královičová J, Christensen MB, Vořechovský I. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005;33:4882–4898. doi: 10.1093/nar/gki811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 14.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spena S, Tenchini ML, Buratti E. Cryptic splice site usage in exon 7 of the human fibrinogen Bβ-chain gene is regulated by a naturally silent SF2/ASF binding site within this exon. RNA. 2006;12:948–958. doi: 10.1261/rna.2269306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornblihtt AR. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratti E, Baralle M, De Conti L, Baralle D, Romano M, Ayala YM, Baralle FE. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 26.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 27.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 28.Lund M, Kjems J. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA. 2002;8:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–6975. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 32.Thanaraj TA. Positional characterisation of false positives from computational prediction of human splice sites. Nucleic Acids Res. 2000;28:744–754. doi: 10.1093/nar/28.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busslinger M, Moschonas N, Flavell RA. β+ thalassemia: aberrant splicing results from a single point mutation in an intron. Cell. 1981;27:289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- 34.Felber BK, Orkin SH, Hamer DH. Abnormal RNA splicing causes one form of α thalassemia. Cell. 1982;29:895–902. doi: 10.1016/0092-8674(82)90451-2. [DOI] [PubMed] [Google Scholar]
- 35.Treisman R, Proudfoot NJ, Shander M, Maniatis T. A single-base change at a splice site in a β0-thalassemic gene causes abnormal RNA splicing. Cell. 1982;29:903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- 36.Vořechovský I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burge CB. In: Computational methods in molecular biology. Salzberg SL, Searls DB, Kasif S, editors. Amsterdam: Elsevier Science; 1998. pp. 129–164. [Google Scholar]
- 38.Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984;12:505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deirdre A, Scadden J, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruis BL, Kivens WJ, Siliciano PG. The interaction between the first and last intron nucleotides in the second step of pre-mRNA splicing is independent of other conserved intron nucleotides. Nucleic Acids Res. 1994;22:5190–5195. doi: 10.1093/nar/22.24.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aebi M, Hornig H, Weissmann C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- 44.Burset M, Seledtsov IA, Solovyev VV. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29:255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O′Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 46.McConville CM, Stankovic T, Byrd PJ, McGuire GM, Yao QY, Lennox GG, Taylor MR. Mutations associated with variant phenotypes in ataxia-telangiectasia. Am. J. Hum. Genet. 1996;59:320–330. [PMC free article] [PubMed] [Google Scholar]
- 47.Rathmann M, Bunge S, Beck M, Kresse H, Tylki-Szymanska A, Gal A. Mucopolysaccharidosis type II (Hunter syndrome): mutation “hot spots” in the iduronate-2-sulfatase gene. Am. J. Hum. Genet. 1996;59:1202–1209. [PMC free article] [PubMed] [Google Scholar]
- 48.Tuffery-Giraud S, Saquet C, Chambert S, Claustres M. Pseudoexon activation in the DMD gene as a novel mechanism for Becker muscular dystrophy. Hum. Mutat. 2003;21:608–614. doi: 10.1002/humu.10214. [DOI] [PubMed] [Google Scholar]
- 49.Hutchinson S, Wordsworth BP, Handford PA. Marfan syndrome caused by a mutation in FBN1 that gives rise to cryptic splicing and a 33 nucleotide insertion in the coding sequence. Hum. Genet. 2001;109:416–420. doi: 10.1007/s004390100573. [DOI] [PubMed] [Google Scholar]
- 50.Nijbroek G, Sood S, McIntosh I, Francomano CA, Bull E, Pereira L, Ramirez F, Pyeritz RE, Dietz HC. Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am. J. Hum. Genet. 1995;57:8–21. [PMC free article] [PubMed] [Google Scholar]
- 51.Celebi JT, Wanner M, Ping XL, Zhang H, Peacocke M. Association of splicing defects in PTEN leading to exon skipping or partial intron retention in Cowden syndrome. Hum. Genet. 2000;107:234–238. doi: 10.1007/s004390000362. [DOI] [PubMed] [Google Scholar]
- 52.Tsou HC, Ping XL, Xie XX, Gruener AC, Zhang H, Nini R, Swisshelm K, Sybert V, Diamond TM, et al. The genetic basis of Cowden′s syndrome: three novel mutations in PTEN/MMACI/TEP1. Hum. Genet. 1998;102:467–473. doi: 10.1007/s004390050723. [DOI] [PubMed] [Google Scholar]
- 53.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 54.Attanasio C, de Moerloose P, Antonarakis SE, Morris MA, Neerman-Arbez M. Activation of multiple cryptic donor splice sites by the common congenital afibrinogenemia mutation, FGA IVS4+1G>T. Blood. 2001;97:1879–1881. doi: 10.1182/blood.v97.6.1879. [DOI] [PubMed] [Google Scholar]
- 55.Cavalcanti DP, Matejas V, Luquetti D, Mello MF, Zenker M. Fraser and Ablepharon macrostomia phenotypes: Concurrence in one family and association with mutated FRAS1. Am. J. Med. Genet. A. 2006;143:241–247. doi: 10.1002/ajmg.a.31426. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Poh-Fitzpatrick M, Carriero D, Ostasiewicz L, Chen T, Taketani S, Piomelli S. A novel mutation in erythropoietic protoporphyria: an aberrant ferrochelatase mRNA caused by exon skipping during RNA splicing. Biochim. Biophys. Acta. 1993;1181:198–200. doi: 10.1016/0925-4439(93)90112-e. [DOI] [PubMed] [Google Scholar]
