Abstract
Postreplicative maintenance of genomic methylation patterns was proposed to depend largely on the binding of DNA methyltransferase 1 (Dnmt1) to PCNA, a core component of the replication machinery. We investigated how the slow and discontinuous DNA methylation could be mechanistically linked with fast and processive DNA replication. Using photobleaching and quantitative live cell imaging we show that Dnmt1 binding to PCNA is highly dynamic. Activity measurements of a PCNA-binding-deficient mutant with an enzyme-trapping assay in living cells showed that this interaction accounts for a 2-fold increase in methylation efficiency. Expression of this mutant in mouse dnmt1−/− embryonic stem (ES) cells restored CpG island methylation. Thus association of Dnmt1 with the replication machinery enhances methylation efficiency, but is not strictly required for maintaining global methylation. The transient nature of this interaction accommodates the different kinetics of DNA replication and methylation while contributing to faithful propagation of epigenetic information.
INTRODUCTION
Genomic DNA in mammalian cells is commonly methylated at position 5 of cytosine residues in CpG sequences. This epigenetic modification plays an important role in the regulation of gene expression and chromatin structure and is essential for normal development, cell differentiation, X chromosome inactivation and genomic imprinting (1,2). Cell-type-specific methylation patterns are established de novo in early developmental stages by the action of Dnmt3a and 3b and are then maintained through subsequent cell generations primarily by the action of Dnmt1 (3,4). In somatic cells, Dnmt1 is the predominant DNA methyltransferase in terms of abundance, contribution to global methyltransferase activity and to genomic methylation levels (2,5). Reduction of Dnmt1 levels leads to hypomethylation, genomic instability and cancer (6). Aberrant genomic methylation is often associated with human disease and tumorigenesis (7).
Due to its strong preference for hemimethylated substrate DNA in vitro (8–10) and its accumulation at replication sites during S-phase (11,12) Dnmt1 is thought to act on hemimethylated CpG sites generated during DNA replication. The association of Dnmt1 with replication factories was proposed as an efficient mechanism for coupling maintenance of genomic methylation patterns to DNA replication (11). Later, direct interaction with the replication processivity factor PCNA was shown and a small PCNA-binding domain (PBD) was mapped to the N-terminal region of Dnmt1 (12,13). This domain was also shown to be necessary for recruitment of Dnmt1 to DNA damage sites, suggesting that it is responsible for coupling DNA repair and restoration of methylation patterns (14). Thus, direct interaction with PCNA would ensure that methylation patterns are faithfully preserved in different situations involving DNA synthesis. However, DNA replication is highly processive taking about 0.035 s per nucleotide (15), while in vitro steady-state kinetic analysis of purified recombinant Dnmt1 revealed rather low turn-over rates of about 70–450 s per methyl group transfer (16). Although DNA methylation by Dnmt1 may be faster in vivo, it is not likely to come close to the 3–4 orders of magnitude faster DNA replication. In fact, cytosine methylation is a highly complex reaction involving recognition of hemimethylated CpG sites, binding of S-adenosylmethionine, flipping of the cytidine base out of the double helix, formation of a covalent bond between the enzyme and the cytidine, transfer of the methyl group and release of the covalent bond by β-elimination (17–19). These considerations leave open the question of how DNA replication and methylation are kinetically and mechanistically coordinated.
In addition to Dnmt1 several other factors directly and indirectly involved in DNA replication, such as DNA Ligase I, Fen1, CAF-1 and Cyclin A have been shown to redistribute to replication foci during S-phase (20–24). Many of these factors have been found to interact directly with PCNA, which forms a homotrimeric ring around the DNA helix and serves as a platform for tethering them to the replication machinery (25). Even taking into account the trivalent nature of the PCNA ring, the sheer number of its potential binding partners during replication makes it clear that they cannot all possibly bind at the same time in a constitutive manner.
The functional relevance of the interaction between Dnmt1 and PCNA and its contribution to the maintenance of epigenetic information after DNA replication, however, remains unclear. We have addressed this question by comparing the kinetics and activity of GFP-tagged wild-type Dnmt1 and PCNA-binding-deficient mutants in live cell assays and Dnmt1 deficient embryonic stem (ES) cells. Our data show that the interaction of Dnmt1 with PCNA is highly transient, increases the efficiency of postreplicative methylation by 2-fold, but is not required for restoring CpG methylation in Dnmt1 deficient ES cells.
MATERIALS AND METHODS
Expression constructs
The expression constructs RFP-PCNA, GFP-Ligase, GFP-Dnmt1wt, GFP-Dnmt1Δ1-171, GFP-Dnmt1C1229W as well as the PBD-GFP construct were described earlier (13,21,26,27). The GFP-Dnmt1Q162E and GFP-Dnmt1F169S expression constructs were derived from the GFP-Dnmt1wt construct by overlap extension PCR mutagenesis (28,29) using the outer forward primer 5′-CAG ATC TCG AGC TCA AGC TTC-3′, the inner reverse primer 5′-GTG TCA AAG CTC TGA TAG ACC AGC-3′, the inner forward primers 5′-GAACCACCAGGGAGACCACCATC-3′ for Q162E and 5′-CACGGCTCACTCCACGAAGG-3′ for F169S and the outer reverse primer 5′-CTGGAATGACCGAGACGCAGTCG-3′. The final PCR fragments containing the mutations were digested with BglII and HindIII and exchanged with the corresponding fragment in the GFP-DNMT1wt construct. Mutations were confirmed by DNA sequencing and molecular size of fusion proteins was tested by expression in HEK 293T cells and western blot analysis. For stable transfections we inserted the cassette containing the wt Dnmt1 cDNA fused to GFP from the GFP-Dnmt1wt construct into the pCAG-IRESblast vector (30).
