Abstract
The specific aminoacylation of tRNA by tyrosyl-tRNA synthetases (TyrRSs) relies on the identity determinants in the cognate tRNATyrs. We have determined the crystal structure of Saccharomyces cerevisiae TyrRS (SceTyrRS) complexed with a Tyr-AMP analog and the native tRNATyr(GΨA). Structural information for TyrRS–tRNATyr complexes is now full-line for three kingdoms. Because the archaeal/eukaryotic TyrRSs–tRNATyrs pairs do not cross-react with their bacterial counterparts, the recognition modes of the identity determinants by the archaeal/eukaryotic TyrRSs were expected to be similar to each other but different from that by the bacterial TyrRSs. Interestingly, however, the tRNATyr recognition modes of SceTyrRS have both similarities and differences compared with those in the archaeal TyrRS: the recognition of the C1-G72 base pair by SceTyrRS is similar to that by the archaeal TyrRS, whereas the recognition of the A73 by SceTyrRS is different from that by the archaeal TyrRS but similar to that by the bacterial TyrRS. Thus, the lack of cross-reactivity between archaeal/eukaryotic and bacterial TyrRS-tRNATyr pairs most probably lies in the different sequence of the last base pair of the acceptor stem (C1-G72 vs G1-C72) of tRNATyr. On the other hand, the recognition mode of Tyr-AMP is conserved among the TyrRSs from the three kingdoms.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) play a central role in the assembly of amino acids into polypeptide chains. They catalyze the specific esterification of a given amino acid to its corresponding tRNA through a two-step reaction (1). In the first step, the specific amino acid and ATP substrates are recognized and then are converted into a reactive aminoacyl-adenylate (aa-AMP) intermediate in the presence of magnesium ions. In the next step, the amino acid moiety from the aa-AMP is transferred to the 3′-CCA terminus of the cognate tRNA. This enzymatic function is crucial for the fidelity of protein synthesis, in which the genetic code is translated to the amino acid sequence. The primary sequence analyses as well as the tertiary structure determinations allowed the partition of the 20 aaRSs into two exclusive classes, I and II, each consisting of 10 enzymes (2). Each class I enzyme has a Rossmann-fold domain as the catalytic domain. In addition, two consensus motifs, HIGH and KMSKS, are conserved among the class I enzymes. The class I enzymes are further divided into three sub-classes: Ia, Ib and Ic (3,4). The class Ic enzymes are aaRSs for tyrosine and tryptophan and are unusual in that they act as dimers, while the other class I (Ia and Ib) enzymes act as monomers.
Tyrosyl-tRNA synthetase (TyrRS) is the first aaRS to have its crystal structure solved (5). The specific aminoacylation of tRNA by TysRS relies on the identity determinants (the anticodon bases, the C1-G72 base pair, and the discriminator base A73) in the cognate tRNATyr (6–8). To date, a number of crystal structures of the TyrRSs have been solved: they are from four bacteria, Bacillus stearothermophilus [BstTyrRS, (9)], Staphylococcus aureus [SauTyrRS, (10)], Thermus thermophilus [TthTyrRS, (11)] and Escherichia coli [EcoTyrRS, (12)]; from eukarya Homo sapiens [HsaTyrRS, (13)]; and from four archaea, Methanococcus jannaschii [MjaTyrRS, (14,15)], Archaeglobus fulgidus, Pyrococcus horikoshi and Aeropyrum pernix [AfuTyrRS, PhoTyrRS, and ApeTyrRS, respectively, (16)]. Because the archaeal/eukaryotic TyrRSs–tRNATyrs pairs do not cross-react with their bacterial counterparts (8), the recognition modes of the identity determinants by the archaeal and eukaryotic TyrRSs were expected to be similar to each other but different from that by the bacterial TyrRSs. Such orthogonality is used for the incorporation of unnatural amino acids into proteins with engineered pairs of TyrRSs and tRNATyrs (17). In such situations, structural information on archaeal, eukaryotic and bacterial TyrRSs complexed with their cognate tRNATyrs has long been awaited. In a half-decade, crystal structure analyses of bacterial (Thermus thermophilus) (11) and archaeal (Methanococcus jannaschii) (14) TyrRSs complexed with their cognate tRNATyrs have been reported. Although previous experiments showed that TyrRS could bind only one tRNA per dimer (18) in solution, the crystal structure analyses (11,14) have shown that two tRNATyrs are bound to each dimer in a symmetrical fashion in the crystal. A plausible explanation for this discrepancy (asymmetry in solution vs symmetry in crystal) has been described by Yaremchuk et al. (11). A structural comparison revealed the structural basis for orthogonal specificities of archaeal and bacterial TyrRSs (14). On the other hand, no structures are available for eukaryotic TyrRSs complexed with their cognate tRNATyrs. To understand the molecular basis for the recognition of their cognate tRNATyrs by eukaryotic TyrRSs, we initiated the structure analysis of TyrRS from Saccharomyces cerevisiae (SceTyrRS), the model organism for lower eukaryotes.
Here we present the crystal structure at 2.4-Å resolution of the ternary complex of SceTyrRS complexed with a Tyr-AMP analog and the native tRNATyr(GΨA). The present structure of SceTyrRS complexed with the cognate tRNATyr and the previously reported structures of bacterial and archaeal TyrRSs (TthTyrRS and MjaTyrRS, respectively) complexed with their cognate tRNATyrs provide a full set of the recognition modes of the identity determinants of tRNATyrs by TyrRSs from three kingdoms.
MATERIALS AND METHODS
Crystallization
Chemicals were purchased from Wako Pure Chemical Co. (Tokyo, Japan). The purification of native modified tRNATyr was performed in a way similar to the method as described (44). A Tyr-AMP analog (O-(adenosine-5′-O-yl) N-(L-tyrosyl)phosphoramidate (Tyr-AMPN), (Figure 1A) was prepared as described (45). We expressed and purified a C-terminally truncated SceTyrRS (hereafter simply SceTyrRS, residues 1–364), which has full TyrRS activity, for the present crystal structure analysis. SceTyrRS was expressed and purified in a way similar to the method described previously for the full-length SceTyrRS (residues 1–394) (34). Crystals of the ternary complex of SceTyrRS were obtained by the hanging-drop vapor diffusion method, as described elsewhere (46). Briefly, a droplet was prepared by mixing an equal volume of a protein solution containing ca. 0.2 mM SceTyrRS, 5 mM Tyr-AMPN (Figure 1A), ca. 0.2 mM tRNATyr, 40 mM KCl in 20 mM Tris buffer at pH 7.5 and a reservoir solution containing 25% (v/v) polyethyleneglycol 400 (PEG400) and 100 mM CaCl2 in 100 mM Tris buffer at pH 7.5. The crystals belong to tetragonal space group P41212 with cell dimensions of a = b = 63.85 Å and c = 330.3 Å (under the cryogenic conditions described below). Assuming one SceTyrRS subunit and one tRNATyr molecule per asymmetric unit, we obtained a VM value of 2.55 Å3/Da, corresponding to a solvent content of 52%. Since the crystallization conditions of SceTyrRS contained 25% (v/v) PEG400 in reservoir solutions, X-ray data collections could be performed under cryogenic conditions without further addition of a cryo-protectant. Crystals were mounted in nylon loops and flash-cooled in a cold nitrogen gas stream at 100 K just before data collection. Crystals of the ternary complex of full-length TyrRS were obtained in a similar condition as described above and had similar morphology and cell dimensions to those of the truncated TyrRS. However, they diffracted quite poor (ca. 10-Å resolution).
Figure 1.