- 57.Williamson D, Brown KP, Langdown JV, Baglin TP. Haemoglobin Dhofar is linked to the codon 29 C>T (IVS-1 nt-3) splice mutation which causes β+ thalassaemia. Br. J. Haematol. 1995;90:229–231. doi: 10.1111/j.1365-2141.1995.tb03409.x. [DOI] [PubMed] [Google Scholar]
- 58.Chao HK, Hsiao KJ, Su TS. A silent mutation induces exon skipping in the phenylalanine hydroxylase gene in phenylketonuria. Hum. Genet. 2001;108:14–19. doi: 10.1007/s004390000435. [DOI] [PubMed] [Google Scholar]
- 59.Aretz S, Uhlhaas S, Sun Y, Pagenstecher C, Mangold E, Caspari R, Moslein G, Schulmann K, Propping P, et al. Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Hum. Mutat. 2004;24:370–380. doi: 10.1002/humu.20087. [DOI] [PubMed] [Google Scholar]
- 60.López-Bigas N, Rabionet R, de Cid R, Govea N, Gasparini P, Zelante L, Arbones ML, Estivill X. Splice-site mutation in the PDS gene may result in intrafamilial variability for deafness in Pendred syndrome. Hum. Mutat. 1999;14:520–526. doi: 10.1002/(SICI)1098-1004(199912)14:6<520::AID-HUMU11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 61.Sidwell RU, Sandison A, Wing J, Fawcett HD, Seet JE, Fisher C, Nardo T, Stefanini M, Lehmann AR, et al. A novel mutation in the XPA gene associated with unusually mild clinical features in a patient who developed a spindle cell melanoma. Br. J. Dermatol. 2006;155:81–88. doi: 10.1111/j.1365-2133.2006.07272.x. [DOI] [PubMed] [Google Scholar]
- 62.Yamada T, Tachibana A, Shimizu T, Mugishima H, Okubo M, Sasaki MS. Novel mutations of the FANCG gene causing alternative splicing in Japanese Fanconi anemia. J. Hum. Genet. 2000;45:159–166. doi: 10.1007/s100380050203. [DOI] [PubMed] [Google Scholar]
- 63.Spingola M, Grate L, Haussler D, Ares, M. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 65.Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in Alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 66.Parker R, Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5′ splice junction uncouples recognition, cleavage, and ligation. Cell. 1985;41:107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- 67.Jacquier A, Rodriguez JR, Rosbash M. A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985;43:423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- 68.Fouser LA, Friesen JD. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- 69.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dou Y, Fox-Walsh KL, Baldi PF, Hertel KJ. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA. 2006;12:2047–2056. doi: 10.1261/rna.151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siliciano PG, Guthrie C. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 73.Séraphin B, Rosbash M. Exon mutations uncouple 5′ splice site selection from U1 snRNA pairing. Cell. 1990;63:619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- 74.Cohen JB, Snow JE, Spencer SD, Levinson AD. Suppression of mammalian 5′ splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl Acad. Sci. USA. 1994;91:10470–10474. doi: 10.1073/pnas.91.22.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawa H, Shimura Y. Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992;6:244–254. doi: 10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- 76.Wassarman DA, Steitz JA. Interactions of small nuclear RNA′s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 77.Kandels-Lewis S, Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 78.Crispino JD, Sharp PA. A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes Dev. 1995;9:2314–2323. doi: 10.1101/gad.9.18.2314. [DOI] [PubMed] [Google Scholar]
- 79.Hwang DY, Cohen JB. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev. 1996;10:338–350. doi: 10.1101/gad.10.3.338. [DOI] [PubMed] [Google Scholar]
- 80.Brackenridge S, Wilkie AO, Screaton GR. Efficient use of a ‘dead-end’ GA 5′ splice site in the human fibroblast growth factor receptor genes. EMBO J. 2003;22:1620–1631. doi: 10.1093/emboj/cdg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ, Schaal H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alvarez CJ, Wise JA. Activation of a cryptic 5′ splice site by U1 snRNA. RNA. 2001;7:342–350. doi: 10.1017/s1355838201002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 84.Cortes JJ, Sontheimer EJ, Seiwert SD, Steitz JA. Mutations in the conserved loop of human U5 snRNA generate the use of novel cryptic 5′ splice sites in vivo. EMBO J. 1993;12:5191–5200. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 86.Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maroney PA, Romfo CM, Nilsen TW. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 88.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic centre of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 89.Valadkhan S, Manley JL. A tertiary interaction detected in a human U2-U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA. 2000;6:206–219. doi: 10.1017/s1355838200992197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vidal V, Verdone L, Mayes AE, Beggs JD. Characterization of U6 snRNA-protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crispino JD, Blencowe BJ, Sharp PA. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 93.Tarn WY, Steitz JA. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 94.Krawczak M, Cooper DN. Single base-pair substitutions in pathology and evolution: two sides to the same coin. Hum. Mutat. 1996;8:23–31. doi: 10.1002/(SICI)1098-1004(1996)8:1<23::AID-HUMU3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.