Abstract
The proposed Drosophila melanogaster L23a ribosomal protein features a conserved C-terminal amino acid signature characteristic of other L23a family members and a unique N-terminal extension [Koyama et al. (Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and l23a, with unique histone-like amino-terminal extensions. Gene 1999; 226: 339–345)], absent from Saccharomyces cerevisiae L25 that nearly doubles the size of fly L23a. The ability of fly L23a to replace the role of yeast L25 in ribosome biogenesis was determined by creating a yeast strain carrying an L25 chromosomal gene disruption and a plasmid-encoded FLAG-tagged L23a gene. Though affected by a reduced growth rate, the strain is dependent on fly L23a-FLAG function for survival and growth, demonstrating functional compatibility between the fly and yeast proteins. Pulse-chase experiments reveal a delay in rRNA processing kinetics, most notably at a late cleavage step that converts precursor 27S rRNA into mature 25S rRNA, likely contributing to the strain's slower growth pattern. Yet, given the essential requirement for L23(a)/L25 in ribosome biogenesis, there is a remarkable tolerance for accommodating the fly L23a N-terminal extension within the structure of the yeast ribosome. A search of available databases shows that the unique N-terminal extension is shared by multiple insect lineages. An evolutionary perspective on L23a structure and function within insect lineages is discussed.
INTRODUCTION
A conserved structural blueprint for building a ribosome might be anticipated given the common function of ribosomes in protein synthesis. Numerous studies have revealed the evolutionarily conserved secondary structure of ribosomal RNA (rRNA) across species [reviewed by (1)]. Among ribosomal protein constituents, several families of highly conserved proteins are recognized between evolutionarily divergent groups [e.g. (2–5)]. Primary rRNA-binding proteins are within the group of structurally conserved proteins that recognize highly conserved features of rRNA (1,6). Other ribosomal proteins are less well-conserved in structure between lineages. Variability in ribosomal protein composition contributes to diversity in ribosome composition observed in many organisms. Thus, within a given species evolutionarily conserved and structurally divergent ribosomal components may comprise the translation apparatus.
An important ribosomal protein family is defined by Escherichia coli protein EL23. Numerous structural equivalents have been recognized in many organisms, identified as ‘L23’ in most prokaryotes, ‘L25’ in yeasts and as ‘L23a’ in most eukaryotes, including insect lineages discussed in this paper. Ribosomal protein L23(a)/L25 is an essential protein, binding to precursor large subunit rRNA early in ribosome assembly (7–11). The multifaceted roles of L23 become increasingly apparent; the protein functions in early assembly events as well as in early and late stages of rRNA processing in yeast (9,12,13). After the completion of ribosome maturation, L23 later assumes a co- and post-translational role as the nascent polypeptide emerges from the ribosome. L23 interacts with several protein folding and targeting apparatus components, including the chaperone trigger factor (TF) in E. coli, signal recognition particle, Sec61 translocation channel, and the nascent polypeptide-associated complex (NAC) (14–20).
Yeast L25 is the most widely studied eukaryotic member of the L23(a)/L25 protein family. Three distinct functional domains, essential for rRNA maturation, have been identified within the yeast protein: an N-terminal region which harbors the nuclear localization signal (NLS) (21), a central domain required for rRNA binding (7,22), and a C-terminal region which is required for 60S large subunit assembly (23). Although L25 is reportedly involved in early and late rRNA processing steps, a specific function for L25 during rRNA processing has yet to be defined.
L23(a)/L25 binds within Domain III of 23S–28S rRNA, near the peptidyl transferase center of the ribosome (10,24,25). Within the large ribosomal subunit, L23 assumes a prominent position on the subunit surface at the exit tunnel for the nascent polypeptide, based on structural studies of the Haloarcula marismortui ribosome (26,27), residing next to proteins L29, L19 and L39e (16,27). The interior surface of the exit channel consists primarily of rRNA domains. The proximity of L23 to the nascent polypeptide and the membrane translocon, and its documented interactions with several protein folding and translocation components (14–20), favor the proposal that L23 also plays a key role in targeting inner membrane proteins (16).
Numerous L23(a)/L25 homologues have been identified, each with the signature rRNA-binding domain and the nearly invariant RNA-binding motif KKAYVRL, found in the C-terminal portion of the protein. Several studies have confirmed the ability of L23 family members to interact with 25S–28S rRNA-binding sites from different organisms, demonstrating that a core of conserved interactions must exist between L23(a)/L25 proteins and the rRNA-binding site [e.g. (9,28)]. In two cases, Arabidopsis L23a (L23aA) (29) and rat L23a (30) have both been confirmed as functional homologues of yeast L25 through their abilities to rescue yeast strains grown under nutritional conditions in which the endogenous L25 gene was not expressed. In each case, the plant and mammalian homologues are structurally similar to yeast L25 (Figure 1).
Figure 1.
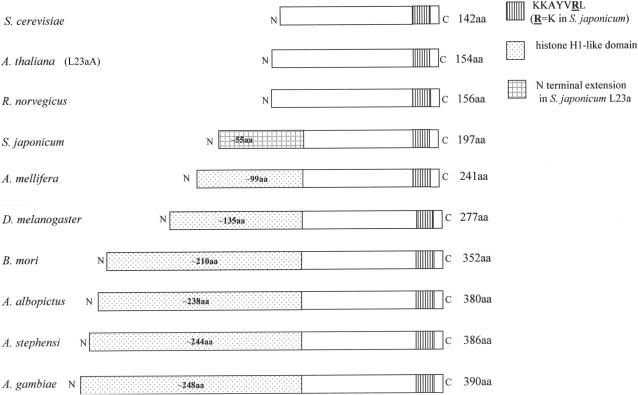
Schematic alignment of L25/L23a ribosomal proteins. L25/L23a ribosomal proteins are shown to illustrate differences in protein size, the N-terminal extensions with similarity to histone H1 within the insect lineages, and the conserved KKAYVRL RNA-binding motif. Accession numbers for L23(a)/L25 sequences are as follows: S. cerevisiae (P04456), A. thaliana (L23aA: AAC27837), R. norvegicus (P62752), S. japonicum (AAP06228), A. mellifera (XP_393135), D. melanogaster (NP_523886), B. mori (AAV34835), A. albopictus (AAY41436) A. stephensi (AAY41435), A. gambiae (XP_316083). A multiple amino acid sequence alignment is represented in Supporting Figure 1.
The Drosophila melanogaster L23 member (called L23a) has been identified as such due to structural similarity in the C-terminal rRNA-binding domain with known L23(a)/L25 family members (Figure 1). Although all known eukaryotic members of this protein family have an N-terminal extension of variable length, harboring the NLS (7,21,31), the proposed D. melanogaster L23a homologue (and several other L23a homologues discussed in this paper) is even more structurally divergent in the N-terminal region. Fruit fly L23a carries an extra domain of approximately 135 amino acids (aa) with similarity to histone H1 (32), extending its overall size to 277 aa (33; accession NP_523886) compared to the yeast L25 protein of 142 aa (34; accession P04456; Figure 1 and Table 1). While it has been presumed that fly L23a is the functional equivalent of other L23(a)/L25 proteins based on C-terminal half homology, no previous studies have confirmed this.
Table 1.
