Abstract
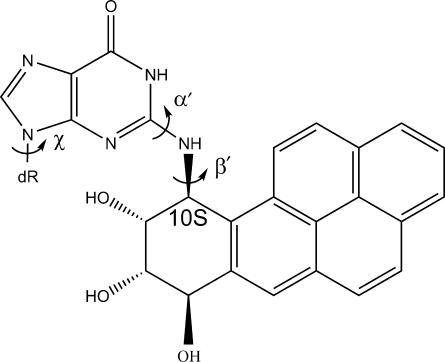
We have investigated how a benzo[a]pyrene-derived N2-dG adduct, 10S(+)-trans-anti-[BP]-N2-dG ([BP]G*), is processed in a well-characterized Pol I family model replicative DNA polymerase, Bacillus fragment (BF). Experimental results are presented that reveal relatively facile nucleotide incorporation opposite the lesion, but very inefficient further extension. Computational studies follow the possible bypass of [BP]G* through the pre-insertion, insertion and post-insertion sites as BF alternates between open and closed conformations. With dG* in the normal B-DNA anti conformation, BP seriously disturbs the polymerase structure, positioning itself either deeply in the pre-insertion site or on the crowded evolving minor groove side of the modified template, consistent with a polymerase-blocking conformation. With dG* in the less prevalent syn conformation, BP causes less distortion: it is either out of the pre-insertion site or in the major groove open pocket of the polymerase. Thus, the syn conformation can account for the observed relatively easy incorporation of nucleotides, with mutagenic purines favored, opposite the [BP]G* adduct. However, with the lesion in the BF post-insertion site, more serious distortions caused by the adduct even in the syn conformation explain the very inefficient extension observed experimentally. In vivo, a switch to a potentially error-prone bypass polymerase likely dominates translesion bypass.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are environmental pro-carcinogens that are produced during combustion of organic materials (1). Benzo[a]pyrene (BP), one of the most extensively studied PAHs, is found in a wide range of substances ingested or inhaled by humans, such as automobile exhaust, tobacco smoke and food (1–3). Several BP metabolic activation pathways have been delineated (4–6). The predominant diol epoxide pathway yields, among others, the major reactive (+)-anti-benzo[a]pyrene diol epoxide (BPDE) (6). The (+)-anti-BPDE enantiomer is highly mutagenic and tumorigenic in mammals (7). This metabolite can react chemically with DNA, forming several bulky BP-base adducts, among which the 10S(+)-trans-anti-[BP]-N2-dG ([BP]G*) adduct (Figure 1) is dominant and is the focus of this study (8–11). This adduct can lead to mutations when replicated in vivo and in vitro (12–24). Although such bulky adducts mainly block replicative DNA polymerases, error-prone bypass can occur in vivo if the blocked replicative polymerase is replaced by one or more bypass polymerases (25–31). Nevertheless, replicative polymerases are also able to bypass such adducts, though rarely, with potentially mutagenic consequences (32,33).
Figure 1.
Structure of the 10S(+)-trans-anti-[BP]-N2-dG ([BP]G*) adduct. Torsion angle definitions are as follows: χ: O4′-C1′-N9-C8; α′: N1-C2-N2-[BP]C10; β′: C2-N2-[BP]C10-C9.
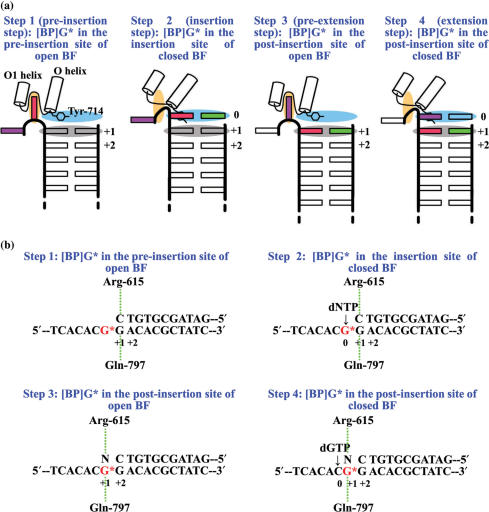
Bacillus fragment (BF) is a Pol I family replicative DNA polymerase large fragment from a thermophilic strain of Bacillus stearothermophilus (34). Our focus on this particular replicative polymerase is motivated by the availability of high-resolution BF crystal structures of a binary complex (PDB ID: 1L3S) with enzyme and primer/template DNA, and a ternary complex (PDB ID: 1LV5) which contains, in addition, an incoming Watson-Crick paired 2′-deoxynucleotide triphosphate (dNTP) (35). BF adopts an open conformation in the binary complex. Placement of the correct dNTP partner opposite the template base causes the fingers domain to close tightly around the active site via an induced-fit mechanism (36–39) to form a reaction-ready ternary complex (35, 40–43). Three important sites have been delineated at and near the active site of BF, namely the pre-insertion, insertion and post-insertion sites. The pre-insertion site, in the fingers domain, is composed of two helices, O and O1, and the loop connecting them. In the open BF, the pre-insertion site is occupied by the base next-to-be replicated, while the insertion site is blocked by a critical polymerase amino acid residue, Tyr-714. When the polymerase closes, the base which was previously in the pre-insertion site moves to the insertion site, where it pairs with the incoming dNTP and the insertion site is no longer blocked by Tyr-714. The pre-insertion site is blocked by the loop connecting the O and O1 helices. After the incoming dNTP is incorporated into the 3′-end of the primer, the polymerase opens again, the DNA translocates, and the new base pair moves to the post-insertion site. The insertion site is now again blocked by Tyr-714, the pre-insertion site is occupied by the next-to-be-replicated template base and the replication cycle continues (35). Thus, the pre-insertion, insertion and post-insertion sites are vital to the functioning of this type of polymerase.
An incompletely resolved open BF crystal structure with the [BP]G* adduct as the base next-to-be-replicated and a well-resolved open BF crystal structure with [BP]G* at the post-insertion site paired with cytosine (PDB ID: 1XC9) (44) provide partial insights into the inhibition of a replicative polymerase by a bulky adduct. These two structures represent the stages before and after nucleotide insertion opposite the adduct, respectively. In the incompletely resolved structure, the adducted guanine, which would occupy the pre-insertion site if not modified by the BP moiety, is disordered, is not located in the pre-insertion site, and is not poised for the next step of nucleotide incorporation. In the well-resolved structure, the modified dG* adopts a normal anti conformation with the BP rings on the DNA minor groove side. Normally on this side there are extensive interactions between DNA and the BF polymerase, constituting a scanning track that is most important in ensuring polymerase fidelity and processivity (45–49). Placement of the BP in this position disrupts the track. In addition, the presence of the adduct distorts the primer end so that the 3′-OH is no longer in an appropriate position for attacking the α-phosphate of the incoming dNTP during nucleotide incorporation. The position of the BP is such that it would interfere with dNTP binding and polymerase closing.
In the present work we investigated, using molecular modeling and dynamics simulations, how the representative replicative polymerase, BF, treats the [BP]G* adduct through an entire replication cycle. The availability of the series of crystal structures with well-characterized sites employed in the replicative cycle presents an excellent opportunity to elucidate molecular details of lesion processing in this model system. We investigated the effect of the [BP]G* adduct on the crystallographically well-determined BF structures, considering for the first time both open binary and closed ternary complexes, as well as the pre-insertion, insertion and post-insertion sites. This work builds on earlier studies involving the [BP]G* adduct in closed ternary complexes of the Pol I family DNA polymerase from phage T7 (50–52). Our primer extension and single dNTP insertion studies provide the experimental foundation for the modeling studies. The experimental data clearly show predominant polymerase blockage with a small amount of lesion bypass and preferential insertion of purines opposite the adduct. Our modeling studies provide molecular views of the various intermediates in the multi-step process of replication past the bulky [BP]G* adduct, enabling us to gain a better understanding of the structural properties leading to predominant polymerase blockage and to rationalize the rare mutagenic lesion bypass. Blockage is engendered by substantial distortions of the active site in the anti glycosidic bond conformation of the dG*, while some bypass appears feasible when this bond is in the less distorting syn conformation; the syn conformation also supports the preferential misincorporation of purines opposite the lesion.