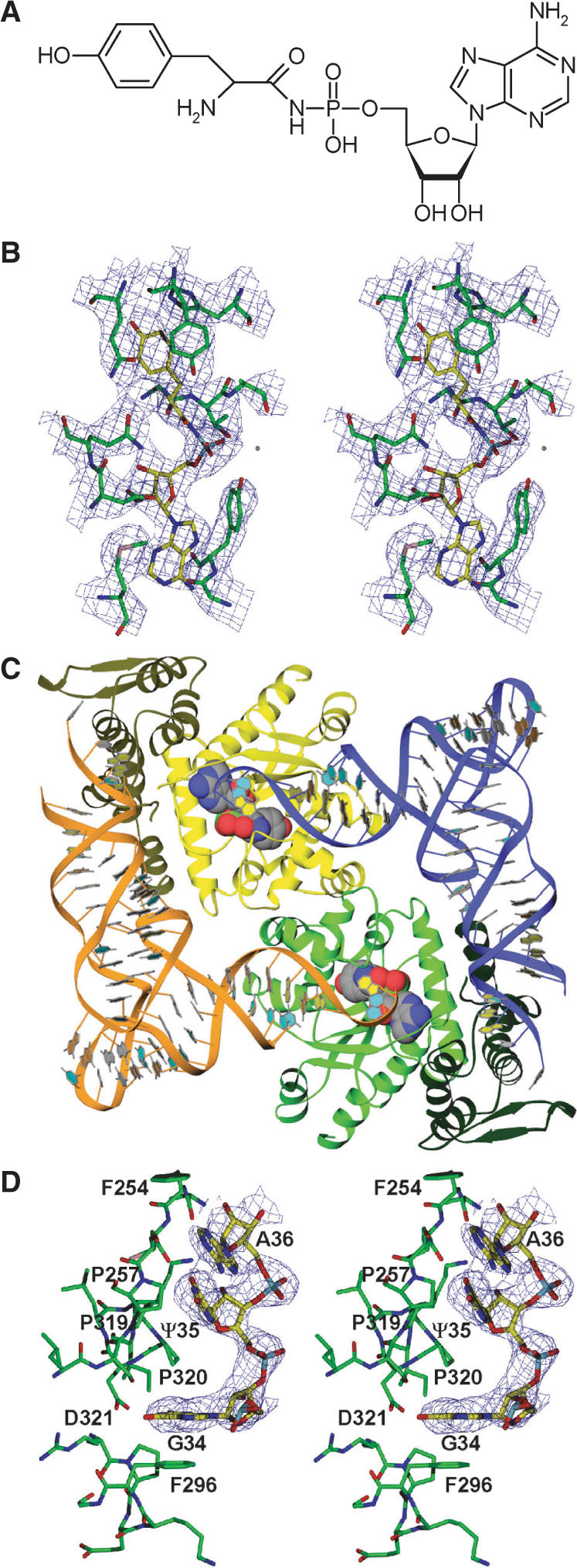
Structure determination of the ternary complex of SceTyrRS. (A) Structure of the Tyr-AMP analog (Tyr-AMPN), having an N-acyl phosphoramidate linkage where the oxygen atom of the mixed anhydride bond (-C-O-P-) of Tyr-AMP was replaced by an amino group (-C-NH-P-), used for the present crystal structure analysis. (B) Stereo diagram of the experimentally phased MAD map at 3.0-Å resolution of the Tyr-AMPN binding site of the ternary complex of SceTyrRS. The contour level is 1.2σ. (C) Ribbon drawing of the dimer of the ternary complex of SceTyrRS. The SceTyrRS (yellow and green) and tRNATyr (blue and orange) molecules are shown as ribbon models. The catalytic and anticodon-binding domains of TyrRS are shown as light and dark colors, respectively. The bound Tyr-AMPN molecules are shown as CPK models. The molecular two-fold axis [coinciding with the (1 1 0) crystallographic two-fold axis] is perpendicular to the plane of the paper. The structural discontinuity in the anticodon-loop of tRNATyr is due to the disordered nucleotides C32–U33. (D) Stereo diagram of the experimentally phased MAD map at 3.0-Å resolution showing the well-ordered anticodon bases (G34-Ψ35-A36) of Sce-tRNATyr. The contour level is 1.0σ.
Data collection
Initially, a native dataset was collected and several attempts were made to solve the structure of SceTyrRS by the molecular replacement techniques. The structures of several TyrRSs complexed with or without cognate tRNATyr and deposited in the Protein Data Bank, having ca. 10–20% sequence identity with SceTyrRS, were used as search models. Secondly, attempts were made to find good heavy-atom derivatives for phasing by the isomorphous replacement techniques. Since both of these attempts failed, we prepared a Se-Met substituted SceTyrRS using LeMaster medium (47) and E. coli B834(DE3) cells for phasing by the multiwavelength anomalous diffraction (MAD) method. The MAD data collection was performed at beamline 38B1, SPring-8. XAFS measurements were carried out around the selenium K absorption edge using an Se-Met SceTyrRS crystal in a cold nitrogen gas stream at 100 K. Subsequently, four datasets were collected from a new single crystal of Se-Met SceTyrRS on and around the selenium K absorption edge at 100 K using an ADSC Quantum-4R CCD detector. All datasets were integrated using the program package DPS (48). Scaling and processing were performed using the CCP4 program suite (49). Thereafter, a high-resolution dataset was collected from a single crystal of native SceTyrRS at beamline 40B2, SPring-8 (λ = 1.00 Å) in a similar way. The data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics for the ternary complex of SceTyrRS
| Data collection statistics | |||||
|---|---|---|---|---|---|
| Data set | Native | Se-MAD (4 wavelengths) | |||
| Peak | Edge | Low-Remote | High-Remote | ||
| X-ray source | SPring-8 BL40B1 | SPring-8 BL38B2 | |||
| Temperature | 100K | 100 K | |||
| Detector | ADSC Q4R | ADSC Q4R | |||
| Resolution (outer shell) (Å) | 2.4 (2.53–2.4) | 3.0 (3.16–3.0) | |||
| Wavelength (Å) | 1.0000 | 0.9798 | 0.9801 | 0.9819 | 0.9727 |
| Unique reflections | 27 897 | 14 733 | 14 738 | 14 737 | 14 741 |
| Multiplicity | 4.1 (4.2) | 19.8 (19.4) | 13.4 (13.1) | 13.4 (13.0) | 13.5 (13.6) |
| Completeness (%) | 99.4 (99.6) | 99.9 (100) | 99.9 (100) | 99.9 (100) | 99.9 (100) |
| I/σ(I) | 11.7 (2.0) | 9.1 (2.5) | 8.9 (2.2) | 8.7 (2.0) | 8.6 (1.9) |
| Rsym (%) | 4.8 (39.6) | 6.8 (29.9) | 7.1 (34.4) | 7.2 (36.4) | 7.3 (38.4) |
| Refinement statistics | |||||
| Resolution range (outer shell) (Å) | 40 – 2.4 (2.46–2.4) | ||||
| No. of reflections: | |||||
| working set test set | 26 411 1403 | ||||
| R-factor | 0.245 (0.347) | ||||
| Free R-factor | 0.289 (0.405) | ||||
| No. of protein atoms [average B-factors (Å2)] | 2699 (33.4) | ||||
| No. of Mg atoms [average B-factors (Å2)] | 1 (42.5) | ||||
| No. of Tyr-AMPN atoms [average B-factors (Å2)] | 35 (48.3) | ||||
| No. of tRNA atoms [average B-factors (Å2)] | 1472 (33.6) | ||||
| No. of water molecules [average B-factors (Å2)] | 57 (55.7) | ||||
| RMSDs: | |||||
| bond lengths (Å) bond angles (deg.) | 0.006 1.124 | ||||
| Ramachandran plot: most favored (%) | 90.2 | ||||
| additional allowed (%) | 9.2 | ||||
| generously allowed (%) | 0.6 | ||||
Structure determination
Initial phase calculation was carried out at 3.0-Å resolution using the program SHARP/autoSHARP (50). Of the nine selenium sites, seven were found. Interpretation of the electron density maps and model-building procedures were carried out on a Linux PC with the aid of the program X-fit as implemented in the program XtalView version 4.0 (51). The obtained model was refined at 2.4-Å resolution with the programs CNS (52) and REFMAC (53). After each refinement calculation, the obtained model was corrected with difference Fourier maps using XtalView. Water molecules were picked by the water-add routine in XtralView. The stereochemistry of the model was verified using the program PROCHECK in the CCP4 program suite. The present model includes residues 8 to 356 of SceTyrRS, one tRNATyr molecule, one Tyr-AMPN molecule, one magnesium ion and 57 water molecules per asymmetric unit. Residues 224–233 of SceTyrRS, the base moieties 16, 20, 31, 40, 41 and 46 of tRNATyr, and the whole nucleotides 17, 20a, 20b, 32 and 33 of tRNATyr, were disordered. The current R-factor is 0.245 (Rfree = 0.289) for the resolution range of 40.0–2.4 Å. The root-mean-square-distances (RMSDs) from ideal values are 0.006 Å for bond lengths and 1.124 for bond angles. The refinement statistics are summarized in Table 1. The atomic coordinates have been deposited in the Protein Data Bank with the entry code 2DLC.