Amino acid similarity among selected L23a proteins
| SPECIES | L23(a)/L25 Accession No. | L23a size (aa) | Similarity with D.m. N-terminal extension (%) | Similarity with D.m. L23a RNA-binding domain (%) |
|---|---|---|---|---|
| Saccharomyces cerevisiae | P04456 | 142 | – | 77 |
| Arabidopsis thaliana | AAC27837 (L23aA) NP_191088 (L23aB) | 154 | – | 78 |
| Rattus norvegicus | P62752 | 156 | – | 83 |
| Drosophila melanogaster | NP_523886 DQ450529 | 277 | 100 | 100 |
| Apis mellifera | XP_393135 | 241 | 37 | 88 |
| Bombyx mori | AAV34835 | 352 | 53 | 88 |
| Aedes albopictus | AAY41436 | 380 | 52 | 93 |
| Anopheles stephensi | AAY41435 | 386 | 49 | 91 |
| Anopheles gambiae | XP_316083 | 390 | 47 | 91 |
| Schistosoma japonicum | AAP06228 | 197 | No significant similarity | 72 |
In order to test the function of D. melanogaster L23a and to gain insight into the interchangeability of L23 ribosomal components, we examined whether D. melanogaster L23a could function in yeast. We have created a yeast strain that is completely dependent on fly L23a for survival, providing the first report of an L23a protein containing a novel domain as a functional member of the L23(a)/L25 ribosomal protein family. Within the yeast strain, pulse-chase experiments demonstrate that the most significant difference in rRNA maturation kinetics is a delay in a late step(s) that converts 27S rRNA into mature 25S rRNA, likely contributing to the slow growth phenotype of the strain.
A search of available databases shows that the extended L23a insertion is not unique to D. melanogaster L23a. Rather, multiple insect species have varying sizes of insertions. Given the functional homology of D. melanogaster L23a to yeast L25, demonstrated in this study, we suspect the other insect genes are functional as well. This would suggest a remarkable tolerance of L23a to novel insertions within this essential ribosomal component.
MATERIALS AND METHODS
Amino acid alignments
L23a amino acid sequences were aligned pairwise with D. melanogaster L23a using the GenBank database and BLASTP 2.2.13 software (35). Following alignment of the more highly conserved C-terminal domain of L23a proteins using default SEG filtering in the BLASTp subprogram, unaligned amino acids in the N-terminal domains of L23a proteins were aligned pairwise by removing default SEG filtering in the BLASTp subprogram, allowing for the greatest degree of similarity between N-terminal domains. Multiple amino acid sequence alignments were performed with Clustal W software (36).
Strains and growth conditions
The yeast strains and oligonucleotides used in this study are detailed in Tables 2 and 3, respectively. For most analyses strains were grown in synthetic complete (SC) medium or rich medium (YP) containing 2% dextrose as a carbon source, as described (37).
Table 2.
Yeast strains used in this study
| Strain | Genotype [plasmid] (plasmid number) | Source | Chromosomal L25 | Plasmid L23a/L25 | Protein expressiona |
|---|---|---|---|---|---|
| YCR9 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pep4Δ::HIS3 prbΔ::his3 prc1Δ::hisG | Inada et al. (37) (YIT617) | L25 | _ | L25 |
| YCR10 | YIT617 rpl25::LEU2 [pRPL25-FH-URA3CEN] (BIT700) | Inada et al. (37) (YIT613) | _ | L25-FLAG | L25-FLAG |
| YCR11 | YCR9 [pL23a-FLAG] (BCR2) | This study | L25 | L23a-FLAG | L25 |
| YCR13 | YCR9 [pRPL25-FH-URA3CEN] (BIT700) | This study | L25 | L25-FLAG | L25; L25-FLAG |
| YCR16 | YCR11 rpl25::LEU2 | This study | _ | L23a-FLAG | L23a-FLAG |
aNote that protein expression profiles were determined by Western blot analyses (see Figures 4a and b).
Table 3.
Oligonucleotides used in this study
| OLIGO | Sequence (5′–3′) |
|---|---|
| OCR1 | GTCACGGATCCATGCCACCCAAAAA GCCAACCGA |
| OCR2 | GTCACGGATCCTTACTTGTCATCG TCATCCTTGTAGTCGCCGCGGCCGATTA TGATGCCGATCTTGTTGGCA |
| OCR3 | CGCTAGAGCTCGGCATGGGTCACTTATTTAA |
| OCR4 | TTTATCTTATTGATCTTCTTTGT |
| OCR5 | CGTCAGAATTCCCTGATGTACAACTCTTTACT |
| OVW1 | CGGGATCCATGCCACCCAAAAAGCCAAC |
| OVW2 | GCATCTCGAGGATGATCTTCTTCTGGACCT |
| Ry25S probe ‘2’ | GAAGAATCCATATCCAGGTTCCGG |
| Ry18S probe ‘1’ | CATGGCTTAATCTTTGAGAC |
| RyPGK1 | CGAAGGCATCGTTGATGTAAACATCAGCC |
Construction of pL23a-FLAG (BCR2)
The pL23a-FLAG plasmid was derived from BIT700 (38), a YCplac33-based plasmid (39) containing yeast RPL25 fused to a FLAG-(His6) epitope (FH), whose expression is controlled by the RPL25 promoter and terminator regions. The RPL25-(FH) gene and 5′ regulatory sequences were excised from BIT700 as a BamHI-BamHI fragment and replaced with a BamHI-BamHI fragment containing the L23a cDNA fused to the FLAG epitope. The BamHI L23a-FLAG fragment was generated by PCR using an L23a cDNA as a substrate, oligonucleotides OCR1 (forward) and OCR2 (reverse) as primers (Table 3) and HotStarTaq polymerase (Qiagen). OCR2 encodes the FLAG sequence fused in frame to the L23a sequence just 5′ of the stop codon. Next, the RPL25 promoter and 5′ regulatory sequences were inserted into the vector's SacI and SmaI sites as a SacI-blunt ended fragment. This promoter fragment, generated by yeast colony PCR on YCR9 (Table 2) using OCR3 (forward) and OCR4 (reverse) as primers (Table 3) and HotStarTaq polymerase (Qiagen), contains the 420 bp sequence immediately 5′ to the translational start codon, and includes both RPG boxes previously reported as required sequence elements for L25 gene transcription (40).
Disruption of the RPL25 locus
A DNA fragment containing RPL25 sequence disrupted by the LEU2 gene was obtained by yeast colony PCR on YCR10 using OCR3 (forward) and OCR5 (reverse) as primers (Table 3) and HotStarTaq polymerase (Qiagen). The resulting fragment is comprised of 420 bp of RPL25 5′ regulatory sequence, the RPL25 coding region disrupted by LEU2, followed by 367 bp of 3′ regulatory sequence. The fragment was introduced into YCR11 (Table 2) through transformation and LEU2 colonies were selected on synthetic complete media lacking leucine (SC-LEU). The colonies were screened for disruption of the RPL25 locus through colony PCR using OCR3 and OCR5 as primers, resulting in strain YCR16 (Table 2).
Growth rate analysis
Overnight cultures of strains YCR10, YCR11 and YCR16 (Table 2) grown in SC-URA media were diluted and added to 100 ml SC-URA in sidearm flasks to give an OD600 between 0.07 and 0.16. OD600 readings were measured spectrophotometrically at approximately half hour intervals for a period of 10 h in order to determine doubling times for each strain during logarithmic growth.
Detection of L23a-FLAG, L25-FLAG and L25 by Western blot analysis
Crude cellular lysates from strains YCR9, 10, 11, 13 and 16 (Table 2) were prepared as previously described (38). Crude lysates were fractionated by 15% SDS–PAGE. Duplicate gels were run: one gel was stained with Coomassie Blue and the other was used for immunoblotting. Arrays of protein markers, including Biorad Precision markers were used. Proteins were transferred by electroblotting onto Schleicher and Schuell Optitran 0.2 μm nitrocellulose membranes. For Western blot analysis, filters were blocked in 1XPBS, 10% non-fat dry milk, 0.3% Tween 20® (Sigma), and then incubated with anti-FLAG M2 mouse monoclonal antibody (Sigma) at a concentration of 10 μg/ml. FLAG-tagged proteins were detected using goat anti-mouse IgG whole molecule alkaline phosphatase conjugate affinity-isolated antibody (Sigma) at a 1:30,000 dilution. Yeast L25 was detected using rabbit anti-yeast L25 (a generous gift from A. Faber and H.A. Raué, Vrije Universiteit, Amsterdam) at a dilution of 1:10,000 and secondary antibody goat anti-rabbit IgG alkaline phosphatase conjugate (Sigma) at a dilution of 1:30,000. Blots were developed using BCIP/NBT (Sigma) as a substrate.