MATERIALS AND METHODS
Chemicals, enzymes and oligonucleotides
[γ-32P] ATP (3000 ci/mmol) was purchased from Perkin-Elmer Life Sciences, Inc. (Boston, MA). The dNTPs, dATP, dCTP, dGTP and dTTP were purchased from New England Biolabs, Inc. (Beverly, MA). BF was prepared as described previously (34). The 11-mer oligonucleotides with a central triple base motif CACACGGACAC were synthesized by standard phosphoramidite methods on a Biosearch Cyclone automated DNA synthesizer (Milligen-Biosearch Corp., San Rafael, CA) and were purified and desalted by standard HPLC protocols. The site-specifically modified oligodeoxynucleotides were ligated to a 13-mer and a 19-mer to form 43-mer template oligonucleotide strands, 5′-GAC TAC GTA CTG TCA CAC G*GA CAC GCT ATC TGG CCA GAT CCG C-3′. The ligated oligonucleotides were further purified and desalted by three successive ethanol precipitations. The quality of the samples was verified by gel electrophoresis; fluorescence and UV absorption spectroscopy were employed to further verify that the BP residue was intact.
Running-start primer extension
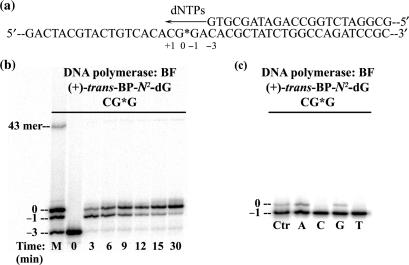
The 43-mer templates were annealed with a 5′-end 32P-labeled 22-mer primer. The BP modified guanine residue was positioned at template position 25 counted from the 3′ end of the template strand (Figure 2a). The time course of primer extension assays of the adducted templates were carried out at 37°C. The standard 30 μl reactions contained 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1 mM DTT, 50 μg/ml BSA, 4% glycerol, 4.5 nM primer:template (1:1.5 ratio) complexes, 200 μM dNTPs and 20 nM of BF. The experiments were stopped after pre-selected time intervals (3, 6, 9, 12, 15 and 30 min) by the addition of a 5 μl stop solution (20 mM EDTA in 95% formamide, 0.05% bromophenol blue, and 0.05% xylene cyanol) to the reaction mixture. The samples were then heated at 90°C for 5 min, chilled on ice and then applied to a 15–20% denaturing polyacrylamide gel containing 7 M urea. The replication products were visualized by autoradiography and quantitatively analyzed by a Storm 840 phosphorimager and the Storm ImageQuant software (Amersham).
Figure 2.
(a) Template-primer strand duplex used in the running start experiment showing the numbering scheme. (b) Typical running start primer extension experiments catalyzed by BF as a function of incubation time (min). Extension of the primer strand, a 22-mer with the terminal 3′-G base opposite the template base C labeled ‘–3’ on the template strand. The position of the [BP]G* is labeled ‘0’ (25-mer). The ‘–1’ position denotes a 24-mer. The lane marked ‘M’ contains 22-mer, 24-mer, 25-mer and 43-mer primer strands (the 43-mer corresponds to the length of a fully extended primer strand). Reactions were initiated with 4.5 nM of the primer/template duplex (1:1.5 ratio), 20 nM of BF, and 200 μM of dNTPs at 37°C. (c) Typical single step 2′-deoxynucleotide triphosphate (dNTP) insertion catalyzed by BF. The primer strand was 24 nucleotides long with the terminal 3′-C positioned opposite the template base labeled ‘–1’ which corresponds to the darkest bands in this figure. Incorporation of each individual dNTP opposite the [BP]G* adduct results in the lighter bands just above the darkest bands, where A = dATP, C = dCTP, G = dGTP, T = dTTP, and Ctr = ‘–1’ and ‘0’ position marker. Reactions were initiated at 2 mM of each dNTP, 15 nM of [DNA], and 2 nM of BF at 37°C for 30 min.
Single dNTP insertion opposite the template lesion site
The 43-mer templates were annealed with a 5′-end 32P-labeled 24-mer primer. The one-step insertion assays of each dNTP (2 mM) opposite the modified guanine were performed utilizing 15 nM primer/template complexes and 2 nM BF concentrations at 37°C for 30 min. The products were visualized and analyzed in the same manner as the running-start primer extension assays.
Molecular modeling
The crystal structures of the open binary complex (PDB ID: 1L3S) and the closed ternary complex (PDB ID: 1LV5) of BF (35) were used as the starting structures for molecular modeling with coordinates obtained from the Protein Data Bank (53). All molecular modeling was carried out using Insight 2000.1 (Accelrys, Inc., a subsidiary of Pharmacopeia, Inc.). The original crystal structures of the ternary and binary complexes were remodeled (full details provided in Supplementary Data). The remodeled complexes were then used to build the initial models for the molecular dynamics (MD) simulations. The [BP]G* was modeled into (i) the pre-insertion site of open BF; (ii) the insertion site of closed BF; (iii) the post-insertion site of open BF; and (iv) the post-insertion site of closed BF, representing four important steps as the adduct threads through the polymerase (Figure 3a). The DNA sequences were remodeled to match that used experimentally. According to the different positions of [BP]G* in BF at the four steps, four corresponding structures were built (Figure 3b). We then made a grid search, at 5° intervals in combination of χ, α′, and β′, for structures with minimal close contacts, within the ranges: χ(anti) = 230 ± 50°, χ(syn) = 40 ± 40°, α′ = 0–360°, and β′ = 270 ± 60° [near the low energy domain of β′, which governs the orientation of this stereoisomer (54)]. Thus we investigated both anti and syn conformations of the glycosidic torsion angle χ (Figure 1) of the BP modified dG* at each of the four steps. Starting from Step 2, we placed the normal partner C opposite the anti-[BP]G* and all four partners opposite the syn-[BP]G*. For the anti case, one [BP]G* conformation with fewest collisions was found for each of Steps 1, 2 and 4; two conformations were found for Step 3, one similar to the available crystal structure and the other different from it. We also simulated the available crystal structure. Therefore, three initial models were investigated for the anti-[BP]G* at Step 3. For the syn case, one [BP]G* conformation was found for each of the four steps. The torsion angles in the initial models for all four steps are given in Table S1. For each step, an unmodified control, containing normal partner C, where appropriate, was also constructed for comparison. All initial models are shown in Figure S2.
Figure 3.
(a) Cartoons showing four consecutive steps as [BP]G* passes through the active site region of BF. Color code: red rectangle, [BP]G*; green rectangle: the partner opposite [BP]G*; purple rectangle: the base to be replicated after [BP]G*; orange ellipse, the pre-insertion site; light blue ellipse, the insertion site; grey ellipse, the post-insertion site. Adapted from (35). (b) [BP]G* modified DNA structures used in the four steps. Minor groove hydrogen bonding interactions between BF and the +1 base pair are indicated. N = A/C/G/T. Base pair numbers given are independent of the position numbers in Figure 2.