Graphics programs
Figures were produced using both the DINO (http://www.dino3d.org) and POV-Ray (http://www.povray.org) programs (Figure 1B and D) or both the Ribbons (54) and POV-Ray programs (Figures 1C, 2, 4, 5, 6 and 7).
Figure 2.
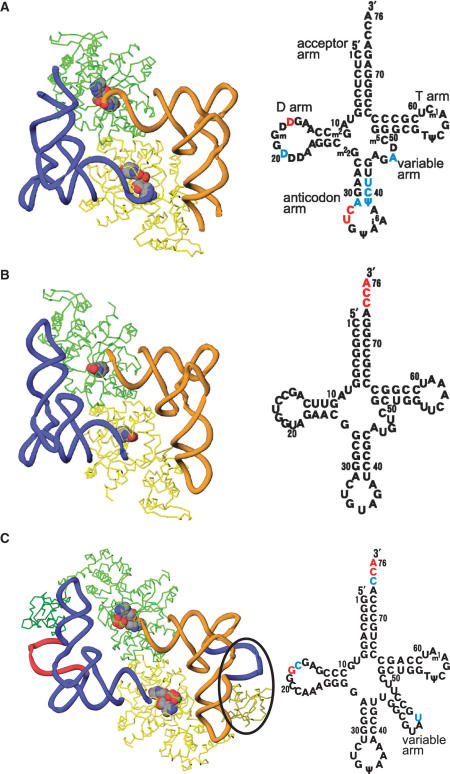
Overall structures of TyrRSs complexed with their cognate tRNATyrs. The TyrRS and tRNATyr molecules are colored as in Figure 1C. Cloverleaf models of tRNATyrs are shown in the right panel. The disordered nucleotides in the crystal structure analyses are shown in blue for the disordered bases (phosphate backbone is visible) and red for the entirely disordered nucleotides. (A) Eukaryotic TyrRS (SceTyrRS). (B) Archaeal TyrRS (MjaTyrRS). (C) Bacterial TyrRS (TthTyrRS). A variable arm of tRNA and an additional C-terminal domain of TyrRS that specifically exist in bacterial systems are indicated by an ellipsoid.
Figure 4.
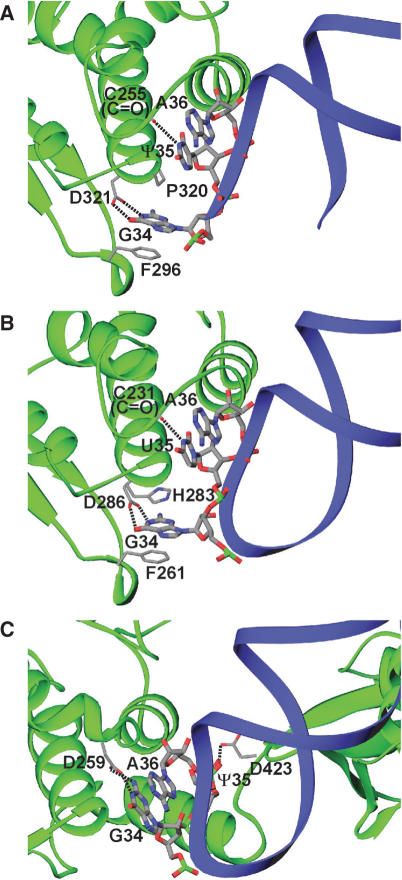
Anticodon recognition by TyrRSs. Protein and tRNA backbones are shown as in Figure 1C. Possible hydrogen bonds are indicated by dashed lines. (A) Eukaryotic TyrRS (SceTyrRS). The structural discontinuity in the anticodon-loop of tRNATyr is due to the disordered nucleotides C32–U33. (B) Archaeal TyrRS (MjaTyrRS). (C) Bacterial TyrRS (TthTyrRS).
Figure 5.
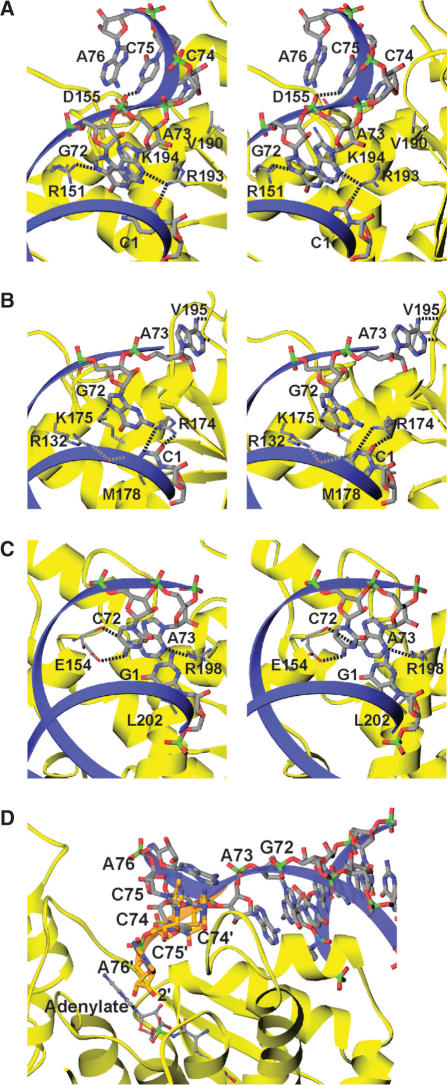
Acceptor arm recognition by TyrRSs. Protein and tRNA backbones are shown as in Figure 1C. Possible hydrogen bonds are indicated by dashed lines. (A) Eukaryotic TyrRS (SceTyrRS). (B) Archaeal TyrRS (MjaTyrRS). (C) Bacterial TyrRS (TthTyrRS). (D) Possible recognition model of the 3′-CCA terminus of tRNATyr by SceTyrRS. The C74-C75-A76 of tRNATyr is modeled (orange) into the active site of SceTyrRS by manually rotating the experimentally determined 3′-CCA terminus of tRNATyr (light gray), which was ordered but flipped out from the active site in the present crystal structure analysis. The backbone conformation of the 3′-CCA terminus and the positions of the bases are by no means necessarily correct. The aim is to show that the observed mode of tRNATyr binding to SceTyrRS allows the positioning of 2′-OH of the ribose of A76 adjacent to the carboxyl group of the Tyr-AMP without steric clashes.
Figure 6.