Affinity purification of yeast ribosomes containing FLAG-tagged L23a
Ribosomes were purified from crude extracts derived from YCR10 (containing plasmid-encoded FLAG-tagged L25 and a disruption of chromosomal L25 by LEU2) and YCR16 (containing plasmid-encoded FLAG-tagged L23a with endogenous L25 disrupted by LEU2) strains using an anti-FLAG M2 affinity gel resin (Sigma) according to the established procedure of Inada et al. (38). Crude extracts from strain YCR9 were used as a negative control for affinity purification as this strain lacks any FLAG-tagged proteins. Carboxyl terminal FLAG bacterial alkaline phosphatase (Sigma) at a concentration of 5 mg/ml was used as a positive control to verify the affinity purification protocol. FLAG peptide (Sigma) at a concentration of 100 μg/ml was used to elute bound protein complexes from the anti-FLAG M2 affinity gel resin. Ribosome-associated proteins from affinity-purified ribosomes were fractionated by 15% SDS–PAGE and visualized by silver staining (Biorad Silver Staining Plus).
RT-PCR analysis
RNA was isolated from mid-log phase yeast YCR cells using a mechanical disruption method detailed in the Qiagen RNAeasy protocol (Qiagen, Valencia, CA). Column-purified RNA was subjected to DNase treatment using RQ1 RNase-free DNase (Promega, Madison, WI) at 37°C for 30 min. Following phenol/chloroform/isoamyl (25:24:1, v/v/v) extraction and ethanol precipitation, RNAs were either resuspended in RNase-free water for immediate use in RT-PCR or were stored as pellets at −70°C for later use. RNA was extracted from Drosophila adult flies using a guanidine hydrochloride procedure (41). Ethanol-precipitated fly RNA was stored at −70°C.
RT-PCR (50 μl) was performed using the Qiagen One-Step RT-PCR kit. Reactions were incubated sequentially for 30 cycles at three specific temperatures in order to achieve template denaturation (94°C), primer annealing (55°C) and primer extension (72°C). Forward OVW1 and reverse OVW2 primers (Table 3) were used to generate an L23a-specific H1 domain PCR product of 460 bp. RT-PCR products were analyzed on 2% agarose gels.
Pulse-chase labeling and rRNA analysis
YCR9 (Table 2) cells were grown at 30°C in 10 ml of SD-ura media, supplemented with uracil (final concentration 200 μg/ml), to an OD600 of ∼0.4–0.5. YCR16 (Table 2) cells were grown at 30°C in 10 ml of SD-ura media to an OD600 of ∼0.4–0.5. Cells were pelleted and then resuspended in 1.5 ml of the appropriate media. To each set of cultures, 200 μCi of [5,6 3H]-uracil (GE Healthcare) was added. Following a 5-min labeling period, 4 ml of SD-ura media supplemented with an excess of uracil (final concentration 20 mg/ml) were added and 0.5 ml samples collected at various time points after the addition of media containing cold uracil.
RNA was isolated using a LiCl extraction method (42) with volumes adjusted to accommodate the smaller culture volumes used in labeling. Approximately equal counts per minute (∼40,000 cpm) per sample (based on liquid scintillation counting) were loaded onto a 1.2% formaldehyde–agarose gel to fractionate newly synthesized RNAs. 3H-labeled RNAs were transferred onto 0.2 μm Optitran nitrocellulose membrane, sprayed with En3Hance (PerkinElmer), and exposed to Kodak BioMax XAR film.
Northern blot analysis of rRNA
Total RNA was extracted from log phase cells, fractionated on a 1.2% formaldehyde–agarose gel and blotted onto a 0.2 μm Optitran nitrocellulose membrane. Filters were hybridized sequentially with 32P-labeled oligonucleotide probes complementary to yeast 25S, 18S or PGK1 (Table 3) used as a control for loading, according to hybridization conditions outlined in Sambrook et al. (41). RNAs were visualized by phosphorimaging.
RESULTS
Structural alignment of proposed L23a proteins in insect lineages
We are generally interested in L23a structural variation and what effect(s), if any, this variation may have on L23a function in a subset of lineages where the protein binding site within 28S rRNA is specifically cleaved (within the D7a expansion segment), producing a ‘hidden break’ or ‘gap’ within the rRNA [e.g. (43–45)]. A large number of L23a protein sequences are now available in databases; however, only a few such sequences are available from organisms where 28S rRNA ‘gap’ processing has been documented.
The proposed D. melanogaster L23a ribosomal protein features the C-terminal rRNA-binding signature KKAYVRL (22) and an N-terminal extension with homology to the C-terminal region of histone H1 (32). Proposed L23a proteins from other organisms where gap processing has been documented [e.g. Bombyx mori: (46; accession AAV34835); Schistosoma sp.: (47; accession AAP06228); Aedes albopictus: (48; accession AAY41436)] or where gap processing is likely to occur based on taxonomic relatedness between organisms [e.g. Apis mellifera (accession XP_393135), Anopheles stephensi (accession AAY41435) and Anopheles gambiae (accession XP_316083; EAA11004)] also show C-terminal amino acid conservation and larger N-terminal extensions than are characteristic for most eukaryotic L23a proteins (Figure 1; Table 1; Supporting Figure 1). Within the N-terminal region (Table 1; Supporting Figure 1), amino acid sequence divergence is more pronounced; yet, some similarity is noted particularly in repeated stretches of basic amino acids within insect lineages but not in S. japonicum, suggesting a common evolutionary origin for the extra protein domain among the insect lineages (Table 1). Interestingly, preliminary analyses suggest a coincident increase in the size of the L23a N-terminal domain in some lineages and the structural complexity of the 28S rRNA D7 expansion segment within the L23a-binding site, suggesting co-evolution of L23a proteins and the 28S D7a expansion segment as well (Ware and Mendelson, in preparation).
Creation and verification of a yeast strain dependent on D. melanogaster L23a for viability
To detect and analyze L23a protein expression in yeast, we designed a construct that expresses L23a with a FLAG epitope tag fused to its C-terminus (pL23a-FLAG). This particular tagging scheme was chosen because it has been shown that the addition of a FLAG tag to the C-terminus of yeast Rpl25p does not inhibit its function (38). We used this L23a-FLAG construct to transform a yeast strain (YCR9) wild-type for genomic RPL25, to generate strain YCR11 (Table 2). Next, we disrupted the genomic RPL25 locus in strain YCR11 through homologous recombination with a DNA fragment containing the LEU2 gene and flanking L25 gene sequences to generate strain YCR16 (Table 2). Colony PCR analysis using OCR3 and OCR5 primers (Table 3), directed against the RPL25 locus confirmed that strain YCR16 carries a disruption of genomic RPL25, as does strain YCR10 (Table 2; see Figure 2).
Figure 2.
PCR analysis of the disruption of RPL25 in yeast containing pL23a-FLAG. YCR11 was transformed with a DNA fragment containing the LEU2 gene flanked by RPL25 sequence. LEU2 colonies were subjected to PCR with OCR3 and OCR5 primers directed against the RPL25 locus. A strain with the RPL25 locus intact generates a 1.6 kbp PCR product (YCR9; lower arrow), while a strain containing the LEU2 disruption generates a 2.8 kbp product (YCR10 and YCR16; upper arrow). A DNA marker ladder is shown.