Cell culture, transfection and FACS-sorting
Human embryonic kidney (HEK) 293T cells and mouse C2C12 myoblasts were cultured in DMEM supplemented with 10% and 20% fetal calf serum, respectively, and 50 µg/ml gentamycine. HEK 293T cells were transfected with polyethylenimine (Sigma) (31). For live cell observations C2C12 myoblasts were grown to 30-40% confluence on Lab-Tek chamber slides (Nunc) or µ-slides (Ibidi) and transfected with TransFectin transfection reagent (Bio-Rad) according to the manufacturer's instructions. Cells were then incubated overnight before performing live cell analysis. Nuclear localization of GFP-Dnmt1wt was identical to endogenous Dnmt1 as determined by immunostaining with an affinity purified polyclonal antiserum against the N-terminal domain of mouse Dnmt1. GFP-Dnmt1 localization was not affected by additional co-expression of RFP-PCNA (controls not shown). Dnmt1 immunostaining showed that typical expression levels of transfected GFP-Dnmt1 constructs in cells selected for live cell imaging were comparable to those of endogenous Dnmt1 protein (Supplementary Figure 1). For stable expression of GFP-Dnmt1wt, C2C12 cells were grown in a p100 tissue culture dish and transfected as described earlier. Cells were then cultured with 10 µg/ml blasticidin for at least 20 days before homogeneity and levels of expression were determined by fluorescence microscopy and western blotting (Supplementary Figure 2). Mouse wild type and dnmt1−/− J1 ES cells (s allele) (5) were cultured without feeder cells in gelatinized flasks as described (27). J1 cells were transfected with Transfectin (BioRad) 3–4 h after seeding and GFP-positive cells were sorted with a FACS Vantage SE cell sorter (Becton–Dickinson).
Co-immunoprecipitation
HEK 293T cells were transiently transfected with expression plasmids as described above. After 48 h about 70–90% of the cells expressed the GFP constructs as determined by fluorescence microscopy. Extracts from ∼1 × 107 cells were prepared in 200 µl of lysis buffer (20 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2 mM PMSF, 0.5% NP40). After centrifugation supernatants were diluted to 500 µl with lysis buffer without NP40. Extracts were incubated with 1 µg of a GFP-binding protein coupled to sepharose (manuscript in preparation) for 1 h at 4°C with constant mixing. Immunocomplexes were pulled down by centrifugation. The supernatant was removed and 50 µl were collected (referred to as non-bound). The beads were washed twice with 1 ml of dilution buffer containing 300 mM NaCl and resuspended in SDS-PAGE sample buffer. Proteins were eluted by boiling at 95°C and subjected to SDS-PAGE followed by immunoblotting. Antigens were detected with a mouse monoclonal anti-GFP antibody (Roche) and a rat monoclonal anti-PCNA antibody (32).
In vitro methyltransferase assay
Extracts from HEK 293T cells expressing the indicated GFP constructs were prepared and immunoprecipitations were performed as described above. After washing with dilution buffer containing 300 mM NaCl the beads were washed twice with assay buffer (100 mM KCl, 10 mM Tris pH 7.6, 1 mM EDTA, 1 mM DTT) and resuspended in 500 µl of assay buffer. After adding 30 µl of methylation mix {[3H]-SAM (S-adenosyl-methionine); 0.1 µCi (Amersham Biosciences), 1.67 pmol/µl hemimethylated ds 35 bp DNA (50 pmol/µl), 160 ng/µl BSA} incubation was carried out for 2.5 h at 37°C. The reactions were spotted onto DE81 cellulose paper filters (Whatman) and the filters were washed 3 times with 0.2 M (NH4)HCO3, once with ddH2O and once with 100% ethanol. After drying, radioactivity was measured by liquid scintillation. Samples without enzyme and with 2 µg of purified human recombinant DNMT1 were used as negative and positive controls, respectively.
Combined bisulfite restriction analysis (COBRA)
Genomic DNA was isolated by the phenol–chloroform method (33) and bisulfite treatment was as described (34) except that deamination was carried out for 4 h at 55°C. Primer sets and PCR conditions for CpG islands of skeletal α-actin, H19 (region A) and dnmt1o promoters, Xist exon 1 and intracisternal type A particle long terminal repeats (IAP LTRs) were as described (35–39). PCR products were digested with the following enzymes: skeletal α-actin and dnmt1o promoters and IAP LTRs with HpyCH4IV (New England BioLabs); H19 with Bsh136I and Xist with TaqI (both from Fermentas). Digests were separated by agarose electrophoresis except for IAP LTR fragments, which were separated in 10% acrylamide gels. Digestion fragments were quantified from digital images using ImageJ software (http://rsb.info.nih.gov/ij/). The results were corrected for PCR bias, which was calculated as described (40). Briefly, COBRA assays were performed on genomic DNA from untransfected Dnmt1−/− J1 cells methylated in vitro with recombinant SssI methyltransferase (New England BioLabs) and mixed in different proportions with unmethylated DNA from the same cells (Supplementary Figure 6). Bias curves and corrections were calculated using WinCurveFit (Kevin Raner Software). For each amplified sequence digestion with restriction enzymes whose recognition sequence includes cytosine residues in a non-CpG context was used to control for complete bisulfite conversion except for the skeletal α-actin promoter, where bisulfite sequencing revealed about 99% conversion efficiency in all samples (Supplementary Figure 5).
Live cell microscopy, FRAP analysis and trapping assay
Live cell imaging and FRAP experiments were performed on a TCS SP2 AOBS confocal laser scanning microscope (Leica) using a 63 × /1.4 NA Plan-Apochromat oil immersion objective. The microscope was equipped with a heated environmental chamber set to 37°C. Fluorophores were excited with the 488 nm line of an argon laser and a 561 nm solid state diode laser. Confocal image series were typically recorded with a frame size of 256 × 256 pixels, a pixel size of 100 nm, and with 150 ms time intervals. The laser power was typically set to 2–4% transmission with the pinhole opened to 3 Airy units. For FRAP analysis, half of the nucleus was photobleached for 300 ms with all laser lines of the argon laser set to maximum power at 100% transmission. Typically 20 prebleach and 400 postbleach frames were recorded for each series. Quantitative evaluation was performed using ImageJ. The mean fluorescence intensities of the bleached and unbleached region for each time point were background subtracted and normalized to the mean of the last 10 prebleach values (single normalized). These values were divided by the respective total nuclear fluorescence in order to correct for total loss of nuclear fluorescence as well as for the gain of nuclear fluorescence due to import from the cytoplasm over the time course (double normalized). Since the fluorescence in the bleached region differed from cell to cell and typically did not reach background level the values were also normalized to zero. Here, only the distal two-thirds of the unbleached part in the first postbleach frame were considered as reference to take into account fast diffusing molecules invading into the bleached half (triple normalized). For each construct and cell cycle stage 6–10 nuclei were averaged and the mean curve as well as the standard error of the mean (SEM) was calculated. Half times of recovery were calculated from the mean curves.