Force field parameterization
Full details are given in Supplementary Data.
MD simulation protocol
MD simulations were carried out using the SANDER module of AMBER 7.0 (55) with the Cornell et al. force field (56) and the parm99.dat parameter set (57). Following equilibration, 2 ns of production MD were carried out. Full details are given in Supplementary Data.
Trajectory analysis
Trajectories were collected for each modified system and its corresponding unmodified control, and were analyzed using the PTRAJ and CARNAL modules of the AMBER package (55) to obtain ensemble average values for properties of interest.
Stability of the MD simulations
The RMSDs (root-mean-square deviations) of the whole structure of each system, calculated relative to the first production frame, are shown in Figure S3. The RMSDs of the active site, composed of all the residues within 5 Å of the nascent base pair relative to the first frame, are also provided (Figure S3). We found that all systems are stable after 800 ps. The following analyses are based on this range. All structural figures were prepared using PyMOL (58).
RESULTS
BF is mainly blocked by [BP]G* but allows some bypass
Running-start primer-extension experiments were carried out with the primer terminus several nucleotides away from the lesion site (Figure 2a). The 22-mer primer strand is quickly extended by two nucleotides up to the nucleotide opposite the template base flanking the adduct [BP]G* on the 3′ side. Insertion of the nucleotide opposite [BP]G* is slowed but is almost complete (90%) after a 30 min reaction time (Figure 2b). In contrast, further extension of the primer strand beyond the [BP]G* adduct on the template strand is significantly inhibited. Nevertheless, a small amount of full extension (∼1%) to a 43-mer is also observed after an incubation time of 30 min. It is evident from Figure 2b that the incorporation of a dNTP opposite the lesion is significantly more effective than primer extension beyond the lesion site.
In order to determine which 2′-deoxyribonucleotides are incorporated opposite [BP]G* in the current sequence context, standing-start single nucleotide insertion assays were carried out using a 24-mer primer strand with its 3′-terminal base paired with the template base flanking [BP]G* on the 3′-side. The experimental conditions were adjusted in order to limit the incorporation of nucleotides to <10%. At a [DNA]/[Enzyme] ratio of 7.5 employed in these experiments, it is evident that purines rather than pyrimidines are preferentially inserted opposite [BP]G* (Figure 2c, and Supplementary Data). The steady-state kcat and Km Michaelis-Menten parameters, determined for dATP and dGTP insertion opposite [BP]G* (Figure S4), are summarized in Table S3. The values of the insertion efficiencies fins = kcat/Km are ∼7000 and ∼20 000 times smaller, respectively, than the value of insertion of the normal dCTP opposite an unmodified G template base in the same sequence context (Table S3).
In order to investigate the structural basis of these observations, especially the relatively facile insertion of dNTP opposite the lesion, and the strong inhibition of primer extension beyond the lesion, we constructed initial models with anti- and syn-[BP]G* at (i) the pre-insertion site of open BF; (ii) the insertion site of closed BF; (iii) the post-insertion site of open BF; and (iv) the post-insertion site of closed BF. These are four consecutive steps as [BP]G* advances through the active site of BF (Figure 3a). For studies of mismatched dNTP insertion, we considered only the syn-[BP]G* case, based on our modeling results that anti-[BP]G* would be totally blocking irrespective of the incoming dNTP. The models were subjected to 2 ns of MD following equilibration, and achieved stability after 800 ps (Figure S3). The structures, after MD, are shown in Figures 4, S5 and S6, and analyses of the trajectory ensembles are provided in Tables S4–S11 and Figures S7–S11.
Figure 4.
Simulated systems after 2 ns MD. Color code: light-pink, protein (ribbon representation); magenta, Tyr-714; grey, template DNA strand; cyan, primer DNA strand; orange, Mg2+ ions (sphere representation); yellow, modified dG*; red, BP moiety; blue, dATP/dA; green, dCTP/dC; light blue, dGTP/dG; chocolate, dTTP/dT. (k–m) are the three simulated systems with [BP]G* at the post-insertion site of open BF: two computer-generated models (k,1) and one crystal structure (m). The arrows in (s) indicate the motion of the Watson-Crick edges of modified guanine and partner C. The arrow in (v) indicates the movement of the phosphate of [BP]G*. Figure 4 is continued on the next page.
Structural analyses
The structural properties relevant to polymerase function were analyzed in each case in the presence of the [BP]G* lesion and compared to the corresponding unmodified control. We chose structural and geometric features of the active site region of the binary polymerase-DNA and ternary polymerase-DNA-dNTP complexes deemed important for faithful and efficient catalysis (35,59,60). Our hypothesis is that distortions to the correct alignment, as observed with unmodified templates, impede or slow polymerase activity. The structural characteristics of the active site we examined are: (i) the width of the pre-insertion site (Figure S7 and Table S4) at Steps 1 and 3 (the open binary complex): the pre-insertion site is open and occupied by the next-to-be-replicated base in the binary complex, but is blocked by a polypeptide loop in the closed ternary complex (35); (ii) the number and occupancies of the hydrogen bonds in the [BP]G*-containing base pair (Figure S8) and its neighboring base pairs; hydrogen bond occupancies are the percent of time during the stable region of the MD trajectory (0.8–2.0 ns) that the hydrogen bond is present according to the following criteria: 3.3 Å between heavy atoms (donor and acceptor) and a hydrogen bonding donor-hydrogen-acceptor angle of 135°; specifically, we monitored the hydrogen bonds at the +1 base pair at Step 1; 0 and +1 base pairs at Step 2; +1 and +2 base pairs at Step 3; and 0, +1 and +2 base pairs at Step 4 (Figure 3b and Table S5); (iii) the stacking interaction between Tyr-714 and the templating base at the post-insertion site (Figure 4) at Steps 1 and 3 (the binary complex): Tyr-714 blocks the insertion site and stacks with the template base at the post-insertion site in the binary complex, but this stacking interaction is not present in the ternary complex (35); (iv) base stacking interactions in the nascent base pair and the DNA duplex region around the [BP]G (Figure 4); (v) minor groove interactions at the post-insertion site (+1 base pair) (Figure 3b and Table S6), where the newly formed base pair first meets the minor groove scanning track; at this site, the N3/O2 of the template base forms hydrogen bonds with Gln-797, and the O2/N3 of the primer base forms hydrogen bonds with Arg-615 (35); (vi) the width of the post-insertion site, measured by the distance between the C1’ atoms of the template and primer strands at this site (Table S7); (vii) hydrogen bonds between amino acid residues and incoming dNTP; these stabilize nucleotide binding at Steps 2 and 4 (the ternary complex) (Table S8); (viii) the active site pocket geometry (defined in Figure 5a and b) at Steps 2 and 4 (the ternary complex) (Figure 5 and Table S9); (ix) blockage by BP of the nucleotide binding pocket at Steps 1 and 3 (the binary complex) (Figure 6); (x) frequency of sampling a near reaction-ready distance, in the range of 3.1–3.5 Å, between Pα of the dNTP and the O3′ of the primer end (59,61) (Figure S9 and Table S10); (xi) the angles formed by the O3′, Pα of the dNTP and the O connecting Pα and Pβ, ideally 180° for in-line attack and chemical reaction (62) (Figure S9 and Table S10); (xii) distance between the two Mg2+ ions (Table S11): normal values are ∼3.5 Å (63); (xiii) octahedral coordination of the Mg2+ ions (Table S11). Properties (x)–(xiii) are for Steps 2 and 4 (the ternary complex).
Figure 5.