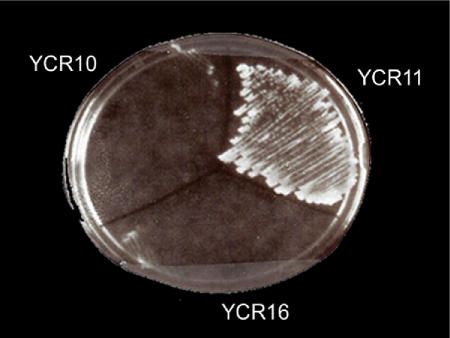
Since L25 is an essential protein in S. cerevisiae, one prediction about YCR16 is that it should require the L23a-FLAG plasmid which contains a URA3 marker for growth. We tested this prediction by plating strains YCR10, YCR11 and YCR16 onto media containing 5-fluoroorotic acid [5-FOA; (49)], which is converted to the toxic product, fluorodeoxyuridine when yeast cells express URA3. Under these plating conditions, growth was inhibited in strains YCR10 and YCR16, indicating that neither strain could lose the plasmid and remain viable (Figure 3).
Figure 3.
YCR16 is dependent on pL23a-FLAG for viability. YCR10, 11 or 16 were streaked from rich medium (YPD) onto synthetic complete plates (SC) containing 0.1% 5-FOA and incubated at 30°C for 4 days.
In order to rule out the possibility that sequence from the RPL25 locus had replaced L23a sequence on pL23a-FLAG through homologous recombination, we rescued the pL23a-FLAG plasmid from YCR16 and analyzed it by digestion with several restriction enzymes. Comparison of restriction fragment sizes with the original vector showed no alteration in the plasmid (data not shown). Sequencing of the L23aFLAG insert within pL23a-FLAG showed three nucleotide changes that alter the deduced amino acid sequence at three positions relative to the GenBank D. melanogaster L23a sequence (accession number NP_523886). All three amino acid changes occur at positions where amino acid variation is present within L23a sequences (Supporting Figure 1). Although this L23a variant may have arisen due to errors induced in RT-PCR by Taq DNA polymerase, it is likely that the variant is a naturally occurring form. From numerous independent isolations of cloned PCR products derived from RT-PCR experiments using different RNA preparations, we have only been able to recover the canonical version of L23a (accession number NP_523886) or the variant described here containing all three amino acid changes (accession number DQ450529). With either explanation, it is clear that the variant encodes a functional protein.
Expression of pL23a-FLAG within strains YCR11 and YCR16
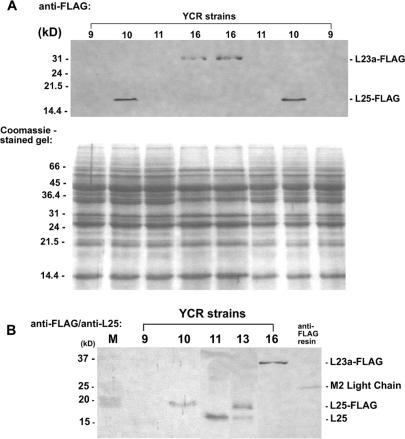
Confirmation that pL23a-FLAG was expressed within strain YCR16 was obtained by Western blot analysis using an anti-FLAG M2 mouse monoclonal primary antibody followed by detection using goat anti-mouse IgG whole molecule alkaline phosphatase conjugate. Initial analyses clearly showed the presence of FLAG-tagged L23a protein within strain YCR16 (Figure 4a and b). FLAG-tagged yeast L25 protein was also readily detected within strain YCR10 (Figure 4a and b). However, no FLAG-tagged L23a protein was detected within strain YCR11 containing the pL23a-FLAG plasmid and an intact chromosomal copy of L25 (Figure 4a and b). Our inability to detect L23a-FLAG protein within strain YCR11 was confirmed in several instances (see Figure 4a and b, for example). Interestingly, yeast FLAG-tagged L25 protein was detected in strain YCR13 that carries a plasmid encoding L25-FLAG along with an intact chromosomal copy of L25 (Figure 4b). Protein expression levels for L25 and L25-FLAG in strain YCR13 are not identical. Within strain YCR13, L25 expression appears to be less than the level that accumulates in strain YCR11; however, in a separate isolate of this strain (carrying a chromosomal copy of L25 and a plasmid-encoded FLAG-tagged L25; called strain YCR14 not described in this study), no differences in L25 and L25-FLAG protein levels were seen, indicating expression variation between strains when both L25 and L25-FLAG genes are present (data not shown).
Figure 4.
Protein expression within YCR strains. (A) Western blot analysis of strains YCR9, 10, 11 and 16 using anti-FLAG M2 antibody, showing L25-FLAG and L23a-FLAG protein expression within strains YCR 10 and 16, respectively. A duplicate gel, stained with Coomassie Blue, is shown to assess loading. (B) Western blot analysis of strains YCR 9, 10, 11, 13 and 16 using anti-FLAG M2 and anti-L25 antibodies. An aliquot of an anti-FLAG M2 affinity gel resin was included to show the position of the M2 light chain (25 kDa) relative to L25, L25-FLAG and L23a-FLAG proteins. Protein markers (M) are indicated on the left. Two lanes from protein samples derived from other strains not discussed in this study have been removed from the blot image. Only a small amount of L25 was detected in this particular blot for strain YCR9.
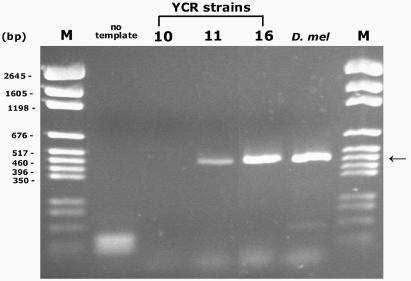
Several possibilities might account for the lack of fly L23a-FLAG protein within strain YCR11. There was no rationale for proposing a negative interaction between chromosomal and plasmid genes since simultaneous expression of both genes was readily detectable within strain YCR13. By restriction analysis of rescued plasmid pL23a-FLAG, we ruled out any gross changes in sequence that might affect plasmid gene expression, although small differences would not be detectable with this method. By RT-PCR analysis, we determined if the plasmid was transcribed within strain YCR11. Using L23a H1 domain-specific primers, L23a-specific RT-PCR products were detected within YCR11, but not in YCR10 where only FLAG-tagged L25 is expressed (Figure 5). The YCR11 product is the same size as the product generated using D. melanogaster adult RNA as a template for amplification. A noticeable reduction in the amount of PCR product for strain YCR11 compared to strain YCR16 (shown in this particular RT-PCR experiment) was not consistently observed in all experiments, likely reflecting experimental variation. Equivalent amounts of PCR product for YCR16 and another strain that is genotypically equivalent to YCR11 (not discussed here, called YCR12; note that L23a-FLAG protein also fails to accumulate in this strain—data not shown) were observed in other RT-PCR experiments (data not shown). Since the analysis was not designed as a quantitative PCR, we cannot confirm if there is reduced L23a plasmid transcription within strain YCR11 compared to strain YCR16. The data implicate a post-transcriptional mechanism to account for the lack of fly L23a-FLAG protein within strain YCR11.
Figure 5.
L23a-FLAG RNA is expressed within strain YCR11. The expected 460 bp PCR product (at the position of the arrow) was generated by RT-PCR using primers specific for the fly L23a H1 domain, indicating the presence of L23a-FLAG-specific RNA transcripts within strains YCR11 and YCR16 and in the D. melanogaster total RNA control. The minor band in the D. melanogaster lane is a non-specific product unrelated to L23a. A lane from an RNA sample derived from another strain not discussed here has been removed from the image. A pGEM DNA marker (M) is shown.