The trapping assay to measure postreplicative methylation efficiency in living cells was previously described (27). 5-Aza-2′-deoxycytidine (Sigma) was added at a final concentrations of 30 µM and cells were incubated for the indicated periods before performing FRAP experiments. Microscope settings were as described above except that a smaller ROI (3 µm × 3 µm) was selected and the time interval was set to 208 ms. FRAP data were double normalized as described above.
For presentation, we used linear contrast enhancement on entire images. For color figures we chose magenta as false color for red fluorescence to accommodate for colorblindness.
RESULTS
Point mutations within the PBD abolish the interaction of Dnmt1 with PCNA
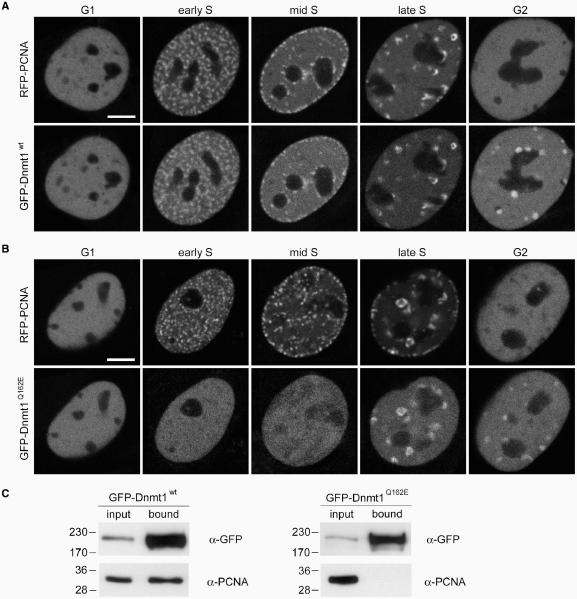
We recently demonstrated a loss of association with replication foci in early and mid S-phase of a GFP-Dnmt1 fusion construct with a deletion of the first 171 amino acids which includes most of the PBD (GFP-Dnmt1Δ1-171) (13). To address the function of the PBD in early and mid S-phase more specifically and to exclude potential misfolding of the protein due to the large deletion we have generated GFP-Dnmt1 constructs bearing single point mutations of highly conserved residues within the PIP (PCNA-interacting peptide)-Box (41) of the PBD (GFP-Dnmt1Q162E and GFP-Dnmt1F169S) (Figure 1). Either of these mutant constructs were expressed in C2C12 mouse myoblasts together with RFP-PCNA, which served as S-phase marker (42). Both Dnmt1 constructs showed a diffuse nuclear distribution in early and mid S-phase cells (Figure 2B and Supplementary Figure 3), in contrast to the wild-type Dnmt1 construct (GFP-Dnmt1wt) which was concentrated at replication foci during early and mid S-phase (Figure 2A). In late S-phase and G2, however, the wild type as well as all the PBD mutant constructs, including GFP-Dnmt1Δ1–171, were similarly concentrated at chromocenters (Figure 2A and B and Supplementary Figure 1), confirming the additional binding to heterochromatin in late S-phase mediated by the TS domain (13). Co-immunoprecipitation experiments confirmed that the Q162E point mutation abolishes the interaction between Dnmt1 and PCNA (Figure 2C). Similar results were obtained with GFP-Dnmt1F169S and GFP-Dnmt1Δ1–171 (Supplementary Figure 3). Thus, both Q162E and F169S point mutations prevent accumulation of Dnmt1 at replication foci during early to mid S-phase, while localization at constitutive heterochromatin in late S-phase and G2 is not affected. These results clearly confirm the role of the PBD in mediating the interaction between Dnmt1 and PCNA in vivo.
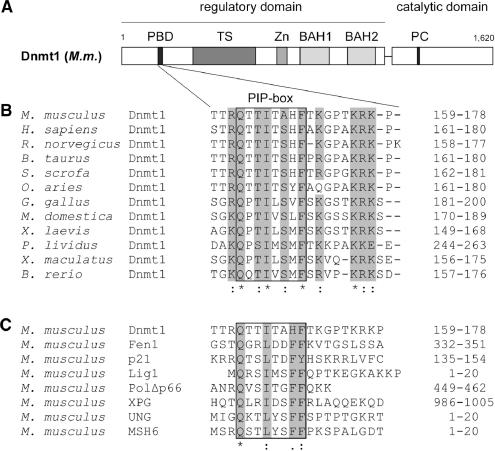
Figure 1.
Structure of Dnmt1 and alignment of PCNA-binding domains (PBD). (A) Schematic representation of the mouse Dnmt1 consisting of an N-terminal regulatory and a C-terminal catalytic domain. The PBD, the TS domain, the Zn-binding domain (Zn), the two bromo-adjacent homology domains (BAH) and the catalytic center (PC motif) are highlighted. (B) Alignment of Dnmt1 PBDs from different species. Highly conserved residues are gray shaded. Accession numbers: Mus musculus P13864; Homo sapiens P26358; Rattus norvegicus Q9Z330; Bos taurus Q6Y856; Sus scrofa Q4TTV6; Ovis aries Q865V5; Gallus gallus Q92072; Monodelphis domestica Q8MJ28; Xenopus laevis Q6GQH0; Paracentrotus lividus Q27746; Xiphophorus maculatus Q9I8X6; Brachydanio rerio Q8QGB8. (C) Alignment of PBDs in different proteins from Mus musculus. Gray shaded amino acids are highly conserved within the PIP-Box. Accession numbers: Dnmt1 P13864; Fen1 P39749; p21 P39689; DNA Ligase1 P37913; Polymerase δ/subunit Δp66 Q9EQ28; XPG P39749; UNG:P97931 MSH6 P54276.
Figure 2.