(a,b) Definition of active site pocket geometry in closed ternary complexes: (a) minor groove side wall (defined by Ile-657, Glu-658, Tyr-714 and Gly-715) and width of pocket (measured by distance between C1′ of templating base and C1′ of incoming dNTP at insertion site); (b) floor (defined by flat face of base pair at post-insertion site) and ceiling (defined by Lys-706 and Phe-710) of pocket, and height of pocket on primer and template sides (measured by distance between Lys-706 Cα and carbon 6 of primer base at post-insertion site, and distance between Phe-710 Cα and carbon 6 of templating base at post-insertion site). (c) pocket wall in anti-[BP]G*:dCTP system at Step 2, deformed by BP (colored in red) on minor groove side. (d) Increase in pocket height on template side due to tilted plane of modified guanine in syn-[BP]G*:dC system at Step 4; syn-[BP]G*:dA and syn-[BP]G*:dT systems are similar. Also shown is an increase in pocket height on primer side in syn-[BP]G*:dC system. The single-head arrow indicates rolling motion in major groove edge of C. The BP moiety is again shown in red.
Figure 6.
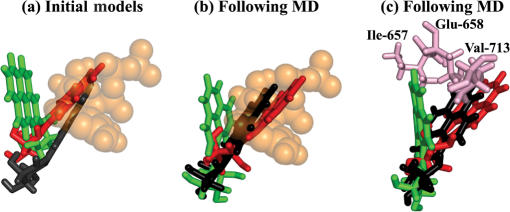
(a) Superposition of the three initial models with anti-[BP]G* at the post-insertion site of open BF (Step 3). Color code: red, anti-1-[BP]G*:dC; green, anti-2-[BP]G*:dC; black, the anti-3-[BP]G*:dC (crystal structure); orange, nucleotide binding pocket; (b) Superposition of the three systems following dynamics. Color code is the same as (a). (c) The amino acids (pink) comprising the barrier preventing convergence of anti-2-[BP]G*:dC with anti-1-[BP]G*:dC and anti-3-[BP]G*:dC. Color code for the three systems is the same as (a). Only the BP moiety is shown in all three figures.
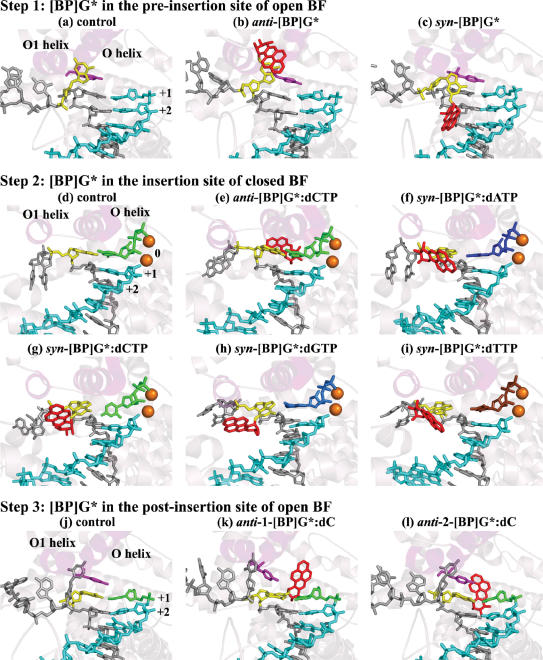
Step 1. [BP]G at the pre-insertion site of open BF: anti-[BP]G* seriously disturbs the pre-insertion site, while syn-[BP]G* does not
When the BP modified dG* adopts the normal anti conformation as in the unmodified control, the BP moiety is deeply imbedded in the pre-insertion site (Figure 4b), significantly enlarging its width (>3 Å) as compared to the unmodified control (Table S4). However, when the adducted dG* adopts the less common syn conformation, the BP is out of the pre-insertion site and the base remains accommodated there (Figure 4c); thus the pre-insertion site width is unaffected (Table S4). The adduct at the pre-insertion site, whether anti or syn, does not affect Watson-Crick hydrogen bonding of the +1 G:C pair (Figure 3b and Table S5). In addition, the following structural features are also not affected significantly: stacking between Tyr-714 and the base at the post-insertion site (Figure 4b and c), base stacking in the DNA duplex region (Figure 4b and c), minor groove contacts at the post-insertion site (Figure 3b and Table S6), the nucleotide binding pocket and the width of the post-insertion site (Table S7).
Step 2. [BP]G* at the insertion site of closed BF: anti-[BP]G* causes more distortion to the active site pocket and minor groove contacts than syn-[BP]G*
anti-[BP]G*:dCTP minor groove
The presence of BP on the minor groove side disrupts the important minor groove interaction on the template side (hydrogen bond occupancy of only 1%) (Figure 3b and Table S6). The BP also significantly enlarges the height of the active site on the template side (Table S9) and pushes the template to the side part of the active site wall (Tyr-714 and Gly-715) away from close contact with the modified guanine (Figure 5c). While the anti modified dG* forms three hydrogen bonds with its partner, dCTP (Figure S8b), one hydrogen bond has reduced occupancy by ∼30% as compared to the control (Table S5).
However, other active site features are near normal as compared to the unmodified control: stacking of the modified guanine with its neighbor (Figure 4e), hydrogen bonding in the +1 G:C pair (Table S5), the post-insertion site width (Table S7), protein-dNTP interactions (Table S8), the Pα-O3′ distance and in-line attack angle (Figure S9 and Table S10), and the distance between the two Mg2+ ions and their octahedral coordination (Table S11). These well-preserved properties are necessary but not sufficient for efficient chemical catalysis, and disturbance to the minor groove contacts and active site pocket suggests impeded enzymatic activity.
syn-[BP]G*:dA/C/G/TTP major groove
The two important hydrogen bonds between the +1 base pair and Arg-615 and Gln-797 on the minor groove side (Figure 3b) are maintained for the four different dNTPs (Table S6). The hydrogen bonds between protein residues and the dNTP have similar occupancies in the unmodified control, except for the hydrogen bond between Lys-706 and O1γ of dNTP in the syn-[BP]G*:dGTP case (occupancy 60% lower than unmodified) (Table S8). The four amino acids comprising the active site wall are tightly wrapped around the nascent base pair, as in the unmodified control. The width and height of the active site are nearly normal (increased by less than 1 Å) (Table S9). The Pα-O3′ distance, in-line attack angle, distance between the two Mg2+ ions and their coordination are normal in all systems (Figure S9, Tables S10 and S11).
The syn-[BP]G* adduct has one hydrogen bond with dATP (occupancy 73%), one hydrogen bond with dCTP (occupancy 96%), and two hydrogen bonds with dGTP (occupancies 99% and 73%, respectively), all formed during MD equilibration. The syn-[BP]G*:dTTP initial model contains no hydrogen bond between the adducted dG* and dTTP and none is formed during MD (Figure S8c–f and Table S5). The three hydrogen bonds in the +1 G:C Watson-Crick pair are maintained in all systems except in the case of syn-[BP]G*:dCTP. In this case, the occupancy of the hydrogen bond between the amino group of G and O2 of C is decreased by ∼30% as compared to the unmodified control (Table S5), because the C plane in dCTP is almost perpendicular to that of this primer terminal C (Figure 4g). The single hydrogen bond between syn-[BP]G* and dCTP is directly opposite the glycosidic bond axis and the active site holds the base pair loosely, allowing relatively free rotation of the C about the χ torsion angle. The syn adducted dG* stacks poorly with its neighboring base on the 3′ side in the four major groove adduct systems (Figure 4f–i).