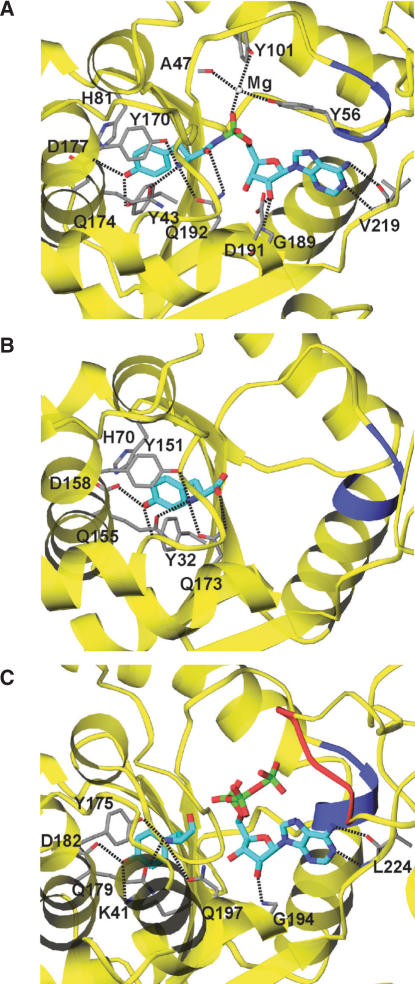
Amino acid and ATP recognition by TyrRSs. The bound substrate analogs are shown as stick models (cyan). Possible hydrogen bonds are indicated by dashed lines. Two consensus motifs, HIGH and KMSKS, conserved among the class I aaRSs are shown in blue and red, respectively. (A) Eukaryotic TyrRS (SceTyrRS). (B) Archaeal TyrRS (MjaTyrRS). (C) Bacterial TyrRS (TthTyrRS).
Figure 7.
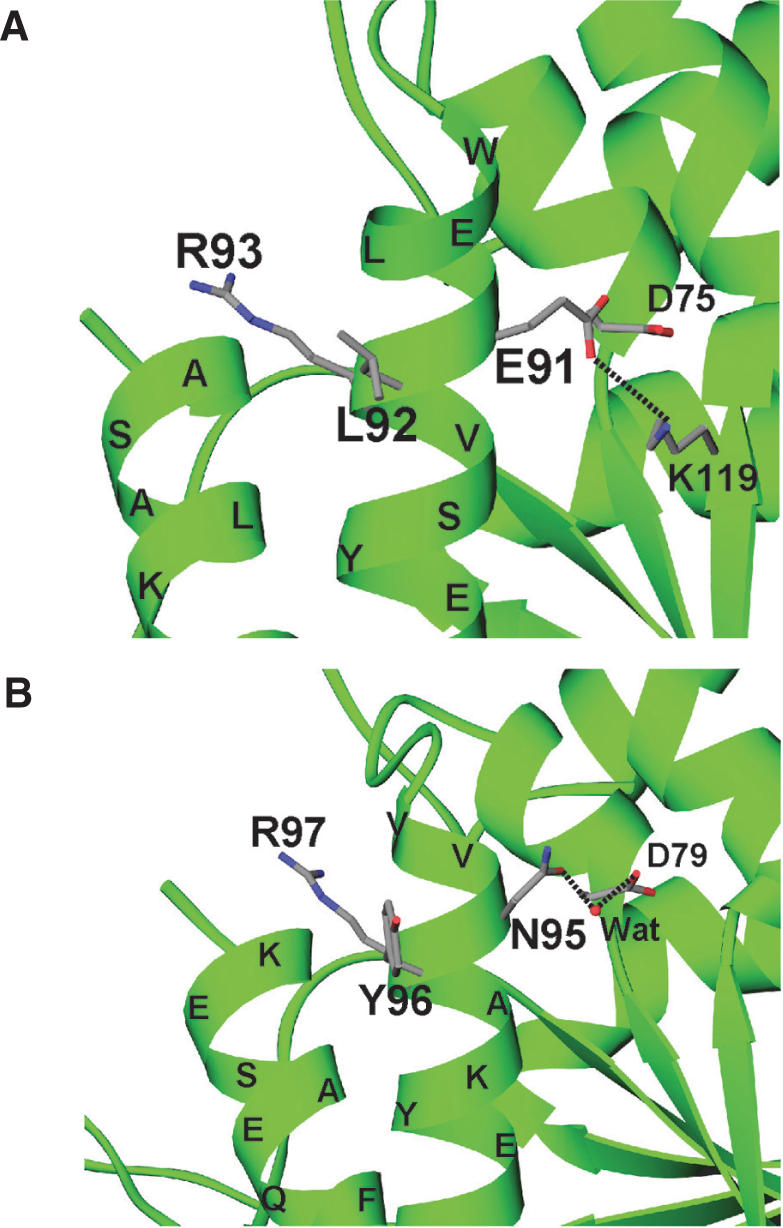
Triplet motifs crucial for the cytokine activity of eukaryotic TyrRSs. (A) The ELR motif of human TyrRS. (B) The NYR motif of yeast TyrRS.
RESULTS
Structure determination
The crystallization trials conducted to date have not successfully obtained crystals of ligand-free SceTyrRS. Fortunately, however, crystals of the ternary (SceTyrRS/Tyr-AMP analog/tRNATyr) complex of SceTyrRS were successfully obtained. For this study, a Tyr-AMP analog having an N-acyl phosphoramidate linkage where the oxygen atom of the mixed anhydride bond (-C-O-P-) of Tyr-AMP was replaced by an amino group (-C-NH-P-) (O-(adenosine-5′-O-yl) N-(L-tyrosyl)phosphoramidate [(hereafter Tyr-AMPN), Figure 1A], was used for crystallization. Initial phase calculation was performed by the MAD method using the Se-Met-substituted SceTyrRS at 3.0-Å resolution (Figure 1B). Further model building and structure refinement were performed using the native SceTyrRS, and we refined resulting model to an R-factor of 0.245 (Rfree of 0.289) at 2.4-Å resolution. The data collection and refinement statistics are summarized in Table 1.
Overall structure
The overall structure of the ternary complex of SecTyrRS is shown in Figure 1C. As observed in the cases of TthTyrRS (11) and MjaTyrRS (14), SecTyrRS forms a homo dimer and two tRNATyrs are bound to each dimer in a symmetrical fashion. The asymmetric unit contains one SceTyrRS subunit and one tRNATyr molecule (one-half of a 2:2 complex). The molecular two-fold axis coincides with a [1 1 0] crystallographic two-fold axis. In addition, a Tyr-AMP analog, Tyr-AMPN, is bound at the active site of SceTyrRS (Figure 1B). A perfectly symmetrical SceTyrRS/tRNATyr/Tyr-AMPN complex in crystal presented here is contrary to the functional asymmetry of TyrRSs in solution (18) that the enzyme exhibit half-of-the-sites’ reactivity with respect to the binding of tyrosine or Tyr-AMP and one tRNA molecule is bound per dimer of TyrRS. As for the explanations for this discrepancy, we completely agree with the notion pointed out by Yaremchuk et al. (11) that perfectly (or nearly) symmetrical structure observed in the crystal structures of TyrRSs is due to the fact that (i) there is ample time for substrate binding to both active sites in a crystallization experiment and there is quite likely preferential crystallization of a symmetrical form and (ii) in the case of tRNA binding, the same arguments hold. In addition, we postulate that considerably higher concentrations (as compared with the previous functional studies for TyrRSs in solution) of SceTyrRS (ca. 0.2 mM), tRNATyr (ca. 0.2 mM) and Tyr-AMPN (5 mM) and a large excess of Tyr-AMPN in the crystallization solution favor the formation of symmetrical dimer in the crystal.