Incorporation of D. melanogaster L23a-FLAG into yeast ribosomes in strain YCR16
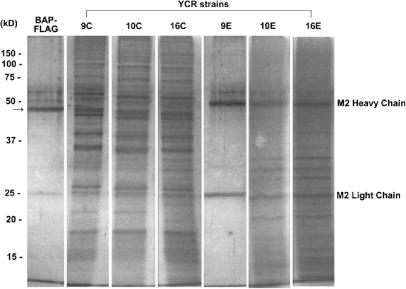
As an essential ribosomal protein L23a-FLAG protein should be a component of the ribosome population within strain YCR16. Inada et al. (38) have previously shown that ribosomes and associated proteins can be purified using a one step affinity purification method where an anti-FLAG M2 antibody resin is used to capture FLAG-tagged L25 protein and associated components. Using an identical strategy, we captured FLAG-tagged L25- and FLAG-tagged L23a-associated proteins from strains YCR10 and YCR16, respectively (Figure 6). Compared to the crude protein extract input for each strain, a subset of proteins was bound and eluted from the resin with a high concentration of FLAG peptide. In the absence of any FLAG-tagged protein in strain YCR9, no proteins were specifically bound to the anti-FLAG resin. Notably the affinity-purified protein patterns from strains YCR10 and YCR16 are nearly identical, indicating that the same subset of proteins is associated with tagged L25 and tagged L23a. Although the identity of the L23a-/L25-FLAG-associated proteins was not confirmed in this analysis, the data are consistent with the conclusion that fly L23a-FLAG protein is a component of ribosomes within strain YCR16 just as FLAG-tagged L25 is a component of ribosomes within strain YCR10 [equivalent to strain YIT613 (38); see Table 2].
Figure 6.
Fly L23a-FLAG is a component of yeast ribosomes in strain YCR16. FLAG-tagged ribosomes were purified from YCR10 and YCR16 crude extracts (C) using an anti-FLAG affinity gel resin according to Inada et al. (38). Crude extracts from strain YCR9 were used as a negative control to confirm that no proteins were bound to the resin in the absence of a FLAG tag. FLAG-tagged bacterial alkaline phosphatase (BAP-FLAG) was used as a positive control for FLAG binding and successful elution from the affinity resin. The arrow on the left marks the position of BAP-FLAG at 49 kDa. Crude (C) and eluted L23a-FLAG-associated proteins (E) were resolved by 15% SDS–PAGE and silver stained. Anti-FLAG affinity resin in the eluates shows the presence of the M2 heavy and light chains at 50 and 25 kDa, respectively. Protein marker sizes are indicated on the left. Duplicate protein sample lanes for each strain were removed from the gel image.
Comparative growth phenotypic characteristics
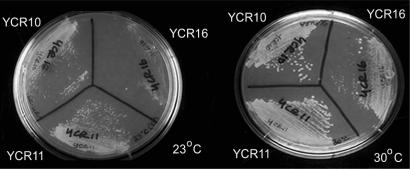
For further phenotypic analysis of YCR strains 10, 11 and 16, we compared strain growth characteristics at different temperatures. At 30°C strain YCR16 grew significantly slower than strains YCR10 and YCR11 (Figure 7). The growth of all strains was retarded at 23°C with strain YCR16 growing much slower relative to strains YCR10 and YCR11 (Figure 7). Doubling times for each strain at 30°C, calculated based on logarithmic growth of strains in liquid culture, were 130 min for YCR10, 140 min for YCR11 and 260 min for YCR16. It is unclear if the difference between the growth rates for strains YCR10 and YCR11 is significant or not (Figure 7). No obvious morphological differences were observed between strains YCR10 and YCR11 (data not shown).
Figure 7.
Growth of strains YCR10, YCR11 and YCR16 at 23°C and 30°C. Strains YCR 10, 11 and 16 were streaked from synthetic complete media lacking uracil (SC-URA) onto SC-URA plates and incubated for 2 days in order to analyze growth characteristics.
Comparative analysis of rRNA processing in strains YCR9 and YCR16
Yeast cells, dependent on fly L23a for survival and growth were significantly delayed in growth relative to cells that were dependent on endogenous L25, requiring twice the amount of time for doubling. Previous studies in which yeast L25 was genetically depleted or mutated have shown that L25 is required but is not sufficient for the removal of ITS2 and is necessary for efficient cleavage at the early sites A0, A1 and A2 [(13); see Figure 8a]. The slow growth pattern of strain YCR16 suggested that rRNA maturation kinetics might be affected. A pulse-chase analysis of pre-rRNA processing was performed to determine if any step(s) in rRNA maturation was affected by fly L23a substitution for yeast L25 in the maturation pathway.
Figure 8.
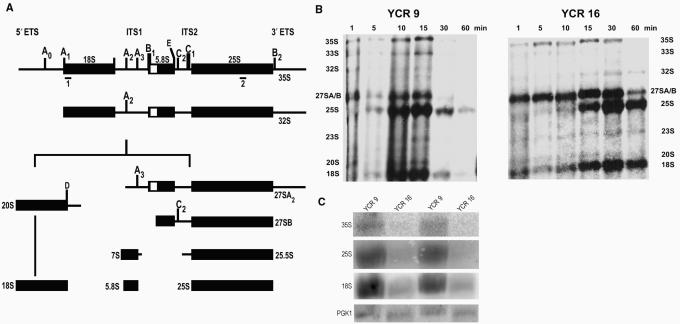
rRNA processing analysis in yeast strains YCR9 and YCR16. (A) Processing scheme for S. cerevisiae pre-rRNA. Note that not all rRNA intermediates are shown here, as this scheme represents a simplified version including rRNAs most relevant for this study. A more complete scheme is included in Raué (62). The nascent pre-rRNA transcript (not shown) is cleaved at site B0 (not shown; located within the 3′ ETS) to produce 35S pre-rRNA. 35S is cleaved at A0 and the resultant 33S rRNA further cleaved at A1, producing 32S pre-rRNA. Cleavage at A2 generates 20S and 27SA2 pre-rRNAs, as precursors to 18S rRNA and 5.8S/25S rRNAs, respectively. Cleavage of 20S pre-rRNA at site D within the cytoplasm yields mature 18S rRNA. 27SA2 pre-rRNA is subject to processing by two pathways (details not shown) to generate 27SBL and 27SBS pre-rRNAs, shown here collectively as 27SB pre-rRNA. Further cleavage of 27SB pre-rRNA at C2 produces 25.5S pre-rRNA and 7SL or 7SS pre-rRNAs (shown as 7S pre-rRNA). The actions of exonucleases on these pre-rRNAs ultimately produce mature 5.8S rRNA and 25S rRNA. The positions of probes used in Northern analysis are shown as ‘1’ and ‘2’, positioned underneath the thick black bars. (B) Pulse-chase analysis of RNA from strains YCR9 and YCR16. Cells were labeled with [5, 6 3H]-uracil as described in ‘Materials and Methods’ section. Samples were collected at 1, 5, 10, 15, 30 and 60 minutes into the chase period. Extracted RNAs were fractionated on a 1.2% formaldehyde–agarose gel, blotted onto a filter, sprayed with En3Hance, and exposed to Xray film for 48 h. YCR9 RNA samples t = 5, 30 and 60 min were underloaded to some extent in this gel. The autoradiogram shown represents one of four independent replicates of the labeling experiment. (C) Northern blot analysis of RNA from YCR9 and YCR16 strains. RNAs from overnight cultures were fractionated onto a 1.2% agarose gel, blotted onto a filter, and probed with antisense oligonucleotide probes. Specific rRNAs were detected as follows: 18S rRNA with probe ‘1’, 25S rRNA with probe ‘2’ and 35S rRNA with both probes.