Step 3. [BP]G* at the post-insertion site of open BF: anti-[BP]G* blocks the nucleotide binding pocket and minor groove contacts are more disrupted by anti- than syn-[BP]G*
anti-[BP]G*:dC minor groove
We simulated three minor groove adduct models at this step, two computer-generated models (anti-1-[BP]G*:dC and anti-2-[BP]G*:dC), and one crystal structure (PDB ID: 1XC9) (44) (anti-3-[BP]G*:dC) (see MATERIALS AND METHODS). The BP orientation in the anti-1-[BP]G*:dC initial model is similar to that in the crystal structure (the anti-3-[BP]G*:dC initial model); the BP in both structures overlaps with the nucleotide binding pocket. However, in the anti-2-[BP]G*:dC initial model, the BP is oriented differently, pointing toward the single stranded region of the template and away from the nucleotide binding pocket (Figure 6a). Following MD, the BP orientation in the anti-1-[BP]G*:dC and anti-3-[BP]G*:dC systems remains essentially unchanged, with the nucleotide binding pocket still blocked by the BP. In the anti-2-[BP]G*:dC system, the BP reorients so that it is closer to the position in the anti-1-[BP]G*:dC and anti-3-[BP]G*:dC systems, where it would collide with the incoming dNTP if BF were to close (Figure 6b). However, the amino acid residues, Ile-657, Glu-658 and Val-713 constitute a barrier preventing convergence of the BP orientation in this anti-2-[BP]G*:dC system with the other two systems (Figure 6c). The RMSDs of the active site regions in the three systems, compared to the crystal structure, are 2.9 Å, 2.5 Å and 1.5 Å, respectively. Minor groove contacts at the post-insertion site are blocked in all three systems with the BP on the minor groove side (Table S6). However, the BP does not affect the width of the pre-insertion and post-insertion sites (Tables S4 and S7), or the hydrogen bonds in the +1 and +2 base pairs (Figure S8h-j and Table S5). The base stacking interactions in the DNA duplex are preserved due to the anti conformation of the modified dG* (Figure 4k–m). The stacking interaction between the Tyr-714 and the modified guanine in the post-insertion site, absent in the original crystal structure (44), remains absent in the anti-3-[BP]G*:dC system, based on the crystal (Figure 4m); it is lost in the anti-1-[BP]G*:dC system (Figure 4k) following dynamics, but remains present in the anti-2-[BP]G*:dC system (Figure 4l). Overall, the three simulated models generally reproduce structural properties in the crystal, but with anti-2-[BP]G*:dC displaying a shifted minor groove BP orientation.
syn-[BP]G*:dA/C/G/T major groove
The BP on the major groove side is too far from the nucleotide binding pocket to block it. However, the syn modified dG* stacks poorly with the Tyr-714 and the base on the 3′-side (Figure 4n–q); also, it cannot hydrogen bond with the Gln-797 (Table S6), since N3 of the guanine is now on the major groove side. The syn-[BP]G* forms one hydrogen bond with dA (occupancy 71%), one hydrogen bond with dC (occupancy 24%) and two hydrogen bonds with dG (occupancies 32% and 77%, respectively) during MD equilibration, but there is no hydrogen bond between syn-[BP]G* and dT (Figure S8k–n and Table S5). The BP does not alter the minor groove contact between the primer terminal base and the Arg-615 (Table S6), the hydrogen bonds in the +2 G:C pair (Table S5), or the pre- and post-insertion site widths (Tables S4 and S7) in the four systems.
Step 4. [BP]G* at the post-insertion site of closed BF: anti-[BP]G* perturbs the hydrogen bond geometry and minor groove contacts more severely than syn-[BP]G*
anti-[BP]G*:dC minor groove
To avoid steric clashes between the benzylic ring of the BP and the Watson-Crick edge of the modified guanine, this edge was 5′-tilted by the ring. Also, the Watson-Crick edge of the partner C was 3′-tilted via a hydrogen bond between O2 of the C and O8 of the BP benzylic ring (Figure 4s). Consequently, Watson-Crick pairing between the bases is impaired, with two hydrogen bonds essentially broken and the third significantly weakened (Figure S8p and Table S5). In addition, the stacking interactions between the modified guanine and its neighboring bases as well as those between the partner C and its neighboring bases are diminished, and the post-insertion site width is significantly enlarged (by >1 Å) (Table S7). The BP blocks the minor groove contacts not only between the modified dG* and Gln-797, but also between the partner C of [BP]G* and Arg-615 (Table S6). The hydrogen bonds stabilizing the dNTP binding have similar occupancies in the modified system as in the unmodified case, except for the bond between Glu-658 and O3′ of dNTP, which is lost (Table S8). The anti-[BP]G* in the post-insertion site does not affect the active site width and height (Table S9), the positioning of the active site wall, or the hydrogen bonds in the neighboring 0 and +2 base pairs (Table S5). The Pα-O3’ distance, in-line attack angle, distance between the two Mg2+ ions and their coordination are normal in this system (Figure S9, Tables S10 and S11).
syn-[BP]G*:dA/C/G/T major groove
The syn-[BP]G* forms, during MD equilibration, one hydrogen bond with dA (occupancy 88%), one hydrogen bond with dC (occupancy 74%), two hydrogen bonds with dG (occupancies 84% and 100%, respectively) and one hydrogen bond with dT (occupancy 88%) (Figure S8q–t and Table S5). In the syn-[BP]G*:dA/dC/dT systems, the base of the modified dG* is tilted to avoid steric clashes between its own phosphate group and the benzylic ring of the BP moiety (Figure 4t, u and w). As a result, the adducted guanine stacks poorly with both its 5′- and 3′- neighboring bases, and the height of the active site on the template side is significantly increased (Figure 5d and Table S9). However, the two hydrogen bonds between [BP]G* and dG hold the bases co-planar in the syn-[BP]G*:dG system, so that the modified guanine is not tilted. The phosphate of the modified dG* is slightly pushed away from the primer strand to make room for the [BP]G* adduct (Figure 4v). As a result, the adducted guanine partially stacks with its neighbors on both 5′- and 3′-sides, although less than in the anti dG unmodified control, and the active site height on the template side is normal (Table S9).
The active site height on the primer side is normal in the syn-[BP]G*:dA/dG/dT systems. However, it is significantly increased in the syn-[BP]G*:dC system (Table S9), because of an unstable orientation of the plane of the C opposite [BP]G* throughout the MD simulation (Figure S10). At times, the C rotates around the glycosidic bond by > 30° so that the C6-containing major groove-side edge rolls toward the 5′-end of the primer (Figure 5d). Thus, the height on the primer side, namely the distance between C6 of dC and Cα of Lys-706, is significantly increased.
The N3 of the syn modified dG*, on the major groove side, cannot hydrogen bond with Gln-797 regardless of the partner base (Table S6). The width of the post-insertion site in the syn-[BP]G*:dA and syn-[BP]G*:dC systems is similar to that in the control, but is significantly increased in the syn-[BP]G*:dG systems and decreased in the syn-[BP]G*:dT system (Table S7). The BP residue does not affect the hydrogen bonds in the 0 and +2 base pairs (Table S5), the active site width (Table S9) or the interactions between amino acids and dNTP (Table S8) in the four systems. The Pα-O3′ distance, in-line attack angle, distance between the two Mg2+ ions and their coordination, are normal in all systems (Figure S9, Tables S10 and S11).