Mutation of the PBD abolishes Dnmt1 interaction with PCNA and prevents accumulation at replication sites in early and mid S-phase. (A, B) Confocal mid sections of living mouse C2C12 myoblasts expressing either GFP-Dnmt1wt (A) or GFP-Dnmt1Q162E (B). Cells were co-transfected with RFP-PCNA to identify replication foci and to distinguish S-phase stages. Scale bars: 5 µm. GFP-Dnmt1wt accumulates at replication sites throughout S phase where it co-localizes with RFP-PCNA. In G2 cells a fraction of Dnmt1 remains associated with the late replicating pericentric heterochromatin. In contrast, GFP-Dnmt1Q162E shows a fully dispersed nuclear distribution in early and mid S-phase stages, whereas in late S-phase and G2 association with centromeric heterochromatin is still observed. (C) Endogenous PCNA efficiently co-immunoprecipitates with GFP-Dnmt1wt but not with GFP-Dnmt1Q162E. Cell extracts were prepared from HEK 293T cells expressing either GFP-Dnmt1wt or GFP-Dnmt1Q162E. Precipitated proteins were detected by immunostaining with antibodies against GFP and PCNA, respectively.
The PBD-mediated interaction of Dnmt1 at replication sites is highly transient
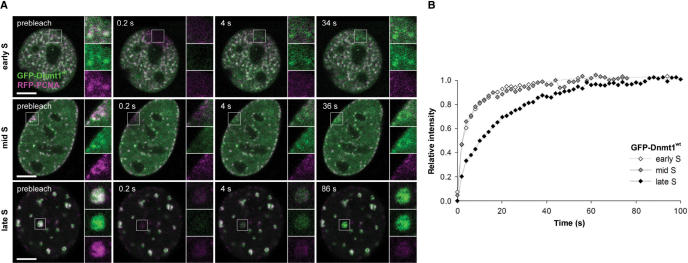
To investigate the dynamics of the PBD-mediated interaction at replication sites we measured fluorescence recovery after photobleaching (FRAP) of GFP-Dnmt1wt throughout S-phase. GFP-Dnmt1wt and RFP-PCNA were co-expressed in C2C12 cells and a small square region of interest (ROI) was bleached (Figure 3). Consistent with earlier observations (26,43) RFP-PCNA showed hardly any recovery within the observation period of 100 s, whereas in the same period GFP-Dnmt1 recovered fully, with very similar kinetics in early and mid S-phase, but notably slower in late S-phase. This result shows that the binding of Dnmt1 at replication sites is more dynamic during early and mid S-phase than in late S-phase, when Dnmt 1 is likely slowed down by the additional interaction with chromatin mediated by the TS domain (13).
Figure 3.
Transient binding of Dnmt1 at replication sites in different S-phase stages. (A) GFP-Dnmt1wt (green) and RFP-PCNA (magenta) co-expressed in mouse C2C12 cells co-localize at replication sites in early, mid and late S-phase. Fluorescence bleaching of a small region of interest reveals fast recovery of GFP-Dnmt1wt, whereas RFP-PCNA shows almost no recovery within the observation period. (B) Quantitative evaluation of the FRAP experiments shown in (A) reveal a significantly faster recovery of GFP-Dnmt1wt in early and mid S-phase compared to late S-phase. Scale bars: 5 µm.
To address the kinetic properties of the PBD-mediated binding to replication sites more specifically, we performed quantitative FRAP analysis of GFP-Dnmt1wt and GFP-Dnmt1Q162E in G1 and early/mid S-phase. As replication sites are not homogeneously distributed in the nucleus, we chose to bleach half nuclei (half-FRAP) to ensure that the bleached region contains a representative number of potential binding sites (Figure 4A). In early/mid S-phase nuclei GFP-Dnmt1wt recovered with a halftime (t1/2) of 4.7 ± 0.2 s and reached complete equilibration (t∞) in about 56 s. These values place Dnmt1 among the more dynamic factors involved in chromatin transactions previously determined with half-FRAP analyses (44). In comparison to GFP-Dnmt1wt, GFP-Dnmt1Q162E showed a slightly increased mobility (t1/2 = 4.4 ± 0.2 s; t∞ ∼45 s), which is likely due to the lack of binding to PCNA rings at replication sites. In the absence of active replication sites in G1, GFP-Dnmt1wt and Dnmt1Q162E showed nearly identical kinetics, which were remarkably similar to the kinetics of Dnmt1Q162E in early/mid S-phase. These data indicate that PCNA binding has only a minor contribution to Dnmt1 kinetics in S phase.
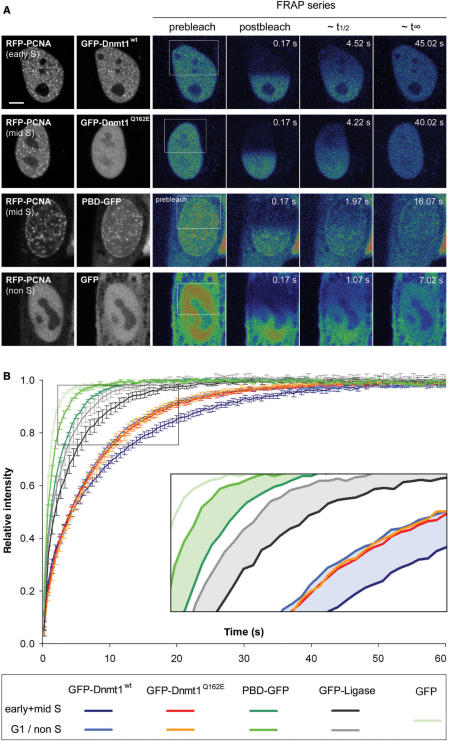
Figure 4.