The subunit of SceTyrRS consists of two domains. One of these, the catalytic domain, provides the groups necessary for converting the substrates Tyr and ATP into reactive intermediate Tyr-AMP (the first step of the aaRS reaction) and for transferring the amino acid moiety from the Tyr-AMP to the 3′-CCA terminus of the cognate tRNATyr (the second step of the aaRS reaction). The other domain is responsible for the recognition of the anticodon bases of the cognate tRNATyr. The two domains are unequal in size; the catalytic domain is somewhat larger and comprises 232 residues, whereas the anticodon-binding domain comprises 117 residues. The catalytic domain comprises residues 8 to 239. The structural core of this domain is an α/β structure (or Rossmann fold) comprised of a six-stranded parallel β-sheet and 10 surrounding α-helices (Figure 1C, light colors). The Tyr-AMPN molecule is bound in the central region of the carboxyl end of the parallel β-sheet in the center of the domain. The anticodon-binding domain comprises residues 240 to 356. The basic element of the secondary structure in this domain consists of six α-helices and a two-stranded anti-parallel β-hairpin (Figure 1C, dark colors). A loop region between the two domains (residues 224–233), including the KMSKS signature motif, which is one of the two consensus motifs conserved among the class I aaRSs, is disordered.
The tRNATyr molecule forms an L-shaped structure. The acceptor stem and anticodon loop of the tRNATyr interact with different subunits of the dimeric TyrRS molecule (Figure 1C). The structural discontinuity in the anticodon-loop of tRNATyr is due to the disordered nucleotides C32–U33. However, the anticodon triplet of tRNATyr (GΨA) was well ordered (Figure 1D). The catalytic domain of one subunit (yellow) recognizes the acceptor stem of a tRNA (blue), while the anticodon-binding domain of the other subunit (green) recognizes the anticodon bases of the same tRNA (blue). The overall structure of the ternary complex of SceTyrRS is similar to that of MjaTyrRS (Figure 2A and B), which is expected from the amino acid sequence similarity (Figure 3).
Figure 3.
Structure-based sequence alignment of TyrRSs. The sequences are derived from eukaryotes, yeast (PDB code: 2DLC) and human (1N3L); in an archaea, M. jannaschii (1J1U); and in bacteria, B. stearothermophilus (1TYD), E. coli (1X8X) and T. thermophilus (1H3E). The secondary structure elements of yeast and T. thermophilus TyrRSs are shown above and below the alignment, respectively. The residues mentioned in the text are shown in red (Tyr-AMP recognition), green (acceptor stem recognition) and blue (anticodon recognition). The residues that are disordered in the crystal structures are represented in italics. Class I signature motifs, HIGH and KMSKS, are highlighted in yellow.
In the TthTyrRS structure, on the other hand, a characteristic long variable arm of bacterial tRNATyr is recognized by an additional C-terminal domain of TthTyrRS (Figure 2C). Amino-acid sequence alignment of the C-terminal domain of bacterial TyrRSs suggested that the conserved sequences of the C-terminal domains determined a conserved secondary structure (19). The structure of the C-terminal domain of BstTyrRS was determined using NMR (20), and was found to have a very similar structure to that of TthTyrRS (11). Recent advances in genome sequencing revealed that bacterial tyrosyl-tRNA synthetases occur in two large subfamilies; TyrRS and TyrRZ that possess about 25% amino-acid sequence identity (21 and references therein). More detailed functional and structural analyses of the TyrRZ–tRNATyr complex are necessary to shed more light on the evolutionary divergence of the enzyme–tRNA interactions of the TyrRS and TyrRZ subfamilies in the bacterial domain.
As observed in the structures of the ternary complexes of TthTyrRS and MjaTyrRS, SceTyrRS has a class II mode of tRNA recognition, i.e. it interacts with tRNATyr from the variable loop and acceptor stem major groove side. This is in strong contrast to canonical class I enzymes, which approach cognate tRNA from the acceptor stem minor groove side.
Anticodon recognition mode
In the present structure analysis, the anticodon triplet of tRNATyr (GΨA) was well ordered (Figure 1D). The first anticodon, G34, is flipped out and base-specifically recognized by SceTyrRS (Figure 4A). The guanine ring moiety of G34 shows a stacking interaction with Phe296. The opposite face of the base has hydrophobic contact with Pro320. The N1 and O6 atoms of G34 have base-specific interactions with Asp321 through bifurcated hydrogen bonds. It is reported that mutation of G34 in yeast tRNATyr impairs aminoacylation by SceTyrRS (7). The second and third anticodon bases, Ψ35 and A36, have fewer base-specific interactions with the enzyme; only N3 of Ψ35 hydrogen bonds with the main chain carbonyl group of Cys255 (Figure 4A). They are accommodated in a hydrophobic patch composed of Phe254, Pro257, Pro319 and Pro320 (Figure 1D). This observation is also consistent with the results of the functional analysis of SceTyrRS by Fechter et al. (7). It is of note here that the tyrosylation activity is compatible with an ‘a+1 shift’ [Figure 6 (7)] of the anticodon in the 3′-direction (G35-U36-A37) but is strongly inhibited in the opposite 5′-direction (G33-U34-A35) (7). This result is explained by the present structure analysis, which shows that the electron density of the anticodon stem region was rather poor: the base moieties 31, 40 and 41 and the whole nucleotides 32 and 33 of tRNATyr were disordered. In addition, our recent biochemical analysis (22) also suggests the flexibility of the anticodon stem region of Sce-tRNATyr. This is consistent with the low G-C content at the anticodon stem region of Sce-tRNATyr as compared with that of Mja–tRNATyr. The flexibility in the 5′-portion of the anticodon loop enables the shift of the anticodon in the 3′-direction, which can be regarded as an insertion of one nucleotide before G34.
The anticodon recognition mode of archaeal MjaTyrRS (Figure 4B) is quite similar to that of eukaryotic SceTyrRS (Figure 4A), except that the hydrophobic interaction between Pro320 and G34 in SceTyrRS is replaced by the stacking interaction between His283 and G34 in MjaTyrRS.
The anticodon recognition mode of prokaryotic TthTyrRS (Figure 4C) is markedly different from those of eukaryotic and archaeal enzymes. In the TthTyrRS complex, the guanine base of G34, base-specifically recognized by Asp259, is stacked with the third anticodon base, A36. The second anticodon base, Ψ35, is oppositely flipped out and base-specifically recognized by Asp423.
Acceptor arm recognition mode
The specific aminoacylation of tRNA by TysRS relies on the identity determinants (the anticodon bases, the C1-G72 base pair, and the discriminator base A73) in the cognate tRNATyr (6–8). A previous observation showed that the strongest determinants are base pair C1-G72 and discriminator base A73 for SceTyrRS, while the three anticodon bases (G34, Ψ35 and A36) contribute to lesser extents (7). A similar observation was reported for MjaTyrRS (8). Before the present crystal structure analysis, the recognition modes of the identity determinants by the archaeal and eukaryotic TyrRSs were expected to be similar to each other but different from that by the bacterial TyrRSs. Interestingly, however, the tRNATyr recognition modes of SceTyrRS have both similarities and differences compared with those in MjaTyrRS: the recognition of the C1-G72 base pair by SceTyrRS is similar to that by MjaTyrRS, whereas the recognition of the A73 by SceTyrRS is different from that by MjaTyrRS but similar to that by TthTyrRS (Figure 5).
The expected feature is that the recognition mode of the identity base pair C1-G72 by SceTyrRS (Figure 5A) is similar to that by MjaTyrRS (Figure 5B). The recognition mode of the C1 base by the side chain of Arg193 in SceTyrRS is equivalent to that by the side chain of Arg174 in MjaTyrRS. The G72 base is recognized by a slightly different manner between SceTyrRS and MjaTyrRS. The 3′-terminal strand of the acceptor stem of SceTyrRS shows a helical conformation. The G72 base in SceTyrRS is recognized by a base-specific hydrogen bond with the side chain of Arg151 and a stacking interaction with the discriminator base A73, which is fixed by a hydrogen bond with Arg193. On the other hand, the 3′-terminal strand of the acceptor stem of MjaTyrRS shows a rather extended conformation. The G72 base is far from the side chain of Arg132, corresponding to Arg151 in SceTyrRS, and is recognized by the side chain of Lys175. The side chain of Arg132 is involved in the recognition of the C1 base, rather than the G72 base, via a water-mediated hydrogen bond (Figure 5B). Although the G72 base recognition mode of SceTyrRS and that of MjaTyrRS have some differences, the archaeal/eukaryotic TyrRSs recognize the identity base pair C1-G72 by conserved residues (Figure 3) and we assume that the recognition mode of the C1-G72 base pair by the archaeal/eukaryotic TyrRSs are essentially conserved.