DISCUSSION
In the present study, we have carried out a computational study, based on key experimental observations, to investigate how the bulky [BP]G* adduct is processed during DNA replication by the polymerase BF. Primer extension and single nucleotide insertion assays reveal that the [BP]G* lesion allows slow incorporation of purine dNTPs when these are paired with the modified guanine on the template strand, but that further extension is strongly inhibited. To account for these observations, we have constructed two sets of initial models with BP-modified dG* in the normal anti and the less prevalent syn conformations. The syn conformation has been observed experimentally by solution NMR methods in the case of an unpaired guanine residue of the (+)-trans-[BP]G* adduct positioned at a template-primer junction (10,64), revealing that the syn conformation is feasible in such structures. We have investigated four consecutive steps in DNA replication as [BP]G* transits through the BF active site region, with polymerase open and closed, in binary and ternary complexes (Figure 3a).
MD simulations were carried out and important structural properties were analyzed in detail and compared to the unmodified control. The MD results structurally interpret our experimental data and are consistent with the available crystal structure (44). We hypothesize that increased distortions at the polymerase active site are a hallmark of reduced polymerase fidelity and efficiency. The distortions for each property were scored, and a composite score was derived for each system (Table 1). A numerically more negative score indicates a more distorted structure. The properties evaluated and the scoring criteria employed were selected based on our current best understanding of proper active site geometry. A recent crystal structure reveals a well-organized active site (59), whose features closely resemble those in our unmodified models, suggesting that our criteria for evaluating distortions are reasonable. However, the scoring function is an evolving one which is being improved as our knowledge base advances. At this stage, the composite scores reflect the degrees of distortion, but the relationship between the scores and the biological outcome is very likely non-linear.
Table 1.
Composite evaluation of structural distortions.
| Step | 1 | 2 | 3 | 4 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BF | Open | Closed | Open | Closed | |||||||||||||||
| [BP]G* site | Pre-insertion | Insertion | Post-insertion | Post-insertion | |||||||||||||||
| dG* Conformation | anti | syn | anti | syn | anti-1 | anti-2 | anti-3 | syn | anti | syn | |||||||||
| [BP]G* Partner | C | A | C | G | T | C | C | C | A | C | G | T | C | A | C | G | T | ||
| Pre-insertion site pocket | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||
| Base pair hydrogen bonding geometry | 0 | 0 | −1 | −2 | −3 | −1 | −3 | 0 | 0 | 0 | −2 | −3 | −2 | −3 | −3 | −2 | −2 | −1 | −2 |
| Stacking interaction | 0 | 0 | 0 | −1 | −2 | −1 | −1 | −1 | 0 | −1 | −2 | −2 | −2 | −2 | −2 | −2 | −2 | −1 | −2 |
| Minor groove contact | 0 | 0 | −2 | 0 | 0 | 0 | 0 | −4 | −4 | −4 | −2 | −2 | −2 | −2 | −4 | −2 | −2 | −2 | −2 |
| Post-insertion site width | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | −1 | −1 |
| Nucleotide binding pocket | 0 | 0 | −5 | −5 | −5 | 0 | 0 | 0 | 0 | ||||||||||
| Protein-dNTP interactions | 0 | 0 | 0 | −1 | 0 | −1 | 0 | 0 | 0 | 0 | |||||||||
| Active site pocket | −3 | 0 | 0 | 0 | 0 | 0 | −1 | −2 | 0 | −1 | |||||||||
| Pα-O3′ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||
| Attack angle | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||
| Mg2+ ions coordination | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||
| Mg2+ ions distance | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||
| Composite score | −1 | 0 | −6 | −3 | −5 | −3 | −4 | −10 | −9 | −10 | −6 | −7 | −6 | −7 | −11 | −7 | −8 | −5 | −8 |
The unmodified controls were assigned a base score of 0 for each structural property. Scoring criteria are given on Page 18 in the Supplementary Data. The more negative the score, the greater the distortion.
The anti-[BP]G* adduct structures are consistent with total blockage while the syn-[BP]G* adducts should allow some bypass
The anti-[BP]G* adduct causes severe distortions to the BF-DNA complex structures at any of the four steps investigated. At Step 1 (Figure 3a), anti-[BP]G* cannot be accommodated in the pre-insertion site without significantly enlarging this site. This is consistent with the partially resolved crystal structure with the BP-modified guanine as the base next to be replicated: the adduct itself is disordered and not positioned in the pre-insertion site (44); the BP modified dG* adduct most likely adopts the preferred anti conformation, and therefore cannot fit into the normal-sized pre-insertion site. At Step 2, the adduct is situated on the minor groove side of the nascent base pair. It disturbs the minor groove scanning track and the active site pocket. The three simulations with anti-[BP]G* at Step 3 reveal structures that have impaired minor groove contacts and impeded nucleotide binding characteristics, quite like the published crystal structure (44) (which provided one of the three initial models). At Step 4, the BP blocks the minor groove interactions and disrupts the primer terminal base pair. These distortions suggest that the anti-[BP]dG* should be totally blocking at any of the four steps.
The syn-[BP]G* disturbs the BF-DNA structure less severely than the anti adduct. At Step 1, the syn-[BP]G* appears to have negligible effect on the BF-DNA structure; the modified dG* residue can be accommodated in the pre-insertion site whose shape is preserved. Starting from Step 2, the BP ring system is positioned on the open major groove side of the DNA duplex where it does not have a significant effect on the minor groove contacts between the DNA and the polymerase. Nor does the [BP]G* adduct in the syn conformation impair the active site pocket at Step 2. Therefore, the syn conformation should allow some bypass. However, the syn adduct distorts other structural properties such as the hydrogen bonding geometry of the [BP]G*-containing base pair and the base stacking interactions, suggesting that polymerase stalling would be prevalent at Steps 2, 3 and 4 (Figure 3a). In a cellular environment, the polymerase would probably stall for a sufficiently long time to allow dissociation. Therefore, a switch to one or more bypass polymerases would likely occur, allowing for possibly error-prone translesion bypass (25–31). The major groove/syn orientation of the [BP]G* adduct for bypass in the Pol I family may be a general theme for this replicative polymerase family; the large open pocket can readily house the bulky adduct in both T7 (50–52) and BF.
The experimental running-start primer extension results (Figure 2a and b) reveal that BF catalyzes the reaction in a quick and efficient manner up to the position right before the lesion site, but further dNTP incorporation is significantly slowed. This is consistent with our modeling studies which suggest that the syn conformation allows for some translesion bypass. Although the syn-[BP]G* adduct at the pre-insertion site (Step 1) does not seem to affect polymerase structure (Table 1), a slowing of the reaction kinetics may be due to the less prevalent occurrence of the [BP]G* in the syn conformation. We note that a conformational rearrangement between syn and anti might take place within the enzyme during a given replication cycle, as observed in BF with another carcinogen-guanine adduct. In a crystal of N-(2′-deoxyguanosin-8yl)-aminofluorene ([AF]G*) in BF, the aminofluorene (AF)-modified dG* was syn at the pre-insertion site of open BF; after one round of replication within the crystal, the modified dG* moved to the post-insertion site of open BF where it was anti (65). In general, the opportunity for a syn-anti or some other rearrangement within the polymerase can explain the possibility for slow bypass past a particularly situated lesion, even in the face of significant distortion. In the case of mismatch structures in BF, a templating base which failed to be housed in the pre-insertion site could still be slowly extended (66), indicating a necessary rearrangement to permit further nucleotide incorporation.