Effect of the PBD on protein mobility in S-phase cells. (A) Half nucleus FRAP series of C2C12 cells expressing GFP-Dnmt1wt and GFP-Dnmt1Q162E, PBD-GFP and GFP alone as indicated in the second column. Cell-cycle stages were identified by the subnuclear pattern of co-expressed RFP-PCNA (first column). Selected pre- and postbleach frames are shown in false color. Lower signal-to-noise ratio of FRAP series images is due to the higher imaging frame rate. Rectangles indicate the bleached ROI. The column marked with ∼t1/2 displays frames corresponding to the half time of fluorescence recovery. The column marked with ∼t∞ displays the frames corresponding to approximately the time points when fluorescence recovery reached the plateau. Bar: 5 µm. (B) Quantitative evaluation of FRAP experiments. Wild-type Dnmt1 shows a small but significant decrease in mobility in early/mid S-phase (dark blue curve) as compared to G1 phase (light-blue curve). A similar mobility shift between early/mid S and non-S-phase cells is also seen for PBD-GFP (dark green and green curve, respectively) and for GFP-Ligase (dark gray and gray curve, respectively). No such shift is observed for the PCNA-binding-deficient Dnmt1 mutant (red and orange curve, respectively). GFP alone (light-green curve) is shown as reference. For clarity only every fourth time point is displayed; data are represented as mean ± SEM. Kinetic shifts are indicated by shadings in the enlarged inset.
The recovery rates measured for the full-length constructs were considerably slower than the rate of GFP alone (t1/2 = 0.8 ± 0.1 s; t∞ ∼10 s), which was used to control for unspecific binding events. As t1/2 is roughly proportional to the cubic root of the molecular mass (45,46), the ∼8-fold size difference of GFP alone to the full-length construct (27 and 210 kDa, respectively) would only account for about a 2-fold slower recovery. Instead, GFP-Dnmt1 full-length constructs recover more slowly, pointing to one (or more) additional and yet uncharacterized cell cycle independent interaction(s).
In order to dissect the PBD-mediated interaction from superimposing effects caused by other potential interactions we assayed the FRAP kinetics of a GFP fusion with the isolated PBD of Dnmt1, i.e. amino acids 159–178 (PBD-GFP). We found that in S–phase the recovery was only about 2 times slower than GFP alone (t1/2 = 1.5 ± 0.1 s; t∞ ∼16 s), thus confirming the highly transient nature of the PBD interaction with PCNA. In non-S-phase cells, PBD-GFP showed an increase in mobility (t1/2 = 1.1 ± 0.1 s; t∞ ∼11 s) similar to that observed with the full-length wild-type construct.
For direct comparison we further analyzed another PCNA-interacting enzyme, DNA Ligase I fused to GFP (GFP-Ligase), and found a similar mobility shift in S phase (t1/2 = 2.4 ± 0.3 s; t∞ ∼27 s) compared to non-S-phase cells (t1/2 = 2.1 ± 0.3 s; t∞ ∼17 s). Thus, with all three PCNA-interacting GFP fusion proteins (GFP-Dnmt1wt, PBD-GFP and GFP-Ligase), but not with the PCNA-binding mutant, the transient association with the replication machinery (PCNA) caused a slower recovery as compared to G1/non-S-phase (Figure 4B, inset).
Together the limited but significant contribution of the PBD-mediated interaction to the kinetic properties of GFP-Dnmt1 and the high mobility of PBD-GFP in S–phase clearly indicate that binding of Dnmt1 to PCNA at replication foci is highly transient.
Interaction with PCNA enhances methylation efficiency in vivo only moderately
Next we investigated the contribution of the highly transient interaction with PCNA to the postreplicative methylation activity of Dnmt1. Earlier it was shown that N-terminal deletions of mouse Dnmt1 comprising the PBD did not alter catalytic activity in vitro (47,48). To test the catalytic activity of the GFP-Dnmt1 constructs used in this study they were expressed in HEK 293T cells, immunopurified and directly assayed for methyltransferase activity in vitro. While the catalytically inactive GFP-Dnmt1C1229W mutant (27) displayed only background activity, GFP-Dnmt1Q162E and GFP-Dnmt1Δ1-171 exhibited enzymatic activity comparable to GFP-Dnmt1wt (Figure 5D), indicating that neither the Q162E point mutation nor deletion of the first 171 amino acids affect the enzymatic activity of Dnmt1.
Figure 5.
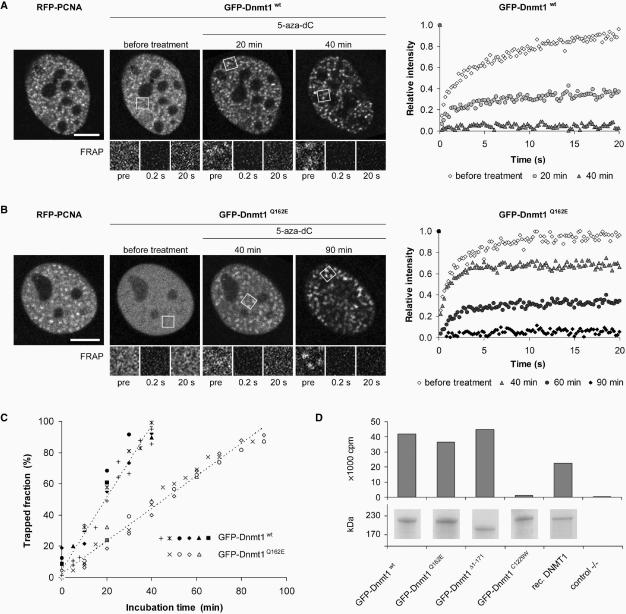
PCNA-binding-deficient mutant Dnmt1 is catalytically active and shows moderately reduced postreplicative methylation activity in vivo. (A–C) Trapping assay for the analysis of postreplicative methylation activity in vivo. (A, B) Confocal mid sections (upper panel) and corresponding FRAP series (lower panel) of a C2C12 mouse myoblast cell co-transfected with RFP-PCNA and either GFP-Dnmt1wt (A) or GFP-Dnmt1Q162E (B) before and at selected time points after incubation with 5-aza-dC. Before drug treatment the observed cells were in early S-phase as indicated by the RFP-PCNA pattern (left). Boxes indicate bleached ROI's, which are shown magnified in the lower panels at indicated time points of the FRAP series. Quantitative FRAP analysis is shown on the right. Scale bars: 5 µm. (C) Time-dependent increase of immobile fractions of GFP-Dnmt1wt and GFP-Dnmt1Q162E. Individual cells assayed at consecutive time points are indicated by closed (GFP-Dnmt1wt) and open (GFP-Dnmt1Q162E) symbols. Character symbols represent different cells each assayed at a single time point. + symbol indicates data points obtained with C2C12 cells stably expressing GFP-Dnmt1wt (C2C12-GMT1) without co-expression of RFP-PCNA. (D) In vitro methyltransferase assay for GFP-Dnmt1wt, GFP-Dnmt1Δ1-171, GFP-Dnmt1Q162E and the catalytically inactive mutant GFP-Dnmt1C1229W. GFP fusions were expressed in HEK 293T cells, immunopurified and the amount of [3H]CH3 transferred to a hemimethylated oligonucleotide substrate was measured. To normalize for the amount of precipitated protein aliquots were analyzed by SDS-PAGE and Coomassie staining (lower panel).