The unexpected feature is that the recognition mode of A73 by SceTyrRS (Figure 5A) is similar to that by TthTyrRS (Figure 5C) but is different from that by MjaTyrRS (Figure 5B). In the case of SceTyrRS (Figure 5A), the N3 of A73 is recognized by Arg193 via a single hydrogen bond, and the discriminator base A73 is stacked with the G72 base. An equivalent recognition mode is observed for the TthTyrRS complex (Figure 5C): the N3 of A73 is recognized by Arg198 via a single hydrogen bond, and the discriminator base A73 is stacked with the C72 base. In the case of MjaTyrRS (Figure 5B), the discriminator base A73 is unstacked with the G72 base and out of the helical continuity of the acceptor stem. The N1 and N6 atoms of A73 are base-specifically recognized by the main-chain amino and carbonyl groups, respectively, of Val195. Since Arg193 in SceTyrRS (Arg198 in TthTyrRS) is also conserved in MjaTyrRS (Arg174), the observation that A73 is bound in a different manner in SceTyrRS and MjaTyrRS may reflect different modes of binding, rather than species-specific difference. However, it should be noted here that the Arg residues are not conserved in BstTyrRS (Trp196) and EcoTyrRS (Trp201) (Figure 3). The different recognition pattern of A73 would be observed in BstTyrRS and EcoTyrRS.
In the present crystal structure of SceTyrRS, the 3′-CCA terminus of tRNATyr is well ordered by a triplex stacking interactions of the C74, C75 and A76 bases, but was flipped-out from the active center. Manual model adjustment of the 3′-CCA terminus allows the 2′-OH of the terminal ribose to be correctly positioned for aminoacylation (Figure 5D). This model is consistent with the fact that TyrRSs preferentially aminoacylate the 2′-OH of A76 in accordance with other class I enzymes, although it can also aminoacylate the 3′-OH (23). Thus we assume that the present structure of the acceptor region of Sce-tRNATyr is not an artifact at least up to the discriminator base A73.
As for the binding mode of the 3′-CCA terminus of tRNATyr, the CCA terminus of MjaTyrRS (14) and that of TthTyrRS (11) were disordered, while that of SceTyrRS was flipped out from the active site. Because neither of the available complex structures (SceTyrRS–tRNATyr-Tyr–AMPN, MjaTyrRS–tRNATyr-tyrosine and TthTyrRS–tRNATyr-tyrosinol–ATP) contains true reactive intermediate, Tyr-AMP, the non-productive binding of the 3′-CCA terminus may occur.
Aminoacyl–AMP recognition mode
The present study successfully revealed the recognition modes of Tyrosyl–AMP, the reactive aminoacyl-adenylate (aa-AMP) intermediate, by SceTyrRS, as a result of our use of a Tyr-AMP analog, Tyr-AMPN (Figure 1A). The Tyr-AMP recognition mode is well conserved among the archaeal, bacterial and eukaryotic TyrRSs as described below. The tyrosine moiety is accommodated in a deep pocket of the enzyme (Figure 6A). The hydroxyl group of the tyrosine moiety makes hydrogen bonds with the side chains of Tyr43 and Asp177. The main-chain amino group of the tyrosine moiety is specifically recognized by three hydrogen bonds: they are from the side chains of Tyr170, Gln174 and Gln192. The carbonyl group of the tyrosine moiety is recognized by the side chain of Gln192. In archaeal and bacterial TyrRSs (Figure 6B and C), the recognition mode of the tyrosine moiety of Tyr-AMP is apparently similar to that of SceTyrRS. Tyr170, Gln174, Asp177 and Gln192 in SceTyrRS are well conserved (Figure 3) and play the same roles in the binding of the tyrosine moiety of Tyr-AMP (or its analog) among TyrRSs. In the case of BstTyrRS/TyrAMP complex (9), however, the side chain of Gln195 (Gln192 in SceTyrRS) does not form a hydrogen bond with the amino group of Tyr-AMP. Instead, the side chain of Asp78 is involved in the hydrogen bond with the amino group of Tyr-AMP together with the side chains of Tyr169 and Gln173 (Tyr170 and Gln174 in SceTyrRS). As for the recognition mode of the AMP moiety of Tyr-AMPN, the adenine ring is base-specifically recognized by the main-chain atoms of Val219 in SceTyrRS [N6(Ade)—O(Val229) and N1(Ade)—N(Val229), Figure 6A]. A similar recognition mode is also found for the main chain atoms of Leu224 in TthTyrRS (Figure 6C). The oxygen atom of the phosphate group of Tyr-AMPN is fixed to the protein surface [Ala47(O), Tyr56(OH), and Tyr101(OH)], via a magnesium ion (Figure 6A).
The conserved HIGH and KMSKS signature motifs
The conserved signature motifs of class I aaRSs, HIGH and KMSKS (blue and red, respectively, in Figure 6), are involved in the catalysis of tyrosine activation with ATP. It is reported that the HIGH motif is involved in binding with the γ-phosphate group of ATP in EcoTyrRS (12). In the present structure analysis, the HIGH motif's important role in tyrosine activation is not observed because of the absence of β- and γ-phosphate groups in Tyr-AMPN (Figure 6A).
The KMSKS motif shows conformational changes in tyrosine activation (12): initially, the KMSKS motif adopts the open form and then, upon binding of the adenosyl moiety of ATP, shifts to the semi-open form before finally assuming the ATP-bound closed form. In that study, Kobayashi et al.(12) assumed that after the amino acid activation, the KMSKS motif adopts the semi-open form to accept the 3′-CCA terminus of tRNA for the aminoacyl transfer reaction. In the present structure of SceTyrRS, residues 224–233, including the KMSAS sequence (Figure 3), are disordered. This flexibility of the loop containing the KMSKS motif would allow the Tyr-AMP to be fully exposed and the 3′-CCA terminus of tRNA to access the aminoacyl transfer center. Unfortunately, however, the 3′-CCA terminus of tRNATyr is ordered but flipped out from the active center in the present crystal structure (Figure 5D). In the case of TthTyrRS (Figure 6C), the loop containing the KMSKS motif interacts with ATP and is structurally well ordered.
DISCUSSION
The structural origin of tyrosine identity and species difference
In the aminoacylation of tRNAs, each amino acid is matched with a tRNA that contains the anticodon that corresponds to that amino acid. Although the anticodons within tRNAs are conserved for a given amino acid throughout evolution, the aaRS from one species does not aminoacylate its cognate tRNA from another species in some cases. Typical example is TyrRSs that exhibit species-specific tRNATyr recognition (8,24–30). The origin of species-specific tRNATyr recognition is the presence of a G1-C72 base pair in bacterial tRNATyr and a C1-G72 pair in archaeal/eukaryotic tRNATyr (29). Because the archaeal/eukaryotic TyrRSs-tRNATyrs pairs do not cross-react with their bacterial counterparts, the recognition modes of the identity determinants by the archaeal and eukaryotic TyrRSs were expected to be similar to each other but different from that by the bacterial TyrRSs. Interestingly, however, a structural comparison between the present crystal structure of the ternary complex of SceTyrRS with the available crystal structures of the ternary complexes of TthTyrRS (11) and MjaTyrRS (14) revealed (i) an unexpected similarity in the recognition mode of the discriminator base A73 between SceTyrRS and TthTyrRS (Figure 5A and C) and (ii) some differences in the recognition mode of the G72-A73 bases between SceTyrRS and MjaTyrRS (Figure 5A and B). These features indicate that the interaction mode between TyrRS and the cognate tRNATyr appears to have evolved separately for the three kingdoms of life, i.e. TyrRSs/tRNATyrs pairs have diverged after the kingdoms separated (31).