Purine over pyrimidine insertion is favored opposite the lesion
Our MD results show that the purine partners causes less distortion opposite the syn-[BP]G* than pyrimidines (Table 1). The purines are more comfortably situated opposite the syn modified guanine, whose shape in the active site is pyrimidine-like (Figure 7). The pseudo pyrimidine–pyrimidine pair fits poorly in the active site, with the greatest disturbances involving hydrogen bonding/stacking interactions (Table 1). This is consistent with the experimentally observed favored insertion of purines opposite the [BP]G* adduct in the present study (Figure 2c). In contrast to our findings that purines are preferentially inserted opposite G* in the CG*G sequence context, Hsu et al. (44) reported that dCTP is preferentially inserted opposite G* in a CG*C sequence context. In general, we find that base sequence effects can have a significant influence on dNTP insertion selectivities, efficiencies and translesion bypass. The DNA sequences used in our work and that of Hsu et al. are different, which can account for the different insertion selectivities. These are issues that are at the forefront of our interests and are presently under investigation in our laboratories.
Figure 7.
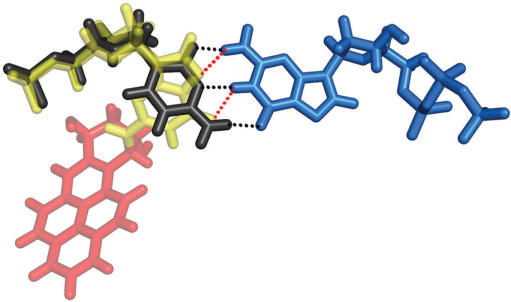
syn-[BP]G*:dGTP base pair overlayed with a normal anti-C:dGTP Watson-Crick base pair. Color code: yellow, modified dG*; red, BP; light blue, dGTP; black, anti-C. Black dashed lines: hydrogen bond between C and dGTP; red dashed lines: hydrogen bond between syn-[BP]G* and dGTP.
Extension beyond the [BP]G* is more impeded than insertion opposite this adduct
Because of differing structural properties, different scoring criteria had to be employed for the ternary and binary complexes. Hence, comparison can only be made between Steps 1 and 3 (binary complexes), and between Steps 2 and 4 (ternary complexes). Table 1 shows the distortion scores. A comparison between Step 1 (pre-insertion step) and Step 3 (pre-extension step) (Figure 3a) indicates that the structure is less perturbed at the pre-insertion step than at the pre-extension step, suggesting that it would be easier to proceed to the insertion than the extension step. In addition, a comparison of Step 2 (insertion step) and Step 4 (extension step) (Figure 3a) shows that the structures are more distorted during extension than during insertion for all partners opposite the adduct. The syn-[BP]G* generally interferes with stacking interactions and minor groove contacts more severely at the pre-extension/extension step than at the pre-insertion/insertion step. These conclusions are consistent with the experimental results since nucleotide incorporation opposite [BP]G* is indeed more probable than extension beyond this adduct. This has been generally found to be the case for the [BP]G* adduct in Pol I type polymerases (14,67–69).
Most of the intermediates investigated by modeling have not been characterized experimentally with the [BP]G* adduct; hence modeling provides the only approach to gaining structure-function understanding of the experimental results. An added advantage of the MD is the dynamic ensemble of structures provided, which cannot be realized experimentally. We note the on-going frontier problems in MD simulations, which are limited by the problem of sampling sufficiency, together with issues relating to force field quality (70–72). It is encouraging that our simulations involving a structure already characterized crystallographically essentially converged with the crystal structure, from a modeled initial position obtained by the same criteria as were the unknown structures.
CONCLUSIONS
Our experimental data show that BF is primarily blocked by the [BP]G* adduct, although a low extent of lesion bypass with full primer extension is also detected; however, nucleotide incorporation opposite [BP]G* is observed with significantly greater efficiency, with purines favored over pyrimidines. Further extension beyond the lesion site is more strongly inhibited than insertion. The results of our structural modeling studies elucidate structural bases for these experimental observations. Our MD simulations show that when BP modified dG* adopts the normal anti conformation at any of the four steps of primer extension investigated, the adduct causes severe distortion to the polymerase structure and thus should totally block nucleotide incorporation/extension. On the other hand, the less prevalent syn conformation of the adducted dG* produces a less distorted polymerase structure, thus explaining how some translesion bypass might occur; however, certain structural disturbances nevertheless suggest that the polymerase would be impeded. When the adducted dG* is in the insertion site of the closed BF and adopts the syn conformation, the purine partners are better accommodated opposite the adduct than pyrimidines, accounting for the preferential insertion of purines; the resulting G:A and G:G mismatches cause G → T and G → C transversion mutations. Furthermore, our MD results show that the adduct disturbs the correct alignment of the active site region more severely at the extension/pre-extension step than at the insertion/pre-insertion step, explaining why extension is more difficult than the successful incorporation of a dNTP opposite the lesion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
This work was supported by NIH Grant 5RO1 CA 28038 (S.B.), 2RO1 CA 099194 (N.E.G), and 5PO1 CA 92584 (L.S.B). Funding to pay the Open Access publication charges for this article was provided by NIH Grant 5RO1 CA 28038. We thank Prof. Robert Shapiro,Chemistry Department, New York University, for helpful discussions. Computational resources supported by the NSF Partnerships for Advanced Computational Infrastructure are gratefully acknowledged.
Conflict of interest statement. None declared.
REFERENCES
- 1.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Fontcuberta M, Arques JF, Martinez M, Suarez A, Villalbi JR, Centrich F, Serrahima E, Duran J, Casas C. Polycyclic aromatic hydrocarbons in food samples collected in Barcelona, Spain. J. of Food Prot. 2006;69:2024–2028. doi: 10.4315/0362-028x-69.8.2024. [DOI] [PubMed] [Google Scholar]
- 3.Tuntawiroon J, Mahidol C, Navasumrit P, Autrup H, Ruchirawat M. Increased health risk in Bangkok children exposed to polycyclic aromatic hydrocarbons from traffic-related sources. Carcinogenesis. 2006;28:816–822. doi: 10.1093/carcin/bgl175. [DOI] [PubMed] [Google Scholar]
- 4.Burczynski ME, Harvey RG, Penning TM. Expression and characterization of four recombinant human dihydrodiol dehydrogenase isoforms: oxidation of trans-7, 8-dihydroxy-7,8-dihydrobenzo[a]pyrene to the activated o-quinone metabolite benzo[a]pyrene-7,8-dione. Biochemistry. 1998;37:6781–6790. doi: 10.1021/bi972725u. [DOI] [PubMed] [Google Scholar]
- 5.Cavalieri EL, Rogan EG. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 6.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 7.Buening MK, Wislocki PG, Levin W, Yagi H, Thakker DR, Akagi H, Koreeda M, Jerina DM, Conney AH. Tumorigenicity of the optical enantiomers of the diastereomeric benzo[a]pyrene 7,8-diol-9,10-epoxides in newborn mice: exceptional activity of (+)-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Proc. Natl Acad. Sci. USA. 1978;75:5358–5361. doi: 10.1073/pnas.75.11.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem. Res. Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 9.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 10.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 11.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo[a]pyrene diol epoxides. Nature. 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 12.Alekseyev YO, Romano LJ. In vitro replication of primer-templates containing benzo[a]pyrene adducts by exonuclease-deficient Escherichia coli DNA polymerase I (Klenow fragment): effect of sequence context on lesion bypass. Biochemistry. 2000;39:10431–10438. doi: 10.1021/bi000552u. [DOI] [PubMed] [Google Scholar]
- 13.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem. Res. Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 14.Zhuang P, Kolbanovskiy A, Amin S, Geacintov NE. Base sequence dependence of in vitro translesional DNA replication past a bulky lesion catalyzed by the exo- Klenow fragment of Pol I. Biochemistry. 2001;40:6660–6669. doi: 10.1021/bi010005o. [DOI] [PubMed] [Google Scholar]