To establish whether the binding to PCNA is required for postreplicative methylation in vivo, we tested the activity of GFP-Dnmt1wt and GFP-Dnmt1Q162E in living cells using a recently developed trapping assay (27). This assay takes advantage of the catalytic mechanism of DNA (cytosine-5) methyltransferases which involves transient formation of a covalent complex with the C6 position of the cytosine residue. When the cytosine analogue 5-aza-2′-deoxycytidine (5-aza-dC) is incorporated into the DNA during replication the covalent complex of Dnmt1 and 5-aza-dC cannot be resolved and Dnmt1 is trapped at the site of action. Time-dependent immobilization, i.e. trapping of GFP-tagged Dnmt1 at replication foci can be visualized and measured by FRAP and reflects enzymatic activity of the fusion protein.
C2C12 cells co-transfected with RFP-PCNA and either GFP-Dnmt1wt or GFP-Dnmt1Q162E as well as C2C12 cells stably expressing GFP-Dnmt1wt were incubated in the presence of 30 µM 5-aza-dC. In early S-phase the focal enrichment of GFP-Dnmt1wt at replication sites increased over time reflecting the accumulation of immobilized enzyme and after 40 min GFP-Dnmt1wt was completely immobilized (Figure 5A). Similar kinetics were observed in mid S-phase (data not shown). As shown above GFP-Dnmt1Q162E displayed a diffuse nuclear distribution in early S-phase cells (Figure 5B). However, with prolonged incubation in the presence of 5-aza-dC an increasing focal accumulation at replication sites was observed. Quantitative FRAP analysis revealed that the immobilization rate of GFP-Dnmt1Q162E, which is a direct measure of its enzymatic activity, was only about 2-fold slower than GFP-Dnmt1wt, resulting in complete trapping after ∼90 min. These results indicate that the PCNA-binding-deficient mutant binds to DNA and is catalytically engaged at hemimethylated sites generated during replication.
Immobilization of GFP-Dnmt1wt at replication sites does not prevent progression of the replication machinery
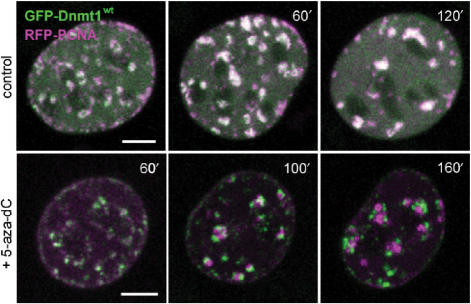
We then probed the stability of the interaction between Dnmt1 and the replication machinery by trapping Dnmt1 with 5-aza-dC and long-term live imaging. In the case of stable interaction, covalent immobilization of Dnmt1 would be expected to stall the progression of the replication machinery. C2C12 cells co-transfected with the GFP-Dnmt1wt and RFP-PCNA constructs were incubated in the presence of 10 µM 5-aza-dC and individual S-phase cells were imaged at consecutive time points for an extended period of time (Figure 6). Progressive separation of GFP-Dnmt1wt and RFP-PCNA foci could be clearly observed over a time period of ∼2 h, indicating that trapping of Dnmt1 did not prevent the progression of replication factories. This result is consistent with the FRAP kinetics of GFP-Dnmt1wt demonstrating the transient nature of the interaction between Dnmt1 and PCNA.
Figure 6.
Immobilization of Dnmt1 does not prevent progression of DNA replication. S-phase C2C12 myoblasts expressing GFP-Dnmt1wt (green) and RFP-PCNA (magenta) were imaged without drug treatment (upper row) and at the indicated time points after addition of 10 µM 5-aza-dC to the medium (lower row). In the control cell the two constructs largely co-localize during transition from mid to late S-phase, whereas in the presence of 5-aza-dC progressive separation of green and red foci indicate immobilization of GFP-Dnmt1wt at postreplicative hemimethylated sites and progression of replication foci containing RFP-PCNA. Projections of confocal mid sections are shown. Scale bar: 5 µm.
GFP-Dnmt1Q162E rescues CpG methylation in Dnmt1−/− ES cells
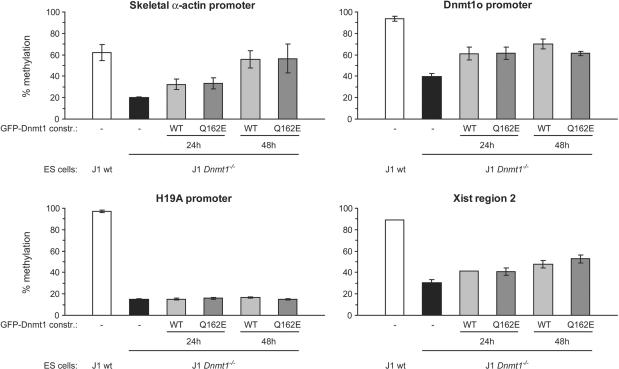
To investigate the contribution to maintenance of methylation patterns by the PBD-mediated interaction with PCNA in vivo we transiently expressed either GFP-Dnmt1wt or GFP-Dnmt1Q162E in Dnmt1 deficient mouse ES cells, which are severely hypomethylated in all genomic compartments (5,49). GFP-positive cells were isolated by FACS sorting 24h and 48 h after transfection and methylation of single-copy sequences and intracisternal type A particle (IAP) interspersed repetitive elements was analyzed by COBRA. An increase in methylation at all tested sites was observed in cells expressing either the wild type or the mutant Dnmt1 constructs already 24 h after transfection (Figure 7 and Supplementary Figure 4A). Further substantial increase of methylation 48 h after transfection was observed only for the skeletal α-actin promoter where the methylation level approached that observed in wild-type cells. The result for the skeletal α-actin promoter were confirmed and extended by bisulfite sequencing (Supplementary Figure 5). It was previously reported that re-expression of wild-type Dnmt1 in dnmt1−/− ES cells does not lead to restoration of methylation at imprinted genes since passage through the germ line is needed for re-establishment of methylation patterns in these sequences (49). We analyzed the promoter of the imprinted H19 gene to control for the specificity of our assay and found that indeed expression of neither GFP-Dnmt1wt nor GFP-Dnmt1Q162E resulted in increased methylation of this sequence (Figure 7). To further evaluate methylation of repetitive sequences we stained transiently transfected dnmt1−/− cells with an antibody against 5-methylcytidine that detects highly methylated satellite repeats present at mouse chromocenters (pericentromeric heterochromatin). Restoration of high DNA methylation levels at chromocenters was observed in both cells expressing GFP-Dnmt1wt and GFP-Dnmt1Q162E (Supplementary Figure 4B).