Figure 8b shows that while there is no measurable delay in the appearance of 27S and 20S rRNA precursors from processing of 35S rRNA, there is an apparent delay in the conversion of 27S rRNA into mature 25S rRNA within strain YCR16. There is no evidence for a delay in 35S rRNA synthesis as this precursor is present at the beginning of the chase period in both strains YCR9 and YCR16 (Figure 8b). Although the products of 35S rRNA processing are present at the beginning of the chase period in both strains, processing of 35S rRNA at early sites A0, A1, or A2 (Figure 8a) may be slightly less efficient in strain YCR16 since a greater amount of 35S rRNA persists in the chase period compared to amounts in strain YCR9 (Figure 8b). A delay is clearly evident at a later processing step (C2; Figure 8a) to form mature 25S rRNA within strain YCR16 compared to strain YCR9 (Figure 8b). At 5 min into the chase period, 25S rRNA is clearly evident within the YCR9 strain (Figure 8b). It is not however until 10–15 min into the chase period that 25S rRNA becomes apparent within YCR16. A delay in 27S rRNA processing was consistently seen in replicates of this experiment. No significant differences in the kinetics of 20S rRNA processing into18S rRNA were detected between strains. While it is unknown if the 27S rRNA processing delay is accompanied by any changes in rRNA synthesis and/or turnover in the YCR16 strain, it is plausible that the slower processing kinetics alone may be sufficient to explain the strain's slower growth rate.
It is possible that a decrease in 25S rRNA maturation kinetics would affect the steady state levels of 25S rRNA in strain YCR16. Northern blot analysis of RNAs isolated from cultures grown overnight shows that the steady state levels of 35S, 25S and 18S rRNAs are diminished compared to levels in strain YCR9 (Figure 8c), even though equal amounts of RNA were loaded onto gels (confirmed by using PGK1 as a loading control). Although the rate of rRNA synthesis within strain YCR16 may be diminished, increased rRNA degradation may be a contributing factor, affecting not only the steady state levels of 25S rRNA, but the levels of 35S and 18S rRNAs as well. In light of the reduced amount of rRNA and yet nearly equivalent amount of L23a-FLAG compared to L25-FLAG detected by Western blot in strains YCR16 and YCR10, respectively (Figure 6a), the level of L23a-FLAG appears to be in excess of the amount of rRNA substrate in strain YCR16. Rapid degradation to eliminate excess yeast L25 protein was previously reported when the L25 gene dosage was increased nearly 50-fold (50). Within strain YCR16, the level of L23a-FLAG protein may be below a threshold required for rapid degradation. Protein turnover may also be affected by slower kinetics of ribosome maturation observed in this strain.
DISCUSSION
Fly L23a-FLAG protein can replace yeast L25 function in yeast ribosome biogenesis and ribosome function
The ability of several heterologous L23a proteins to replace the function of yeast L25 has been documented previously [e.g. (28,29)]; however, in no case has the disparity in L23a protein structure been as remarkable as is the case for D. melanogaster L23a compared to S. cerevisiae L25. Translation function within strain YCR16 is sufficient to maintain cell viability although there are clearly phenotypic deficiencies highlighted by the slow growth pattern in the strain at 23°C and at 30°C.
Several factors may account for the growth deficiency in strain YCR16, including inefficient utilization of the fly L23a NLS by the yeast transport machinery affecting the rate of L23a-FLAG protein nuclear transport or the presence of the small FLAG tag on L23a interfering with interactions required for ribosome synthesis or for post-ribosome maturation steps. Although neither of these possibilities can be completely excluded, the N-terminal extension itself may impede the kinetics of ribosome assembly or post-ribosome maturation processes at crucial steps, ultimately affecting the rate of growth. Earlier studies in which yeast L25 was either mutated (12) or genetically depleted (13) revealed early and late rRNA processing defects. Our studies suggest that 35S rRNA processing (A0–A2) to produce pre-27S rRNAs is not severely compromised, but that the efficiency of late steps (including C2) to produce mature 25S rRNA is affected in strain YCR16. This may affect the ribosome pools available for translation (as suggested by the diminished steady state levels of mature rRNAs), even if translation and post-translation processes proceed normally.
Lack of accumulation of fly L23a-FLAG protein within strain YCR11 that also expresses yeast L25
Although the plasmid-encoded L23a-FLAG gene is transcribed within strain YCR11, no L23a-FLAG protein accumulates. In principle, the difference in L25 gene dosage between strains YCR11 and YCR16 (Table 2) might elevate protein levels beyond what is required for pre-rRNA binding and large ribosomal subunit assembly, resulting in degradation of excess protein in a mechanism previously described in other studies [e.g. (50)]. If so, we would then expect both L25 and L23a-FLAG proteins to be represented in the YCR11 ribosome population, with excess protein degraded. Yet, lack of any detectable accumulation of L23a-FLAG suggests that an alternative explanation must account for the strain's protein accumulation pattern. A reasonable hypothesis is that yeast L25 competes more efficiently for nuclear import and/or for incorporation into yeast 60S ribosomal subunits than fly L23a-FLAG in this strain, ultimately excluding L23a-FLAG protein from being assembled and leading to its rapid turnover. On the other hand, the impact of protein competition is not a factor within strain YCR16 because the fly L23a-FLAG gene provides the sole source of this essential protein. Although the rate of L23a-FLAG protein turnover may be affected due to a change in ribosome maturation kinetics overall, it is clear that fly L23a-FLAG protein accumulates, ribosome assembly proceeds and growth is supported, albeit at a slower rate.
Our results differ from those of Jeeninga et al. (30) in which rat L23a competed efficiently with yeast L25 for incorporation into yeast ribosomes, even in the presence of endogenous L25. Rat L23a shows 62% sequence identity with yeast L25 (30), is comparable in size to yeast L25, and lacks the extended N-terminal domain. This general structural similarity may be sufficient to minimize binding affinity differences that would affect ribosome assembly.
Structural and evolutionary considerations
How does the yeast ribosome accommodate the extra domain of fly L23a-FLAG? To propose a hypothesis for positioning the fly L23a-FLAG extra domain within a chimeric yeast ribosome, we have considered several L23 and protein chaperone interactions as well as the spatial organization of other ribosomal proteins at the exit tunnel. Along with proteins L19, L22, L24, L29 and L31e, L23 surrounds the base of the exit channel (27) and interacts with the protein folding apparatus (see Supporting Figure 2 for the position of L23a on the H. marismortui 50S subunit). High-resolution structural studies of the 50S subunit from D. radiodurans (51) and H. marismortui (26) have revealed L23 structural differences between organisms. All L23 protein family members have a conserved globular domain, but unlike eubacterial L23, archaea and eukaryotic members lack an internal loop that extends into the exit tunnel interior cavity [reviewed by (52)]. The other end of L23 is positioned on the solvent side, close to the opening of the tunnel (53–55).
Forming a hydrophobic cradle to nestle the emerging peptide just outside the exit tunnel (56), TF binds to the ribosome on a composite surface formed from two separate exposed regions in the D. radiodurans L23 globular domain, a single exposed region of L29, and regions of Domain III of 23S rRNA (57). One exposed region in D. radiodurans L23 includes glutamate-14 (Glu-18 in E. coli L23) and the other region includes C-terminal amino acid positions 92–94 (Supporting Figure 1). Glutamate-18 is located in the center of the binding surface, with 23S rRNA interactions on one side of the binding surface and interactions with L29 and the C-terminal portion of L23 on the other side (57). Although TF is only found in eubacteria and chloroplasts, yeast NAC reportedly binds to E. coli and yeast ribosomes, suggesting that even in the absence of a direct test of which eukaryotic L23a residues are involved in binding, it is likely that NAC interacts through an identical binding surface that includes yeast L25 (20).