- 15.Seo KY, Nagalingam A, Tiffany M, Loechler EL. Mutagenesis studies with four stereoisomeric N2-dG benzo[a]pyrene adducts in the identical 5′-CGC sequence used in NMR studies: G->T mutations dominate in each case. Mutagenesis. 2005;20:441–448. doi: 10.1093/mutage/gei061. [DOI] [PubMed] [Google Scholar]
- 16.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst) 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Shukla R, Jelinsky S, Liu T, Geacintov NE, Loechler EL. How stereochemistry affects mutagenesis by N2-deoxyguanosine adducts of 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene: configuration of the adduct bond is more important than those of the hydroxyl groups. Biochemistry. 1997;36:13263–13269. doi: 10.1021/bi971195z. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez H, Loechler EL. Are base substitution and frameshift mutagenesis pathways interrelated? An analysis based upon studies of the frequencies and specificities of mutations induced by the (+)-anti diol epoxide of benzo[a]pyrene. Mutat. Res. 1995;326:29–37. doi: 10.1016/0027-5107(95)00149-d. [DOI] [PubMed] [Google Scholar]
- 19.Moriya M, Spiegel S, Fernandes A, Amin S, Liu T, Geacintov N, Grollman AP. Fidelity of translesional synthesis past benzo[a]pyrene diol epoxide-2′-deoxyguanosine DNA adducts: marked effects of host cell, sequence context, and chirality. Biochemistry. 1996;35:16646–16651. doi: 10.1021/bi9608875. [DOI] [PubMed] [Google Scholar]
- 20.Jelinsky SA, Liu T, Geacintov NE, Loechler EL. The major, N2-Gua adduct of the (+)-anti-benzo[a]pyrene diol epoxide is capable of inducing G–>A and G–>C, in addition to G–>T, mutations. Biochemistry. 1995;34:13545–13553. doi: 10.1021/bi00041a034. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, Sayer JM, Kroth H, Ponten I, O'Donnell M, Woodgate R, Jerina DM, Goodman MF. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J. Biol. Chem. 2002;277:5265–5274. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (-)-cis-anti-benzo[a]pyrene diol epoxide-N2-2'-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 23.Zhao B, Wang J, Geacintov NE, Wang Z. Poleta, Polzeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (-)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 2006;34:417–425. doi: 10.1093/nar/gkj446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 25.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 26.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr. Opin. Genet. Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 31.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 32.Lenne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP. The processing of a Benzo[a]pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol. Microbiol. 2000;38:299–307. doi: 10.1046/j.1365-2958.2000.02116.x. [DOI] [PubMed] [Google Scholar]
- 33.Seo KY, Jelinsky SA, Loechler EL. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 34.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 35.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci. USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beard WA, Wilson SH. Structural insights into DNA polymerase β fidelity: hold tight if you want it right. Chem. Biol. 1998;5:13. doi: 10.1016/s1074-5521(98)90081-3. [DOI] [PubMed] [Google Scholar]
- 37.Frieden M, Pedroso E, Kool ET. Tightening the belt on polymerases: evaluating the physical constraints on enzyme substrate size. Angew. Chem. Int. Ed. Engl. 1999;38:3654–3657. [PubMed] [Google Scholar]
- 38.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 39.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 40.Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 43.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 44.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 45.Beard WA, Bebenek K, Darden TA, Li L, Prasad R, Kunkel TA, Wilson SH. Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase. Molecular nature of polymerase-nucleic acid interactions. J. Biol. Chem. 1998;273:30435–30442. doi: 10.1074/jbc.273.46.30435. [DOI] [PubMed] [Google Scholar]
- 46.Bebenek K, Beard WA, Darden TA, Li L, Prasad R, Luton BA, Gorenstein DG, Wilson SH, Kunkel TA. A minor groove binding track in reverse transcriptase. Nat. Struct. Biol. 1997;4:194–197. doi: 10.1038/nsb0397-194. [DOI] [PubMed] [Google Scholar]
- 47.Latham GJ, Forgacs E, Beard WA, Prasad R, Bebenek K, Kunkel TA, Wilson SH, Lloyd RS. Vertical-scanning mutagenesis of a critical tryptophan in the ‘minor groove binding track’ of HIV-1 reverse transcriptase. Major groove DNA adducts identify specific protein interactions in the minor groove. J. Biol. Chem. 2000;275:15025–15033. doi: 10.1074/jbc.M000279200. [DOI] [PubMed] [Google Scholar]
- 48.Morales J, Kool ET. Minor groove interactions between polymerase and DNA: more essential to replication than Watson-Crick hydrogen bonds. J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 50.Perlow RA, Broyde S. Evading the proofreading machinery of a replicative DNA polymerase: induction of a mutation by an environmental carcinogen. J. Mol. Biol. 2001;309:519–536. doi: 10.1006/jmbi.2001.4674. [DOI] [PubMed] [Google Scholar]
- 51.Perlow RA, Broyde S. Toward understanding the mutagenicity of an environmental carcinogen: structural insights into nucleotide incorporation preferences. J. Mol. Biol. 2002;322:291–309. doi: 10.1016/s0022-2836(02)00751-9. [DOI] [PubMed] [Google Scholar]
- 52.Perlow RA, Broyde S. Extending the understanding of mutagenicity: structural insights into primer-extension past a benzo[a]pyrene diol epoxide-DNA adduct. J. Mol. Biol. 2003;327:797–818. doi: 10.1016/s0022-2836(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 53.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie XM, Geacintov NE, Broyde S. Stereochemical origin of opposite orientations in DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides with different tumorigenic potentials. Biochemistry. 1999;38:2956–2968. doi: 10.1021/bi9825605. [DOI] [PubMed] [Google Scholar]
- 55.Case DA, Pearlman DA, Caldwell JW, Cheatham TEI, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, et al. AMBER 7. San Francisco, CA: University of California; 2002. [Google Scholar]
- 56.Cornell W, Cieplak P, Bayly C, Gould I, Merz K, Ferguson D, Spellmeyer D, Fox T, Caldwell J, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 57.Cheatham T.E. 3rd, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 58.Delano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 59.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 61.Jordan F. Lennard-Jones potential calculations of the barrier to rotation around the glycosidic C-N linkage in selected purine nucleosides and nucleotides. A direct comparison of the results of 6-12 potential calculations with results of semiempirical molecular orbital studies. J. Theor. Biol. 1973;41:375–395. doi: 10.1016/0022-5193(73)90125-2. [DOI] [PubMed] [Google Scholar]
- 62.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 63.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Cosman M, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignments of (+)- and (-)-trans-anti-benzo[a]pyrene-dG adducts positioned at a DNA template-primer junction. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 65.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 66.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 67.Hruszkewycz AM, Canella KA, Peltonen K, Kotrappa L, Dipple A. DNA polymerase action on benzo[a]pyrene-DNA adducts. Carcinogenesis. 1992;13:2347–2352. doi: 10.1093/carcin/13.12.2347. [DOI] [PubMed] [Google Scholar]
- 68.Lipinski LJ, Ross HL, Zajc B, Sayer JM, Jerina DM, Dipple A. Effect of single benzo[a]pyrene diol epoxide-deoxyguanosine adducts on the action of DNA polymerases in vitro. Int. J. Oncol. 1998;13:269–273. doi: 10.3892/ijo.13.2.269. [DOI] [PubMed] [Google Scholar]
- 69.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 70.Hansson T, Oostenbrink C, van Gunsteren W. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2002;12:190–196. doi: 10.1016/s0959-440x(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 71.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl Acad. Sci. USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.