Figure 7.
PCNA-binding-deficient mutant Dnmt1 restores methylation of single-copy sequences in dnmt1−/− ES cells. Mouse dnmt1−/− ES cells were transiently transfected with either the GFP-Dnmt1wt or GFP-Dnmt1Q162E constructs and FACS-sorted 24h and 48 h after transfection. Methylation of single-copy sequences was assayed by COBRA. Shown are methylation percentages at assayed CpG sites with respect to genomic DNA from dnmt1−/− ES cells methylated in vitro with the recombinant methyltransferase SssI (100% methylation; see also Supplementary Figure 6). Mean values and standard errors of duplicate amplifications from each of two independent experiments are indicated.
The experimental procedures employed did not allow detection of significant differences in remethylation kinetics between the two constructs. Nevertheless, these results show that the PCNA-binding-deficient mutant is, like wild-type Dnmt1, able to rescue methylation of both single copy and repetitive sequences in vivo.
DISCUSSION
Faithful replication of genetic and epigenetic information is crucial to ensure the integrity and identity of proliferating cells. Earlier work has demonstrated that the maintenance methyltransferase Dnmt1 binds to the replication processivity factor PCNA and is thus recruited to replication sites (12). The interaction between Dnmt1 and the replication machinery was proposed as a mechanism for coupling maintenance of genomic methylation to DNA replication (11). By traveling along with the replication machinery Dnmt1 would be able to restore symmetrical methylation as soon as hemimethylated CpG sites are generated. The estimated kinetics of DNA replication in vivo and DNA methylation by Dnmt1 in vitro, however, differ by 3–4 orders of magnitude arguing against a stable coupling. Although DNA methylation may be faster in vivo it is not likely to come much closer to the DNA replication rate. In other words, it is reasonable to assume that methylating a cytosine takes far longer than to incorporate it. Also, DNA replication is essentially a continuous process, while methylated CpG sites are encountered discontinuously in vertebrate genomes. Stable binding of Dnmt1 to the replication machinery would stall replication at each hemimethylated CpG site. Here we show that the interaction of Dnmt1 with the replication machinery in vivo is highly dynamic and that immobilization of Dnmt1 at postreplicative hemimethylated sites does not prevent the progression of DNA replication. The transient nature of the interaction between Dnmt1 and the replication machinery allows rapid release and transfer of Dnmt1 to hemimethylated substrate sites, reconciling the paradox of the relative rates of DNA replication and methylation. According to basic principles of enzyme kinetics the local enrichment of Dnmt1 at replication foci would increase the postreplicative methylation rate. At the same time, transient binding of Dnmt1 enables also other replication factors to interact with PCNA. Similar binding dynamics have been shown for the interaction of PCNA with DNA Ligase I and Fen1 (26) and may thus represent a common feature of PBD-containing factors.
Interestingly, the interaction between Dnmt1 and PCNA is believed to be a major mechanism for the methylation maintenance activity of Dnmt1, but its functional relevance had never been tested experimentally. Here we show two lines of evidence that this interaction is not crucial for the maintenance of methylation patterns in mammalian cells. First, postreplicative methylation rate of wild-type Dnmt1 measured in vivo is only 2-fold faster than that of a PCNA-binding-deficient mutant. Second, methylation of both single copy and repetitive sequences in Dnmt1 deficient ES cells was restored by this PCNA-binding-deficient Dnmt1 mutant with efficiency comparable to wild-type Dnmt1. The maintenance of DNA methylation without direct coupling to the replication machinery could in part be explained by the preference of Dnmt1 for hemimethylated sites (8–10). Also, genetic manipulation in the mouse indicate that Dnmt1 is at least 5-fold more abundant than necessary for maintaining normal methylation levels (6,50). Thus, the combined effect of the affinity for hemimethylated sites, relative abundance and simple diffusion could explain the relatively fast immobilization of PCNA-binding-deficient mutants in the presence of 5-aza-dC. In addition, the ability of Dnmt1 to methylate nucleosomal DNA in vitro (51–53), suggests that maintenance of DNA methylation is not necessarily restricted to the short time window before nucleosome assembly.
Recent structural data on Ligase I:PCNA and FEN-1:PCNA complexes indicate that for these enzymes PCNA does not simply serves as a loading platform for the replication machinery, but also causes allosteric activation (56,57). The data presented here cannot rule out a similar mechanism for Dnmt1, but clearly show that interaction with PCNA is not a prerequisite for enzyme activity in vivo. The transient nature of this interaction also argues against PCNA as a classic processivity factor for postreplicative DNA methylation. The major role of the PCNA interaction most likely is to increase the local Dnmt1 concentration and thereby enhance methylation efficiency at replication sites.
Notably, it is still unclear whether the role and regulation of Dnmt1 is similar in different cell types and species. While Dnmt1 is clearly essential for maintenance of DNA methylation in mouse cells (5,54), it seemed dispensable in human tumor HCT116 cells (32). However, two reports recently showed that DNMT1 is essential for maintenance of DNA methylation also in these human tumor cells (32,55). It turned out that genomic methylation was maintained by a residual, truncated DNMT1 form lacking the PBD, arguing that PCNA binding is not strictly required in these cells (32).