The present crystal structure analysis of the eukaryotic SceTyrRS and structural comparisons strongly support the notion pointed out by Wakasugi et al. (29) that the lack of cross-reactivity between archaeal/eukaryotic and bacterial TyrRS-tRNATyr pairs most probably lies in the different sequence of the last base pair of the acceptor stem (C1-G72 vs G1-C72) of tRNATyr.
On the other hand, the recognition modes of Tyr-AMP are conserved among the TyrRSs from all three kingdoms (Figure 6). In the class I aaRSs, the amino acid binding pocket lies at the bottom of an active site cleft in the Rossmann-fold domain. In general, the class I aaRSs use a lock-and-key mechanism to recognize the side chains of their amino acid substrates, although some exceptions exist. Detailed sequence alignment of TyrRSs and TrpRSs as well as the crystal structure analyses of human TyrRS (HsaTyrRS) and human TrpRS (HsaTrpRS) by Yang et al. (32) provided a unique example for amino acid discrimination by TyrRS and TrpRS. An environment for recognition of the hydroxyl group of Tyr side chain is provided by the universal presence of aspartate (Asp173 in HsaTyrRS) and the presence of either tyrosine (Tyr39 in HsaTyrRS) or lysine (Lys41 in TthTyrRS) (Figures 3 and 6). Interestingly, TrpRS uses the structurally equivalent residues (either Pro287 or Tyr159, respectively, in HsaTrpRS, but not both) to hydrogen bond to the indole nitrogen of tryptophan. In the case of yeast system [Asp177 and Tyr43 in SceTyrRS (Figure 6A) and Thr233 and Tyr106 in SceTrpRS(GI:51013347)], the same arguments appear to hold, although crystal structure analysis of SceTrpRS has not yet reported. The present crystal structure analysis of SceTyrRS and structural comparisons of the amino acid binding site of TyrRSs (Figure 6) are consistent with the structural and phylogenetic studies of TyrRS and TrpRS by Yang et al. (32).
Expanding the genetic code
The archaeal/eukaryotic TyrRSs-tRNATyrs pairs do not cross-react with their bacterial counterparts (8). Such orthogonality is used for the incorporation of unnatural amino acids into proteins with engineered pairs of TyrRSs and tRNATyrs (15,17,33–38). We have been trying to utilize the yeast TyrRS/tRNATyr pair as a candidate for the carrier of unnatural amino acid in the E. coli translation system in vivo (39) or in vitro (40). We previously showed that the substitution of tyrosine at position 43 to glycine (Y43G mutation) in SceTyrRS led to a drastic change in amino acid specificity. The Y43G mutant was found to be able to utilize several 3-substituted L-tyrosine analogs, rather than L-tyrosine, as substrates for aminoacylation (34). Used together with yeast amber suppressor tRNATyr, the Y43G mutant should serve as an effective tool for site-specific incorporation of 3-substituted tyrosine analogs into proteins in an appropriate E. coli translation system (41). The present crystal structure analysis can explain the structural basis for recognition of 3-substituted tyrosine analogs by the Y43G mutant SceTyrRS. Since the side chain of Tyr43 is directly involved in binding with the tyrosine moiety of Tyr-AMPN (Figure 6A), substitution of the tyrosine residue to a smaller residue creates a space to accommodate an extra functional group at position 3 of the substrate. Similar replacements are also reported for EcoTyrRS (17,38). The proteins containing unnatural amino acids will be used as molecular switches for signaling pathways, as photocrosslinkers, fluorescently labeled probes, or heavy-atom derivatives for phasing in X-ray structure determination.
The cytokine activity of TyrRS
It is reported that human TyrRS is secreted during apoptosis in cell culture and is cleaved with an extracellular elastase, and the two released fragments (the N-terminal ‘mini-TyrRS’ and the EMAP II-like C-terminal domain) are active cytokines (42). The mini-TyrRS has an ELR motif in the Rossmann-fold domain. This motif is responsible for IL-8-like cytokine activity and is conserved among segmented animals, whereas it is absent in yeast and lower eukaryote. In the case of yeast, SceTyrRS is inactive as a cytokine and has a NYR motif instead of an ELR motif. Interestingly, Liu et al. (43) reported that substitution of the tripeptide NYR to ELR in SceTyrRS resulted in mutant TyrRS with cytokine activity. This result suggests that it is the E and L that are strong candidates for a direct involvement in cytokine receptor binding. However, the Arg side chain appears to be also important for the cytokine activity of TyrRS, because an Arg93-to-Gln mutation in human mini-TyrRS abolished cytokine activity (42). A structural comparison between the ELR motif of human mini-TyrRS and the NYR motif of SceTyrRS (Figure 7) reveals that the overall structures around the motifs are similar to each other. Since the side chains of the ELR motif of human mini-TyrRS and the NYR motif of SceTyrRS are exposed, it is quite reasonable that substitution of the tripeptide NYR to ELR in SceTyrRS resulted in mutant TyrRS with cytokine activity.
ACKNOWLEDGEMENTS
We thank Drs N. Matsugaki, N. Igarashi and M. Suzuki of Photon Factory, and Drs H. Sakai, M. Kawamoto and K. Miura of SPring-8 for their help with the data collection at synchrotron facilities. This work was supported in part by Grants-in Aid for Scientific Research No. 14704067 (to T.Y.) and a grant for the Protein 3000 Project (to N.T.) from the MEXT of Japan. Funding to pay the Open Access publication charges for this article was provided by Showa University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S. Eleven down and nine to go. Nat. Struct. Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 4.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 5.Irwin MJ, Nyborg J, Reid BR, Blow DM. The crystal structure of tyrosyl-transfer RNA synthetase at 2.7 Å resolution. J. Mol. Biol. 1976;105:577–586. doi: 10.1016/0022-2836(76)90236-9. [DOI] [PubMed] [Google Scholar]
- 6.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fechter P, Rudinger-Thirion J, Theobald-Dietrich A, Giege R. Identity of tRNA for yeast tyrosyl-tRNA synthetase: tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry. 2000;39:1725–1733. doi: 10.1021/bi992276t. [DOI] [PubMed] [Google Scholar]
- 8.Fechter P, Rudinger-Thirion J, Tukalo M, Giege R. Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNATyr are conserved but expressed differently. Eur. J. Biochem. 2001;268:761–767. doi: 10.1046/j.1432-1327.2001.01931.x. [DOI] [PubMed] [Google Scholar]
- 9.Brick P, Bhat TN, Blow DM. Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J. Mol. Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- 10.Qiu X, Janson CA, Smith WW, Green SM, McDevitt P, Johanson K, Carter P, Hibbs M, Lewis C, et al. Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors. Protein Sci. 2001;10:2008–2016. doi: 10.1110/ps.18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T, Takimura T, Sekine R, Kelly VP, Kamata K, Sakamoto K, Nishimura S, Yokoyama S. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase. J. Mol. Biol. 2005;346:105–117. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 13.Yang XL, Skene RJ, McRee DE, Schimmel P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl Acad. Sci. USA. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang L, Schultz PG, Wilson IA. Crystal structure of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Protein Sci. 2005;14:1340–1348. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuratani M, Sakai H, Takahashi M, Yanagisawa T, Kobayashi T, Murayama K, Chen L, Liu ZJ, Wang BC, et al. Crystal structures of tyrosyl-tRNA synthetases from archaea. J. Mol. Biol. 2006;355:395–408. doi: 10.1016/j.jmb.2005.10.073. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of E. coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 18.Bedouelle H. Recognition of tRNATyr by tyrosyl-tRNA synthetase. Biochimie. 1990;72:589–598. doi: 10.1016/0300-9084(90)90122-w. [DOI] [PubMed] [Google Scholar]