Based on amino acid alignment of D. melanogaster L23a with E. coli and D. radiodurans L23 (Supporting Figure 1), critical residues required for TF (and NAC) binding are found at fly L23a amino acid positions 196–200 (20; also Supporting Figure 1), within the conserved C-terminus. Thus, within the chimeric ribosomes of strain YCR16, binding surface residues for NAC would include fly L23a positions 196–200 and additional residues at the C-terminal end.
Taken together, we speculate that the N-terminal extra domain of fly L23a forms an exposed ‘appendage’ on the chimeric ribosome, distal to the C-terminal portion of L23a itself and projecting away from the peptide tunnel and ribosomal proteins L29 and L19. Other ribosome interactions may stabilize the position of the proposed appendage. This configuration is favored over a proposal in which the extra domain assumes an internal position within the tunnel where it could sterically occlude the tunnel in a fashion similar to some macrolide antibiotics (53,58) and is consistent with structural models in which an internal L23a loop is absent from eukaryotic L23a (52). Whether or not the ‘surface appendage’ proposal can be extrapolated to fly ribosomes remains to be determined.
From an evolutionary perspective, the coding capacity for the L23a extra domain in insect lineages may have been acquired through an insertion into the 5′ half of an ancestral L23a gene at a position that did not pre-empt existing L23a function(s). Alternatively, other groups may have lost the extra sequence over time with insect lineages maintaining the ancestral L23a gene. A more definitive answer to this question will require a broad comparative survey of L23(a)/L25 sequences, as yet unavailable.
Over time the extra domain may have acquired additional functions within insect lineages. Whatever novel interactions may have evolved, it would be expected that they are congruent with other L23a functions. One interesting possibility is that the L23a extra domain may have a role in processing reactions that create the ‘gap’ within 28S rRNA in several insect lineages. This hypothesis is currently under investigation.
Interactions between L23a and poly-ADP ribose polymerase (PARP), mediated through the novel H1-like domain, have already been identified (32), but not fully explored. PARP plays an important role in many cellular processes, including DNA repair, transcription and apoptosis (59,60). The possibility that L23a may function along with PARP in non-ribosomal pathways is intriguing. In fact, a recent finding that fly L22 co-purifies with histone H1 and is involved in chromatin interactions required for transcriptional repression broadens the concept of ribosomal protein function in general (61). Both fly L22 and L23a carry the histone H1-like N-terminal extra domain (32). A possible role for the L22 N-terminal extension along with PARP in mediating these novel interactions with chromatin was not investigated (61). It is unknown if similar chromatin-L23a interactions occur. Other L23a extra domain interactions, yet to be discovered, may contribute further to the multifunctional capacity of the L23a ribosomal protein family.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported in part by a Block Grant from the Pennsylvania Department of Health and research funds provided by Lehigh University. R. Patel was supported as a Lehigh University Undergraduate Pool Scholar. Dr J. van't Riet is acknowledged for stimulating discussions that resulted in our pursuit of these experiments. The authors thank Dr T. Inada for vectors and strains, Drs A. Faber and H.A. Raué for the ‘well-preserved’ L25 antibody and Dr J. Marzillier for sequencing. We also thank Drs R. Skibbens and A. Brands for discussions about the morphological characteristics of strains created here and Dr M. Itzkowitz for discussions about evolutionary theory. K. Synder contributed to the growth rate analysis of YCR strains. We thank Drs R. Gates and M. Kuchka for comments about the manuscript. We are extremely grateful to M. Brace for assistance with figure preparation. Funding to pay the Open Access publication charges for this article was provided by Lehigh University's Incentive Fund Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Raué HA, Klootwijk J, Musters W. Evolutionary conservation of structure and function of high molecular weight ribosomal RNA. Prog. Biophys. Mol. Biol. 1988;51:77–129. doi: 10.1016/0079-6107(88)90011-9. [DOI] [PubMed] [Google Scholar]
- 2.Wittmann-Liebold B, Kopke A.KE, Arndt E, Kromer W, Hatakeyama T, Wittmann H.-G. Sequence comparison and evolution of ribosomal proteins and their genes. In: Hill WE, Dahlberg A, Garrett RA, Moore PB, Schlessinger D, Warner JR, editors. The Ribosome: Structure, Function, and Evolution. Washington, DC: American Society for Microbiology; 1990. pp. 598–616. [Google Scholar]
- 3.Alksne LE, Anthony RA, Liebman SW, Warner JR. An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl Acad. Sci. USA. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell. Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 5.Manuell AL, Yamaguchi K, Haynes PA, Milligan RA, Mayfield SP. Composition and structure of the 80S ribosome from the green alga Chlamydomonas reinhardtii: 80S ribosomes are conserved in plants and animals. J. Mol. Biol. 2005;351:266–279. doi: 10.1016/j.jmb.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Raué HA, Musters W, Rutgers CA, van’ t Riet J, Planta RJ. rRNA: from structure to function. In: Hill WE, Dahlberg A, Garrett RA, Moore PB, Schlessinger D, Warner JR, editors. The Ribosome: Structure, Function, and Evolution. Washington, DC: American Society for Microbiology; 1990. pp. 217–235. [Google Scholar]
- 7.Rutgers CA, Schaap PJ, van‘t Riet J, Woldringh CL, Raué HA. In vivo and in vitro analysis of structure-function relationships in ribosomal protein L25 from Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1990;1050:74–79. doi: 10.1016/0167-4781(90)90144-q. [DOI] [PubMed] [Google Scholar]
- 8.El-Baradi TT, Raué HA, De Regt CH, Planta RJ. Stepwise dissociation of yeast 60S ribosomal subunits by LiCl and identification of L25 as a primary 26S rRNA binding protein. Eur. J. Biochem. 1984;144:393–400. doi: 10.1111/j.1432-1033.1984.tb08477.x. [DOI] [PubMed] [Google Scholar]
- 9.El-Baradi TT, Raué HA, De Regt V.CH, Verbree EC, Planta RJ. Yeast ribosomal protein L25 binds to an evolutionary conserved site on yeast 26S and E.coli 23S rRNA. EMBO J. 1985;4:2101–2107. doi: 10.1002/j.1460-2075.1985.tb03898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vester B, Garrett RA. Structure of a protein L23-RNA complex located at the A-site domain of the ribosomal peptidyl transferase centre. J. Mol. Biol. 1984;179:431–452. doi: 10.1016/0022-2836(84)90074-3. [DOI] [PubMed] [Google Scholar]
- 11.Yeh LC, Lee JC. Yeast ribosomal proteins L4, L17, L20 and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA. Biochim. Biophys. Acta. 1998;1443:139–148. doi: 10.1016/s0167-4781(98)00202-4. [DOI] [PubMed] [Google Scholar]
- 12.van Beekvelt CA, Kooi EA, de Graaff-Vincent M, van't Riet J, Venema J, Raué HA. Domain III of Saccharomyces cerevisiae 25S ribosomal RNA: its role in binding of ribosomal protein L25 and 60S subunit formation. J. Mol. Biol. 2000;296:7–17. doi: 10.1006/jmbi.1999.3432. [DOI] [PubMed] [Google Scholar]
- 13.van Beekvelt CA, de Graaff-Vincent M, Faber AW, van't Riet J, Venema J, Raué HA. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:5001–5008. doi: 10.1093/nar/29.24.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 15.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 16.Ullers RS, Houben E.NG, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buskiewicz I, Deuerling E, Gu SQ, Jockel J, Rodnina MV, Bukau B, Wintermeyer W. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl Acad. Sci. USA. 2004;101:7902–7906. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 20.Wegrzyn R, Hofmann D, Merz F, Nikolay P, Rauch T, Graf C, Deuerling E. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- 21.Schaap PJ, van't Riet J, Woldringh CL, Raué HA. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J. Mol. Biol. 1991;221:225–237. doi: 10.1016/0022-2836(91)80216-h. [DOI] [PubMed] [Google Scholar]