Also, the requirement of Dnmt1 for cell viability remains unsettled. In mouse fibroblasts inactivation of the dnmt1 gene caused a continuous loss of genomic methylation leading to apoptotic cell death after several cell division cycles (54). Similar results were obtained after depletion of DNMT1 activity by RNA interference in human cells (32). Surprisingly, conditional deletion of the DNMT1 locus in the same cells caused immediate apoptotic cell death long before substantial passive loss of genomic methylation could occur (58) arguing for additional roles of DNMT1. In this regard, the association of Dnmt1 with heterochromatin in G2 phase and occasionally in mitosis in mouse cells (13) would fit well with the mitotic catastrophe observed upon deletion of the DNMT1 gene in HCT116 cells. In addition, the PBD-mediated association of Dnmt1 with repair sites (14) may indicate a direct role in the maintenance of genome integrity (59). Clearly, more experiments are required to resolve the species and cell-type-specific role and regulation of Dnmt1.
In summary, we demonstrate that the association of Dnmt1 with the replication machinery is not strictly required for maintaining global methylation but still enhances in vivo methylation efficiency by 2-fold. Whereas the benefit of Dnmt1 to be directly recruited to replication foci seems subtle in short-term cell culture experiments, it may be more relevant in long-lived organisms and in situations where the nuclear concentration of Dnmt1 is limiting. Indeed, Dnmt1 levels vary considerably in different tissues and developmental stages (60,61). Based on sequence features of the dnmt1 gene a modular structure was proposed to originate from an ancestral DNA methyltransferase that evolved by stepwise acquisition of new domains (47). Thus, the improved efficiency of postreplicative methylation achieved by the PBD-mediated transient binding to PCNA likely represents an additional safety mechanism, which was acquired in the course of evolution and contributes to the faithful maintenance of epigenetic information over the entire lifespan of complex organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Markus Moser and Reinhard Fässler (MPI for Biochemistry, Martinsried) for introducing us to ES cell culture, Hans-Peter Rahn (MDC, Berlin) and Michaela Feuring-Buske (LMU, Munich) for assistance with FACS-sorting, Gernot Längst (University of Regensburg) for help with the methyltransferase activity assay and En Li (Novartis Institutes for Biomedical Research) for providing the pCAG-IRESblast vector. We thank Anja Gahl, Kourosh Zolghadr and Jonas Helma for technical help and Akos Dobay for helpful discussion. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to HL. Open Access publication charges for this article were covered by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 5.Lei H, Oh S, Okano M, Juttermann R, Goss K, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 6.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 8.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl Acad. Sci. USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pradhan S, Talbot D, Sha M, Benner J, Hornstra L, Li E, Jaenisch R, Roberts RJ. Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase. Nucleic Acids Res. 1997;25:4666–4673. doi: 10.1093/nar/25.22.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 11.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 12.Chuang L.S.-H, Ian H.-I, Koh T.-W, Ng H.-H, Xu G, Li BFL. Human DNA-(Cytosine-5) Methyltransferase-PCNA Complex as a Target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 13.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 17.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 19.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 20.Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, Villa A, Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. Embo J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 23.Sobczak-Thepot J, Harper F, Florentin Y, Zindy F, Brechot C, Puvion E. Localization of cyclin A at the sites of cellular DNA replication. Exp. Cell Res. 1993;206:43–48. doi: 10.1006/excr.1993.1118. [DOI] [PubMed] [Google Scholar]
- 24.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 25.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 26.Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods. 2005;2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 28.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 30.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 34.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 35.Warnecke PM, Clark SJ. DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol. Cell. Biol. 1999;19:164–172. doi: 10.1128/mcb.19.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warnecke PM, Biniszkiewicz D, Jaenisch R, Frommer M, Clark SJ. Sequence-specific methylation of the mouse H19 gene in embryonic cells deficient in the Dnmt-1 gene. Dev. Genet. 1998;22:111–121. doi: 10.1002/(SICI)1520-6408(1998)22:2<111::AID-DVG1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Ko Y.-G, Nishino K, Hattori N, Arai Y, Tanaka S, Shiota K. Stage-by-stage change in DNA methylation status of Dnmt1 locus during mouse early development. J. Biol. Chem. 2005;280:9627–9634. doi: 10.1074/jbc.M413822200. [DOI] [PubMed] [Google Scholar]
- 38.McDonald LE, Paterson CA, Kay GF. Bisulfite genomic sequencing-derived methylation profile of the xist gene throughout early mouse development. Genomics. 1998;54:379–386. doi: 10.1006/geno.1998.5570. [DOI] [PubMed] [Google Scholar]
- 39.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 40.Warnecke P, Stirzaker C, Melki J, Millar D, Paul C, Clark S. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Easwaran HP, Leonhardt H, Cardoso MC. Cell cycle markers for live cell analyses. Cell Cycle. 2005;4 doi: 10.4161/cc.4.3.1525. [DOI] [PubMed] [Google Scholar]
- 43.Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- 44.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 47.Margot JB, Aguirre-Arteta AM, Di Giacco BV, Pradhan S, Roberts RJ, Cardoso MC, Leonhardt H. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J. Mol. Biol. 2000;297:293–300. doi: 10.1006/jmbi.2000.3588. [DOI] [PubMed] [Google Scholar]
- 48.Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- 49.Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 50.Gaudet F, Rideout W.M., 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuwaki M, Verreault A. Maintenance DNA methylation of nucleosome core particles. J. Biol. Chem. 2004;279:2904–2912. doi: 10.1074/jbc.M310111200. [DOI] [PubMed] [Google Scholar]
- 52.Robertson AK, Geiman TM, Sankpal UT, Hager GL, Robertson KD. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun. 2004;322:110–118. doi: 10.1016/j.bbrc.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 53.Gowher H, Stockdale CJ, Goyal R, Ferreira H, Owen-Hughes T, Jeltsch A. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005;44:9899–9904. doi: 10.1021/bi047634t. [DOI] [PubMed] [Google Scholar]
- 54.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 55.Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl Acad. Sci. USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 57.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 58.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 59.Brown KD, Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 60.Ratnam S, Mertineit C, Ding F, Howell CY, Clarke HJ, Bestor TH, Chaillet JR, Trasler JM. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 2002;245:304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- 61.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.