- 19.Jermutus L, Guez V, Bedouelle H. Disordered C-terminal domain of tyrosyl-tRNA synthetase: secondary structure prediction. Biochimie. 1999;81:235–244. doi: 10.1016/s0300-9084(99)80057-1. [DOI] [PubMed] [Google Scholar]
- 20.Guijarro JI, Pintar A, Prochnicka-Chalufour A, Guez V, Gilquin B, Bedouelle H, Delepierre M. Structure and dynamics of the anticodon arm binding domain of Bacillus stearothermophilus tyrosyl-tRNA synthetase. Structure. 2002;10:311–317. doi: 10.1016/s0969-2126(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 21.Salazar JC, Zuniga R, Lefimil C, Soll D, Orellana O. Conserved amino acids near the carboxy terminus of bacterial tyrosyl-tRNA synthetase are involved in tRNA and Tyr-AMP binding. FEBS Lett. 2001;491:257–260. doi: 10.1016/s0014-5793(01)02214-1. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga J, Ohno S, Nishikawa K, Yokogawa T. A base pair at the bottom of the anticodon stem is reciprocally preferred for discrimination of cognate tRNAs by Escherichia coli lysyl- and glutaminyl-tRNA synthetases. Nucleic Acids Res. 2006;34:3181–3188. doi: 10.1093/nar/gkl414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer F, Faulhammer H, von der Haar F, Sprinzl M, Stembach H. Aminoacyl-tRNA synthetases from baker's yeast: reacting site of aminoacylation is not uniform for all tRNAs. FEBS Lett. 1975;56:212–214. doi: 10.1016/0014-5793(75)81093-3. [DOI] [PubMed] [Google Scholar]
- 24.Clark J.M., Jr, Eyzaguirre JP. Tyrosine activation and transfer to soluble ribonucleic acid. I. Purification and study of the enzyme of hog pancreas. J. Biol. Chem. 1962;237:3698–3702. [PubMed] [Google Scholar]
- 25.Doctor BP, Mudd JA. Species specificity of amino acid acceptor ribonucleic acid and aminoacyl soluble ribonucleic acid synthetases. J. Biol. Chem. 1963;238:3677–3681. [PubMed] [Google Scholar]
- 26.Chow CM, RajBhandary UL. Saccharomyces cerevisiae cytoplasmic tyrosyl-tRNA synthetase gene. Isolation by complementation of a mutant Escherichia coli suppressor tRNA defective in aminoacylation and sequence analysis. J. Biol. Chem. 1993;268:12855–12863. [PubMed] [Google Scholar]
- 27.Quinn CL, Tao N, Schimmel P. Species-specific microhelix aminoacylation by a eukaryotic pathogen tRNA synthetase dependent on a single base pair. Biochemistry. 1995;34:12489–12495. doi: 10.1021/bi00039a001. [DOI] [PubMed] [Google Scholar]
- 28.Kleeman TA, Wei D, Simpson KL, First EA. Human tyrosyl-tRNA synthetase shares amino acid sequence homology with a putative cytokine. J. Biol. Chem. 1997;272:14420–14425. doi: 10.1074/jbc.272.22.14420. [DOI] [PubMed] [Google Scholar]
- 29.Wakasugi K, Quinn CL, Tao N, Schimmel P. Gene code in evolution: switching species-specific aminoacylation with a peptide transplant. EMBO J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnefond L, Giege R, Rudinger-Thiron J. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie. 2005;87:873–883. doi: 10.1016/j.biochi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Ribas de Pouplana L, Frugier M, Quinn CL, Schimmel P. Evidence that two present-day components needed for the genetic code appeared after nucleated cells separated from eubacteria. Proc. Natl Acad. Sci. USA. 1996;93:166–170. doi: 10.1073/pnas.93.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowal AK, Kohrer C, RajBhandary UL. Twenty-first aminoacyl-tRNA synthetase-supressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc. Natl Acad. Sci. USA. 2001;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno S, Yokogawa T, Nishikawa K. Changing the amino acid specificity of yeast tyrosyl-tRNA synthetase by genetic engineering. J. Biochem. 2001;130:417–423. doi: 10.1093/oxfordjournals.jbchem.a003001. [DOI] [PubMed] [Google Scholar]
- 35.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okumi T, Nakayama H, Takio K, Yabuki T, et al. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 36.Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eykaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA. 2002;99:9715–9720. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi T, Sakamoto K, Takimura T, Sekine R, Vincent K, Kamata K, Nishimura S, Yokoyama S. Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion. Proc. Natl Acad. Sci. USA. 2005;102:1366–1371. doi: 10.1073/pnas.0407039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno S, Yokogawa T, Fujii I, Asahara H, Inokuchi H, Nishikawa K. Co-expression of yeast amber suppressor tRNATyr and tyrosyl-tRNA synthetase in Escherichia coli: possibility to expand the genetic code. J. Biochem. 1998;124:1065–1068. doi: 10.1093/oxfordjournals.jbchem.a022221. [DOI] [PubMed] [Google Scholar]
- 40.Ohno S, Matsui M, Yokogawa T, Nakamura M, Hosoya T, Hiramatsu T, Suzuki M, Hayashi N, Nishikawa K. Site-selective post-translational modification of proteins using an unnatural amino acid, 3-azidotyrosine. J. Biochem. 2007;141:335–343. doi: 10.1093/jb/mvm036. [DOI] [PubMed] [Google Scholar]
- 41.Fukunaga J, Yokogawa T, Ohno S, Nishikawa K. Misacylation of yeast amber suppressor tRNATyr by E. coli lysyl-tRNA synthetase and its effective repression by genetic engineering of the tRNA sequence. J. Biochem. 2006;139:689–696. doi: 10.1093/jb/mvj078. [DOI] [PubMed] [Google Scholar]
- 42.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Yang X.-L, Ewalt KL, Schimmel P. Mutational switching of yeast tRNA synthetase into a mammalian-like synthetase cytokine. Biochemistry. 2002;41:14232–14237. doi: 10.1021/bi0205395. [DOI] [PubMed] [Google Scholar]
- 44.Ohyama T, Nishikawa K, Takemura S. Studies on T. utils tRNATyr variants with enzymatically altered D-loop sequences. I. Deletion of the conserved sequence Gm-G and its effects on aminoacylation and conformation. J. Biochem. 1985;97:29–36. doi: 10.1093/oxfordjournals.jbchem.a135053. [DOI] [PubMed] [Google Scholar]
- 45.Moriguchi T, Yanagi T, Kunimori M, Wada T, Sekine M. Synthesis and properties of aminoacylamido-AMP: chemical optimization for the construction of an N-acyl phosphoramidate linkage. J. Org. Chem. 2000;65:8229–8238. doi: 10.1021/jo0008338. [DOI] [PubMed] [Google Scholar]
- 46.Kusakabe Y, Ohno S, Tanaka N, Nakamura M, Tsunoda M, Moriguchi T, Asai N, Sekine M, Yokogawa T, et al. Crystallization and preliminary X-ray crystallographic analysis of yeast tyrosyl-tRNA synthetase complexed with its cognate tRNA. Protein Peptide Lett. 2006;13:417–419. doi: 10.2174/092986606775974465. [DOI] [PubMed] [Google Scholar]
- 47.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossmann MG, van Beek CG. Data processing. Acta Crystallogr. 1999;D55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 49.Collaborative Computational project number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. 2003;D59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 51.McRee DE. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Carson M. Ribbons. Methods in Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]