- 22.Rutgers CA, Rientjes JM, van't Riet J, Raué HA. rRNA binding domain of yeast ribosomal protein L25. Identification of its borders and a key leucine residue. J. Mol. Biol. 1991;218:375–385. doi: 10.1016/0022-2836(91)90719-m. [DOI] [PubMed] [Google Scholar]
- 23.Kooi EA, Rutgers CA, Kleijmeer MJ, van't Riet J, Venema J, Raué HA. Mutational analysis of the C-terminal region of Saccharomyces cerevisiae ribosomal protein L25 in vitro and in vivo demonstrates the presence of two distinct functional elements. J. Mol. Biol. 1994;240:243–255. doi: 10.1006/jmbi.1994.1438. [DOI] [PubMed] [Google Scholar]
- 24.Weitzmann CJ, Cooperman BS. Reconstitution of Escherichia coli 50S ribosomal subunits containing puromycin-modified L23: functional consequences. Biochemistry. 1990;29:3458–3465. doi: 10.1021/bi00466a006. [DOI] [PubMed] [Google Scholar]
- 25.Thiede B, Urlaub H, Neubauer H, Grelle G, Wittmann-Liebold B. Precise determination of RNA-protein contact sites in the 50 S ribosomal subunit of Escherichia coli. Biochem. J. 1998;334:39–42. doi: 10.1042/bj3340039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 27.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 28.El-Baradi TT, de Regt VC, Planta RJ, Nierhaus KH, Raué HA. Interaction of ribosomal proteins L25 from yeast and EL23 from E. coli with yeast 26S and mouse 28S rRNA. Biochimie. 1987;69:939–948. doi: 10.1016/0300-9084(87)90227-6. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh KB, Bonham-Smith PC. Establishment of Arabidopsis thaliana ribosomal protein RPL23A-1 as a functional homologue of Saccharomyces cerevisiae ribosomal protein L25. Plant Mol. Biol. 2001;46:673–682. doi: 10.1023/a:1011612329398. [DOI] [PubMed] [Google Scholar]
- 30.Jeeninga RE, Venema J, Raué HA. Rat RL23a ribosomal protein efficiently competes with its Saccharomyces cerevisiae L25 homologue for assembly into 60S subunits. J. Mol. Biol. 1996;263:648–656. doi: 10.1006/jmbi.1996.0605. [DOI] [PubMed] [Google Scholar]
- 31.Metzenberg S, Joblet C, Verspieren P, Agabian N. Ribosomal protein L25 from Trypanosoma brucei: phylogeny and molecular co-evolution of an rRNA-binding protein and its rRNA binding site. Nucleic Acids Res. 1993;21:4936–4940. doi: 10.1093/nar/21.21.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koyama Y, Katagiri S, Hanai S, Uchida K, Miwa M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and l23a, with unique histone-like amino-terminal extensions. Gene. 1999;226:339–345. doi: 10.1016/s0378-1119(98)00529-0. [DOI] [PubMed] [Google Scholar]
- 33.Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0084. RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leer RJ, van Raamsdonk-Duin MM, Hagendoorn MJ, Mager WH, Planta RJ. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984;12:6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusova TA, Madden TL. Blast 2 sequences - a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 36.Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guthrie C, Fink GR. In Methods in Enzymology. Vol. 169. San Diego: Academic Press; 1991. Guide to yeast genetics and molecular biology. [PubMed] [Google Scholar]
- 38.Inada T, Winstall E, Tarun S.Z., Jr, Yates 3rd JR, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 40.Woudt LP, Smit AB, Mager WH, Planta RJ. Conserved sequence elements upstream of the gene encoding yeast ribosomal protein L25 are involved in transcription activation. EMBO J. 1986;5:1037–1040. doi: 10.1002/j.1460-2075.1986.tb04319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 43.Basile-Borgia AE, Dunbar DA, Ware VC. Heterologous rRNA gene expression: internal fragmentation of Sciara coprophila 28S rRNA within microinjected Xenopus laevis oocytes. Insect Mol Biol. 2005;14:523–536. doi: 10.1111/j.1365-2583.2005.00583.x. [DOI] [PubMed] [Google Scholar]
- 44.DeLanversin G, Jacq B. Sequence of the central break region of the precursor of Drosophila 26S ribosomal RNA. C. R. Seances Acad. Sci. III. 1983;296:1041–1044. [PubMed] [Google Scholar]
- 45.Ware VC, Renkawitz R, Gerbi SA. rRNA processing: removal of only nineteen bases at the gap between 28Sα and 28Sβ rRNAs in Sciara coprophila. Nucleic Acids Res. 1985;13:3581–3597. doi: 10.1093/nar/13.10.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara H, Ishikawa H. Molecular mechanism of introduction of the hidden break into the 28S rRNA of insects: implication based on structural studies. Nucleic Acids Res. 1986;14:6393–6401. doi: 10.1093/nar/14.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Keulen H, Mertz PM, LoVerde PT, Shi H, Rekosh DM. Characterization of a 54-nucleotide gap region in the 28S rRNA gene of Schistosoma mansoni. Mol. Biochem. Parasitol. 1991;45:205–214. doi: 10.1016/0166-6851(91)90087-m. [DOI] [PubMed] [Google Scholar]
- 48.Kjer KM, Baldridge GD, Fallon AM. Mosquito large subunit ribosomal RNA: simultaneous alignment of primary and secondary structure. Biochim. Biophys. Acta. 1994;1217:147–155. doi: 10.1016/0167-4781(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 49.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 50.El-Baradi TT, van der Sande CA, Mager WH, Raué HA, Planta RJ. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr. Genet. 1986;10:733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]
- 51.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi R, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic bacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 52.Baram D, Yonath A. From peptide-bond formation to cotranslational folding: dynamic, regulatory and evolutionary aspects. FEBS Lett. 2005;579:948–954. doi: 10.1016/j.febslet.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 53.Berisio R, Schluenzen F, Harms J, Bashan A, Auerbach T, Baram D, Yonath A. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 2003a;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 54.Berisio R, Harms J, Schluenzen F, Zarivach R, Hansen HA, Fucini P, Yonath A. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 2003b;185:4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen HA, Harms J, et al. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Eur. J. Biochem. 2003;270:2543–2556. doi: 10.1046/j.1432-1033.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 56.Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- 57.Baram D, Pyetan E, Sittner A, Auerbach-Nevo T, Bashan A, Yonath A. Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc. Natl Acad. Sci. USA. 2005;102:12017–12022. doi: 10.1073/pnas.0505581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Auerbach T, Bashan A, Yonath A. Ribosomal antibiotics: structural basis for resistance, synergism and selectivity. Trends Biotechnol. 2004;22:570–576. doi: 10.1016/j.tibtech.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 59.de Murcia G, Shall S. Poly (ADP-ribosylation) Reactions: From DNA Damage and Stress Signaling to Cell Death. Oxford: Oxford University Press; 2000. [Google Scholar]
- 60.Ziegler M, Oei SL. A cellular survival switch: Poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001;23:543–548. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]
- 61.Ni J.-Q, Liu L.-P, Hess D, Rietdorf J, Sun F-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raué HA. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson M, editor. The Nucleolus. New York: Kluwer Academy/Plenum Publishers; 2003. pp. 1–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.